Abstract
The Rieske FeS protein, an essential catalytic subunit of the mitochondrial cytochrome bc1 complex, is encoded in yeast by the nuclear gene RIP1, whose deletion leads to a respiratory-deficient phenotype. By using biolistic transformation, we have relocated the nuclear RIP1 gene into mitochondria. To allow its expression within the organelle and to direct its integration downstream of the cox1 gene, we have fused the 3′ end of the Saccharomyces douglasii cox1 gene upstream of the mitochondrial copy of RIP1 (RIP1m) flanked by the Saccharomyces cerevisiae cox1 promoter and terminator regions. We show that RIP1m integrated between the cox1 and atp8 genes is mitotically stable and expressed, and it complements a deletion of the nuclear gene. Immunodetection experiments demonstrate that the mitochondrial genome containing RIP1m is able to produce the Rip1 protein in lower steady-state amounts than the wild type but still sufficient to maintain a functional cytochrome bc1 complex and respiratory competence to a RIP1-deleted strain. Thus, this recombined mitochondrial genome is a fully functional mitochondrial chromosome with an extended gene content. This successful mitochondrial expression of a nuclear gene essential for respiration can be viewed at the evolutionary level as an artificial reversal of evolutionary events.
According to the endosymbiotic hypothesis, genetic information has escaped from mitochondria and migrated to the nucleus during the evolution of eukaryotes (1). This hypothesis is supported by the observation that short sequences originating from the mtDNA are present in the nuclear genome of eukaