Abstract
The Snf1/AMP-activated protein kinase (AMPK) family plays fundamental roles in cellular responses to metabolic stress in eukaryotes. In humans, AMPK regulates lipid and glucose metabolism and has been implicated in such metabolic disorders as diabetes and obesity and in cardiac abnormalities. Snf1 and AMPK are the downstream components of kinase cascades, but the upstream kinase(s) have remained elusive. We have here identified three yeast kinases, Pak1p, Tos3p, and Elm1p, that activate Snf1 kinase in vivo. Triple deletion of the cognate genes causes a Snf– mutant phenotype and abolishes Snf1 catalytic activity. All three kinases phosphorylate recombinant Snf1p on the activation-loop threonine. Moreover, Tos3p phosphorylates mammalian AMPK on the equivalent residue and activates the enzyme, suggesting functional conservation of the upstream kinases between yeast and mammals. We further show that the closely related mammalian LKB1 kinase, which is associated with Peutz–Jeghers cancer-susceptibility syndrome, phosphorylates and activates AMPK in vitro. Thus, the identification of the yeast upstream kinases should facilitate identification of the corresponding, physiologically important mammalian upstream kinases.
The Snf1/AMP-activated protein kinase (AMPK) family of kinases is important for metabolic stress responses in eukaryotes (reviewed in refs. 1 and 2). In mammals, AMPK is activated by multiple stresses that lead to an increase in the cellular AMP:ATP ratio and is also activated by leptin (3) and metformin, a drug used to treat type 2 diabetes (4). AMPK has a central role in coordinating energy homeostasis and is a major regulator of lipid metabolism, glucose transport (5), and glycogen storage (6). In humans, AMPK has been implicated in the development and treatment of metabolic disorders, including obesity and type 2 diabetes (2), and mutations in AMPK cause cardiac abnormalities (7).
In the yeast Saccharomyces cerevisiae, the Snf1 kinase is also required for stress responses, notably the adaptation of cells to carbon stress. Snf1 regulates the transcription of many genes in response to glucose limitation, and Snf1 is required for utilization of alternate carbon sources (1). Like AMPK, Snf1 also regulates the activity of metabolic enzymes involved in fatty acid and glycogen metabolism (8–10). The signals that control Snf1 activity in response to glucose levels are not understood, although the AMP:ATP ratio may have a role under some conditions (11). Snf1 also affects meiosis and sporulation, filamentous invasive growth (12–14), and aging (15).
The Snf1 kinase comprises the catalytic subunit Snf1p (α subunit of AMPK), Snf4p (γ subunit of AMPK), and one of three β subunits. Snf4p stimulates kinase activity by counter-acting autoinhibition by the Snf1p regulatory domain (16). The β subunit regulates the subcellular localization of the kinase (17) and mediates interactions with downstream targets (18). Snf1 is activated by phosphorylation of the activation-loop threonine residue (8, 11, 19, 20), as is also the case for AMPK (21).
Despite intensive efforts, the identities of the upstream kinases that phosphorylate Snf1 and AMPK have proved elusive. In yeast, genetic approaches have failed to yield mutations in the cognate gene, suggesting that multiple kinases activate Snf1. Recently, mass spectrometric analysis of yeast protein complexes indicated that Tos3p (YGL179C) copurifies with Snf4p (22), and Pak1p (unrelated to p21-activated kinase) copurifies with Snf1p and with Snf4p (23). Tos3p and Pak1p are closely related, but their functions are unknown, except that overexpression of Pak1p suppresses DNA polymerase mutations (24).
We have explored the possibility that Tos3p and Pak1p activate Snf1 kinase. We present evidence that Tos3p and Pak1p, together with a third closely related kinase, Elm1p, are required for Snf1 function in vivo and show that these three kinases phosphorylate and activate Snf1 in vitro. We further show that Tos3p phosphorylates and activates AMPK in vitro, suggesting that the mammalian upstream kinase(s) are related to these yeast kinases. We therefore searched for closely related mammalian kinases and found that the LKB1 tumor suppressor kinase (25, 26) activates AMPK in vitro.
Materials and Methods
Yeast Strains and Genetic Analysis. S. cerevisiae strains used here were TAT7 (MATa ade2 his3 leu2 trp1 ura3::lexAop-lacZ LYS2::lexAop-HIS3 gal80), MCY2649 (MATα his3 leu2 ura3), and derivatives of W303-1A (MATa ura3 his3 leu2 trp1 ade2 can1). All mutants were constructed in the W303 genetic background. The tos3Δ::kanMX4 and pak1Δ::kanMX4 alleles were recovered from genomic DNA of mutant strains (Research Genetics, Huntsville, AL) by PCR and introduced into W303 by transformation. The elm1Δ::URA3 allele was similarly obtained from a mutant strain (27). Double mutants used for tetrad analysis were MCY5117 (MATα tos3Δ::kanMX4 pak1Δ::kanMX4 ura3 his3 leu2 trp1 ade2 can1) and MCY5122 (MATa tos3Δ::kanMX4 elm1Δ::URA3 ura3 his3 leu2 trp1 ade2 can1). Genotypes of segregants were determined by PCR analysis of genomic DNA. Rich medium was yeast extract/peptone/2% glucose (YPD), and synthetic complete (SC) medium lacking appropriate supplements was used to select for plasmids (28).
Immunoprecipitation, Kinase Assays, and Immunoblot Analysis. Preparation of protein extracts from yeast cells, immunoprecipitation, kinase assays, and immunoblot analysis were carried out as described (18). The extraction buffer was 50 mM Hepes (pH 7.5)/150 mM NaCl/0.1% Triton X-100/1 mM DTT/10% glycerol, containing 2 mM phenylmethylsulfonyl fluoride and Complete protease inhibitor mixture (Roche). The kinase activity of immunoprecipitated LexA-Snf1p was assayed in 50 mM Tris·HCl (pH 7.5)/10 mM MgCl2/1 mM DTT, containing 20 μCi (1 Ci = 37 GBq) of [γ-32P]ATP (3000 Ci/mmol; Perkin–Elmer). The phosphorylation of Snf1 catalytic domain by bead-bound GST-kinases was assayed in 50 mM Tris·HCl (pH 7.5)/2 mM MgCl2/1 mM MnCl2/1mMDTT/5 μM ATP, containing 20 μCi of [γ-32P]ATP. Antibodies were anti-LexA (Invitrogen) or anti-Snf1 (29).
Expression and Purification of GST-Tagged Yeast Kinases and Isolated Snf1 Catalytic Domain. GST fusions to Tos3p (31), Pak1p (31), and Elm1p (32) were expressed in yeast cells from the indicated libraries. In addition, plasmids expressing GST or GST-Tos3p, -Pak1p, or -Elm1p from a copper-inducible promoter (pOV85, pRH95, pRH98, and pRH94, respectively) were isolated from one of the libraries (31) and used to transform MCY2649 for expression of the GST protein. Extracts were prepared from yeast as above, and the GST-kinase was purified on glutathione-Sepharose (Amersham Biosciences).
His-tagged mutant Snf1p catalytic domains, Snf1KD-K84R and -T210A, were expressed in Escherichia coli from pRH89 and pRH90, derivatives of pRJ224 and pRJ226, respectively (33), in pET32c (Novagen). Proteins were purified by cobalt affinity chromatography on TALON resin (BD Biosciences) and eluted with buffer containing 150 mM imidazole, according to the manufacturer's instructions.
Assay of Snf1 Kinase Activity by Phosphorylation of SAMS Peptide. The assay was as described (8, 30). Extracts were prepared in duplicate from yeast cells grown in YPD and harvested by centrifugation. Cells were broken in buffer A [50 mM Tris·HCl, pH 7.5/50 mM NaF/5 mM sodium pyrophosphate/1 mM EDTA/1 mM DTT/0.1 mM phenylmethylsulfonyl fluoride/10% (vol/vol) glycerol]. Snf1 kinase was partially purified by chromatography on DEAE-Sepharose (Amersham Biosciences), and Snf1 activity was eluted from the column with buffer A containing 0.2 M NaCl in 0.5-ml fractions. Peak fractions (2 ml) were pooled and concentrated to ≈0.5 ml by using a Centricon-30 microconcentrator (Amicon). Pooled fractions were assayed in triplicate for phosphorylation of the SAMS peptide (HMRSAMSGLHLVKRR) in the presence of [γ-32P]ATP (specific activity, ≈300,000 cpm/nmol) in reaction buffer [50 mM Hepes, pH 7.5/5 mM MgCl2/1 mM EDTA/0.2 mM ATP/10% (vol/vol) glycerol] containing 0.2 mM SAMS peptide. Kinase activity is expressed in nmol of phosphate incorporated into peptide per minute.
Expression and Purification of Mammalian Kinases. Bacterially expressed AMPK (α1β1γ1) was purified by chromatography using Ni-NTA agarose (Qiagen, Valencia, CA) (34). Plasmid DNA encoding FLAG-tagged mouse LKB1 (gift of A. Ashworth, Institute of Cancer Research, London) was transfected into COS7 cells by using lipofectamine reagent. Cells were harvested 48 h posttransfection, and LKB1 protein was purified by binding to EZview Red ANTI-FLAG M2 affinity gel (Sigma).
Phosphorylation and Assay of AMPK. Bacterially expressed AMPK was incubated with 100 μM ATP, 5 mM MgCl2, 200 μM AMP, and 1 mM DTT in 50 mM Hepes (pH 7.4) in the presence or absence of upstream kinase for 30 min at 30°C. After brief centrifugation to remove the resin, AMPK activity in the supernatant was measured by using the SAMS peptide assay (30). Phosphorylation of T172 was determined by immunoblotting using an antibody that specifically recognizes phosphothreonine 172 within the α subunit of AMPK (Cell Signaling Technologies, Beverly, MA). 32P-phosphate labeling of AMPK was analyzed by incubating a catalytically inactive form of AMPK (2 μg), harboring a D157A substitution in the α subunit (35), with GST-Tos3p or GST bound to glutathione-Sepharose beads in the presence of [γ-32P]ATP for 30 min at 30°C. The beads were removed by brief centrifugation, and proteins in the supernatant were analyzed by SDS/PAGE and autoradiography.
Results
Tos3p and Pak1p Are Functionally Related to Snf1 Kinase. To confirm that Tos3p interacts with Snf1p, we expressed GST-Tos3p and LexA-tagged Snf1p in yeast and demonstrated that the two proteins copurify on glutathione-Sepharose (data not shown). We then introduced tos3Δ and pak1Δ mutations (tos3Δ::kanMX4 and pak1Δ::kanMX4) into yeast cells and tested for phenotypes characteristic of a snf1Δ mutant. The single and double mutants showed no defect in growth on raffinose or glycerol/ethanol (data not shown; Fig. 1B). We therefore sought other genetic evidence that Tos3p and Pak1p are functionally related to the Snf1 kinase. We reasoned that if they activate Snf1, their overexpression might compensate for the absence of the stimulatory subunit Snf4p. Overexpression of GST-Tos3p or -Pak1p partially suppressed snf4Δ mutant defects in SUC2 (invertase) gene expression, and GST-Tos3p restored growth on raffinose in a snf4Δ mutant but did not bypass the requirement for Snf1 (data not shown). We also used an assay in which transcription of a lexAop-lacZ reporter depends on the catalytic activity of LexA-Snf1p bound to the promoter (36). Overexpression of GST-Tos3p or -Pak1p stimulated β-galactosidase synthesis in response to glucose limitation, implying a positive effect on LexA-Snf1p catalytic activity (Fig. 1 A).
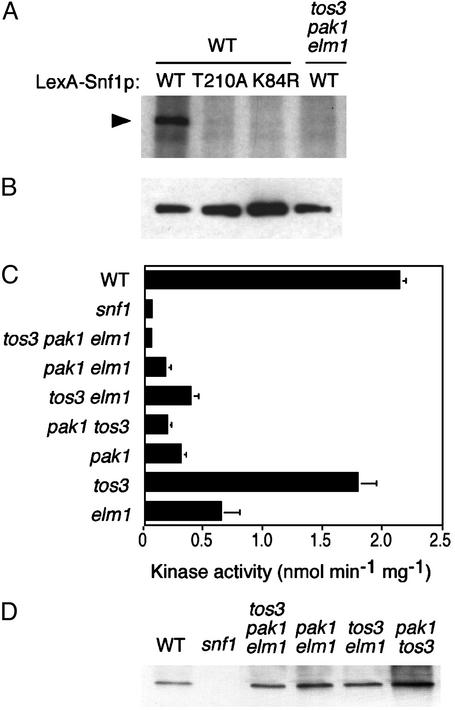
Fig. 1.
Overexpression and mutant phenotypes. (A) Overexpression of Tos3p, Pak1p, and Elm1p stimulates Snf1 function in a reporter assay. Transformants of a yeast strain (TAT7) carrying a lexAop-lacZ reporter expressed GST or GST-Tos3p, -Pak1p, or -Elm1p from a copper-inducible promoter (pOV85, pRH95, pRH98, and pRH94, respectively) and LexA-Snf1p (pOV8; ref. 37). Synthesis of β-galactosidase depended on catalytic activity of LexA-Snf1p (36). Transformants (n = 3) were grown to mid-log phase in selective SC plus 2% glucose, shifted to medium containing 0.5 mM CuSO4 in the presence of 2% glucose (open bars) or 0.05% glucose (filled bars) for 3 h, and assayed for β-galactosidase activity (36). Control transformants expressing LexA with each GST-kinase gave values <0.3 units. (B) Triple tos3Δ pak1Δ elm1Δ mutants exhibit growth defects. The tos3Δ pak1Δ and tos3Δ elm1Δ::URA3 double mutants (both carrying ura3) were crossed and subjected to tetrad analysis. Segregants were replicated from rich medium to SC-Ura plus 2% glucose and SC plus 2% glycerol/3% ethanol. Five tetrads are shown. The same pattern of growth was observed on SC plus 2% raffinose containing antimycin A (1 μg/ml). Asterisks indicate triple mutant segregants. Control strains: WT, snf1Δ mutant, and parental double mutants.
Triple tos3Δ pak1Δ elm1Δ Mutation Causes a Snf– Mutant Phenotype. The lack of phenotype in the double mutant suggested that another kinase provides redundant function. The kinase most closely related to Pak1p and Tos3p is Elm1p (38), which was identified by a mutation that causes elongated cell morphology and affects pseudohyphal development (27). Elm1p has been shown to have roles in the control of bud growth and cytokinesis (39, 40). We introduced elm1Δ::URA3 (27) into the above mutant strains by gene disruption. The tos3Δ elm1Δ and pak1Δ elm1Δ strains grew on raffinose and glycerol/ethanol, but the triple tos3Δ pak1Δ elm1Δ mutant did not.
To confirm this result, we crossed tos3Δ pak1Δ and tos3Δ elm1Δ strains and carried out tetrad analysis. Fourteen triple mutant segregants from thirteen tetrads were defective in growth on glycerol/ethanol and raffinose (Fig. 1B; data not shown). Assays of invertase activity showed that SUC2 expression was abolished (95 units in derepressed WT cells and <1unit in snf1Δ and triple mutant cells). The triple mutant also manifested a defect in glycogen accumulation, another snf1Δ mutant phenotype (data not shown). These genetic findings support the view that a function provided redundantly by Tos3p, Pak1p, and Elm1p is required for Snf1 kinase function in vivo.
Triple tos3Δ pak1Δ elm1Δ Mutation Abolishes Snf1 Kinase Activity in Vitro. To determine whether these three kinases activate Snf1, we used an in vitro kinase assay. Protein extracts were prepared from WT and triple-mutant cells expressing LexA-Snf1p. LexA-Snf1p was immunoprecipitated with anti-LexA and incubated in the presence of [γ-32P]ATP. When immunoprecipitated from the WT extract, LexA-Snf1p was phosphorylated in vitro, and controls with catalytically inactive LexA-Snf1K84R and LexA-Snf1T210A (carrying substitutions of the ATP-binding site lysine and the activation-loop threonine, respectively) confirmed that Snf1 kinase activity was responsible. In contrast, when LexA-Snf1p was precipitated from the triple mutant, no phosphorylation was detected (Fig. 2A), despite equivalent protein levels (Fig. 2B).
Fig. 2.
Assays of Snf1 kinase activity. (A and B) WT and triple mutant cells expressing LexA-Snf1p or its T210A and K84R mutant derivatives [pRJ55, pRJ217, and pRJ215 (16)] were grown in selective SC plus 2% glucose. Proteins were immunoprecipitated from extracts (200 μg) with anti-LexA. (A) Immunoprecipitates were incubated in kinase buffer containing [γ-32P]ATP and analyzed by SDS/PAGE and autoradiography. (B) Immunoprecipitates were analyzed by immunoblotting with anti-LexA. (C) Snf1 kinase activity was assayed by determining phosphorylation of the SAMS peptide (8, 30). A snf1-K84R mutant extract also showed no activity. (D) The assayed fractions were immunoblotted with anti-Snf1.
We also assayed Snf1 catalytic activity in vitro by phosphorylation of the SAMS synthetic peptide substrate (8, 30). Snf1 was partially purified from cell extracts, under conditions that activate the kinase (8, 11), and was incubated with SAMS peptide in the presence of [γ-32P]ATP. The peptide was phosphorylated in assays of WT but not snf1Δ extracts. No activity was detected in the tos3Δ pak1Δ elm1Δ mutant (Fig. 2C); immunoblot analysis confirmed the presence of Snf1p (Fig. 2D). These results indicate that in the absence of Tos3p, Pak1p, and Elm1p, Snf1 kinase remains inactive.
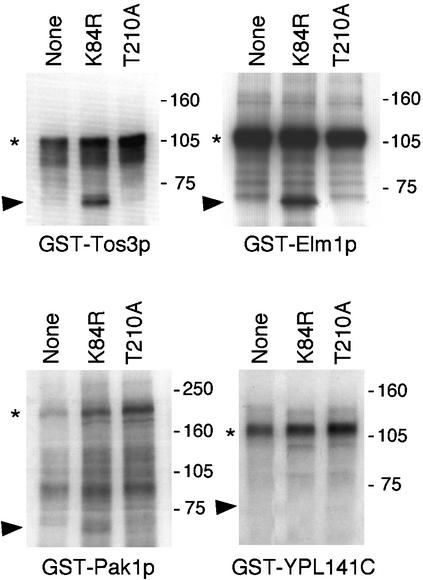
Tos3p, Pak1p, and Elm1p Phosphorylate Snf1p on the Activation-Loop Threonine Residue. To determine whether these three kinases directly phosphorylate Snf1p, we used as a substrate an inactive form of the isolated Snf1 catalytic domain, designated Snf1KD-K84R (Fig. 3). GST-Tos3p, -Pak1p, and -Elm1p were purified from yeast and incubated with bacterially expressed Snf1KD-K84R in the presence of [γ-32P]ATP. GST-Tos3p and -Elm1p both phosphorylated Snf1KD-K84R. GST-Pak1p phosphorylated the substrate weakly, perhaps because only minimal amounts of full-length protein were recovered; autophosphorylation of the full-length GST-Pak1p was also weak. Incubation with an unrelated GST-kinase, YPL141C, confirmed that Snf1KD-K84R was not autophosphorylated. To determine whether Tos3p, Pak1p, and Elm1p phosphorylate the activation-loop threonine, we used a mutant substrate lacking T210, Snf1KD-T210A. No phosphorylation was detected (Fig. 3).
Fig. 3.
Tos3p, Pak1p, and Elm1p phosphorylate Snf1p on T210 in vitro. GST-Tos3p (31), -Pak1p (31), and -Elm1p (32) were purified from yeast cells. The GST-kinase, immobilized on beads, was incubated with bacterially expressed Snf1KD-K84R or -T210A (0.2 μg), or no substrate, in the presence of [γ-32P]ATP for 20 min at 25°C. Proteins were analyzed by SDS/PAGE and autoradiography. An autoradiogram is shown. An arbitrary GST-kinase, YPL141C, served as a control. Arrowheads, mutant Snf1KD; asterisks, autophosphorylated full-length GST-kinase. Molecular mass markers (kDa) are indicated.
Tos3p Phosphorylates and Activates Mammalian AMPK in Vitro. We next examined the ability of the purified yeast GST-kinases to phosphorylate and activate mammalian AMPK. Recombinant AMPK (α1β1γ1) expressed in bacteria (34) was incubated with each of the kinases and MgATP, and AMPK activity was determined by using the SAMS peptide assay. Incubation with GST-Tos3p led to a marked increase in AMPK activity, whereas GST-Elm1p and -Pak1p caused little or no increase (Fig. 4A). GST-Tos3p phosphorylated the α (catalytic) subunit of a catalytically inactive form of AMPK, but not the β or γ subunit (Fig. 4B). Immunoblot analysis showed that GST-Tos3p phosphorylates the α subunit on the activation-loop threonine, T172 (Fig. 4C), as expected for an activating upstream kinase (21). These findings suggest functional conservation of the upstream kinases between yeast and mammals.
Fig. 4.
Tos3p and LKB1 phosphorylate and activate AMPK in vitro.(A) Bacterially expressed AMPK (34) was incubated with MgATP and immobilized GST-kinase (expressed from pRH95, pRH98, and pRH94), and AMPK activity was measured by using the SAMS peptide assay (30). (B) A catalytically inactive mutant AMPK (D157A substitution) was incubated with GST-Tos3p or GST bound to glutathione-Sepharose beads in the presence of [γ-32P]ATP, and proteins were analyzed by SDS/PAGE and autoradiography. Molecular mass markers (kDa) are indicated. (C) Bacterially expressed AMPK (0.15 μg) was incubated with GST-Tos3p, GST, or partially purified rat liver AMPKK (21) and analyzed by immunoblotting with antibody specific to phosphothreonine 172. (D and E) Purified, immobilized FLAG-tagged LKB1, either WT or a catalytically inactive mutant (D194A), was incubated with bacterially expressed AMPK. The empty vector is pCDNA3 (Invitrogen) lacking LKB1 insert. (D) AMPK activity was measured (30). (E) Phosphorylation of T172 was determined by immunoblotting as in C.
Mammalian LKB1 Kinase Phosphorylates and Activates AMPK in Vitro. The catalytic domains of Tos3p, Pak1p, and Elm1p are most closely related to mammalian Ca2+/calmodulin-dependent protein kinase kinase (CaMKK), although they have diverged from the yeast group of CaMKs (38). Previous studies showed that mammalian CaMKK weakly phosphorylates and activates AMPK in vitro, consistent with the idea that the upstream kinases are conserved between yeast and mammals; however, the major AMPK kinase (AMPKK) is distinct from CaMKK and is not Ca2+/calmodulin-stimulated (41). These findings suggest that AMPKK corresponds to another CaMK-related kinase(s).
The catalytic domains of Tos3p, Pak1p, and Elm1p are also very similar to that of the LKB1 (STK11) tumor suppressor kinase. The LKB1 gene is mutated in Peutz–Jeghers syndrome (25, 26), an autosomal-dominant intestinal polyposis syndrome that is associated with substantially increased risk of various other cancers (reviewed in ref. 42). LKB1 is essential for mammalian embryonic development, as shown by the finding that Lkb1-deficient mice die at midgestation with vascular abnormalities and other defects (43).
To examine the possibility that LKB1 functions as an upstream kinase for AMPK, we purified FLAG-tagged LKB1 from mammalian cells. LKB1 was incubated with bacterially expressed AMPK, and AMPK activity was measured. LKB1 strongly activated recombinant AMPK (Fig. 4D), and immunoblot analysis showed that the activation-loop threonine T172 was phosphorylated (Fig. 4E). In control experiments, the catalytically inactive mutant LKB1(D194A) did not activate or phosphorylate AMPK. These in vitro findings suggest that LKB1 is a promising candidate for an upstream kinase for AMPK in vivo.
Discussion
We have identified three kinases, Tos3p, Pak1p, and Elm1p, that function as upstream kinases in the Snf1 kinase cascade. We present genetic and biochemical evidence that these kinases phosphorylate and activate Snf1 kinase in vitro and in vivo. Mutant yeast cells lacking these three kinases are defective in the utilization of carbon sources other than glucose, a signature function of the Snf1 kinase pathway. This report is the first identification of upstream kinases in the Snf1/AMPK kinase cascade with demonstrated physiological roles.
These three kinases clearly exhibit significant functional redundancy, because all three must be mutated to confer the Snf– phenotypes examined here. However, it seems likely that they will prove to have some distinct functions with respect to regulation of Snf1. Snf1 kinase exists in the cell in three different forms, each containing a different β subunit isoform (Sip1p, Sip2p, or Gal83p), and considerable evidence indicates that the different isoforms have distinct functional roles (15, 17, 18). It is tempting to speculate that each of the three upstream kinases exhibits some preference for Snf1 kinase containing a specific β subunit isoform. Another possibility is that the three kinases respond preferentially to different kinds of metabolic stress or to different signals resulting from a single stress, such as glucose limitation. Further studies of the phenotypes of single and double tos3Δ, pak1Δ, and elm1Δ mutants will help address some of these issues.
It is also worth noting that at least one of these kinases, Elm1p, has regulatory functions involving cell morphology, filamentous invasive growth, and control of bud growth and cytokinesis (12, 27, 39, 40) that may have no connection to the Snf1 pathway. In the case of haploid invasive growth, genetic evidence indicates that Elm1p has functions that are clearly separate from its role as an upstream kinase for Snf1 (12). In addition, it is not evident that the suppression of DNA polymerase mutations by overexpression of Pak1p (24) has any relation to the Snf1 pathway.
The identification of the upstream kinases of the Snf1 kinase cascade opens up new possibilities for understanding the regulation of the Snf1 pathway and, more broadly, for understanding metabolic stress responses in yeast. However, the identification of these kinases also has significance beyond yeast, in providing clues for the search for upstream kinases in the AMPK cascade. An initial examination of one closely related mammalian kinase, the LKB1 tumor suppressor kinase, yielded in vitro evidence that LKB1 is a promising candidate for an upstream kinase for AMPK; moreover, the mass of LKB1 is close to that of the catalytic subunit of AMPKK purified from rat liver, which was estimated as 58 kDa (21). These findings further suggest the possibility of a role for AMPK in regulation of growth and transformation, as well as metabolism. We are currently investigating the physiological role of LKB1 in the regulation of AMPK in vivo. Other mammalian kinases that are related to the yeast upstream kinases also are candidates for bona fide kinases of the AMPK cascade. These mammalian upstream kinases are potential therapeutic targets for the treatment of human metabolic disorders and cancers.
Acknowledgments
We thank S. Kuchin for tetrad analysis; E. Phizicky and M. Snyder for libraries; D. Neumann, U. Schlattner, and T. Wallimann for AMPK; and A. Ashworth for plasmids encoding LKB1. This work was supported by National Institutes of Health Grant GM34095 (to M.C.) and the Medical Research Council (D.C.).
Abbreviations: AMPK, AMP-activated protein kinase; SC, synthetic complete.
References
- 1.Hardie, D. G., Carling, D. & Carlson, M. (1998) Annu. Rev. Biochem. 67, 821–855. [DOI] [PubMed] [Google Scholar]
- 2.Kemp, B. E., Stapleton, D., Campbell, D. J., Chen, Z. P., Murthy, S., Walter, M., Gupta, A., Adams, J. J., Katsis, F., Van Denderen, B., et al. (2003) Biochem. Soc. Trans. 31, 162–168. [DOI] [PubMed] [Google Scholar]
- 3.Minokoshi, Y., Kim, Y. B., Peroni, O. D., Fryer, L. G., Muller, C., Carling, D. & Kahn, B. B. (2002) Nature 415, 339–343. [DOI] [PubMed] [Google Scholar]
- 4.Zhou, G., Myers, R., Li, Y., Chen, Y., Shen, X., Fenyk-Melody, J., Wu, M., Ventre, J., Doebber, T., Fujii, N., et al. (2001) J. Clin. Invest. 108, 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mu, J., Brozinick, J. T. J., Valladares, O., Bucan, M. & Birnbaum, M. J. (2001) Mol. Cell 7, 1085–1094. [DOI] [PubMed] [Google Scholar]
- 6.Milan, D., Jeon, J.-T., Looft, C., Amarger, V., Robic, A., Thelander, M., Rogel-Gaillard, C., Paul, S., Iannuccelli, N., Rask, L., et al. (2000) Science 288, 1248–1251. [DOI] [PubMed] [Google Scholar]
- 7.Arad, M., Benson, D. W., Perez-Atayde, A. R., McKenna, W. J., Sparks, E. A., Kanter, R. J., McGarry, K., Seidman, J. G. & Seidman, C. E. (2002) J. Clin. Invest. 109, 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woods, A., Munday, M. R., Scott, J., Yang, X., Carlson, M. & Carling, D. (1994) J. Biol. Chem. 269, 19509–19516. [PubMed] [Google Scholar]
- 9.Hardy, T. A., Huang, D. & Roach, P. J. (1994) J. Biol. Chem. 269, 27907–27913. [PubMed] [Google Scholar]
- 10.Mitchelhill, K. I., Stapleton, D., Gao, G., House, C., Michell, B., Katsis, F., Witters, L. A. & Kemp, B. E. (1994) J. Biol. Chem. 269, 2361–2364. [PubMed] [Google Scholar]
- 11.Wilson, W. A., Hawley, S. A. & Hardie, D. G. (1996) Curr. Biol. 6, 1426–1434. [DOI] [PubMed] [Google Scholar]
- 12.Cullen, P. J. & Sprague, G. F., Jr. (2000) Proc. Natl. Acad. Sci. USA 97, 13619–13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuchin, S., Vyas, V. K. & Carlson, M. (2002) Mol. Cell. Biol. 22, 3994–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen, P. J. & Sprague, G. F., Jr. (2002) Mol. Biol. Cell 13, 2990–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashrafi, K., Lin, S. S., Manchester, J. K. & Gordon, J. I. (2000) Genes Dev. 14, 1872–1885. [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang, R. & Carlson, M. (1996) Genes Dev. 10, 3105–3115. [DOI] [PubMed] [Google Scholar]
- 17.Vincent, O., Townley, R., Kuchin, S. & Carlson, M. (2001) Genes Dev. 15, 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vincent, O. & Carlson, M. (1999) EMBO J. 18, 6672–6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estruch, F., Treitel, M. A., Yang, X. & Carlson, M. (1992) Genetics 132, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCartney, R. R. & Schmidt, M. C. (2001) J. Biol. Chem. 276, 36460–36466. [DOI] [PubMed] [Google Scholar]
- 21.Hawley, S. A., Davison, M., Woods, A., Davies, S. P., Beri, R. K., Carling, D. & Hardie, D. G. (1996) J. Biol. Chem. 271, 27879–27887. [DOI] [PubMed] [Google Scholar]
- 22.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. (2002) Nature 415, 180–183. [DOI] [PubMed] [Google Scholar]
- 23.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., et al. (2002) Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 24.Hovland, P. G., Tecklenberg, M. & Sclafani, R. A. (1997) Mol. Gen. Genet. 256, 45–53. [DOI] [PubMed] [Google Scholar]
- 25.Jenne, D. E., Reimann, H., Nezu, J., Friedel, W., Loff, S., Jeschke, R., Muller, O., Back, W. & Zimmer, M. (1998) Nat. Genet. 18, 38–43. [DOI] [PubMed] [Google Scholar]
- 26.Hemminki, A., Markie, D., Tomlinson, I., Avizienyt, E., Roth, S., Loukola, A., Bignell, G., Warren, W., Aminoff, M., Hoglund, P., et al. (1998) Nature 391, 184–187. [DOI] [PubMed] [Google Scholar]
- 27.Blacketer, M., Koehler, C., Coats, S., Myers, A. & Madaule, P. (1993) Mol. Cell. Biol. 13, 5567–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose, M. D., Winston, F. & Hieter, P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 29.Celenza, J. L. & Carlson, M. (1986) Science 233, 1175–1180. [DOI] [PubMed] [Google Scholar]
- 30.Davies, S. P., Carling, D. & Hardie, D. G. (1989) Eur. J. Biochem. 186, 123–128. [DOI] [PubMed] [Google Scholar]
- 31.Martzen, M. R., McCraith, S. M., Spinelli, S. L., Torres, F. M., Fields, S., Grayhack, E. J. & Phizicky, E. M. (1999) Science 286, 1153–1155. [DOI] [PubMed] [Google Scholar]
- 32.Zhu, H., Klemic, J. F., Chang, S., Bertone, P., Casamayor, A., Klemic, K. G., Smith, D., Gerstein, M., Reed, M. A. & Snyder, M. (2000) Nat. Genet. 26, 283–289. [DOI] [PubMed] [Google Scholar]
- 33.Ludin, K., Jiang, R. & Carlson, M. (1998) Proc. Natl. Acad. Sci. USA 95, 6245–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann, D., Schlattner, U. & Wallimann, T. (2003) Biochem. Soc. Trans. 31, 169–174. [DOI] [PubMed] [Google Scholar]
- 35.Stein, S. C., Woods, A., Jones, N. A., Davison, M. D. & Carling, D. (2000) Biochem. J. 345, 437–443. [PMC free article] [PubMed] [Google Scholar]
- 36.Kuchin, S., Treich, I. & Carlson, M. (2000) Proc. Natl. Acad. Sci. USA 97, 7916–7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vincent, O., Kuchin, S., Hong, S.-P., Townley, R., Vyas, V. K. & Carlson, M. (2001) Mol. Cell. Biol. 21, 5790–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunter, T. & Plowman, G. D. (1997) Trends. Biochem. Sci. 22, 18–22. [DOI] [PubMed] [Google Scholar]
- 39.Sreenivasan, A. & Kellogg, D. (1999) Mol. Cell. Biol. 19, 7983–7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouquin, N., Barral, Y., Courbeyrette, R., Blondel, M., Snyder, M. & Mann, C. (2000) J. Cell Sci. 113, 1435–1445. [DOI] [PubMed] [Google Scholar]
- 41.Hawley, S. A., Selbert, M. A., Goldstein, E. G., Edelman, A. M., Carling, D. & Hardie, D. G. (1995) J. Biol. Chem. 270, 27186–27191. [DOI] [PubMed] [Google Scholar]
- 42.Yoo, L. I., Chung, D. C. & Yuan, J. (2002) Nat. Rev. Cancer 2, 529–535. [DOI] [PubMed] [Google Scholar]
- 43.Ylikorkala, A., Rossi, D. J., Korsisaari, N., Luukko, K., Alitalo, K., Henkemeyer, M. & Makela, T. P. (2001) Science 293, 1323–1326. [DOI] [PubMed] [Google Scholar]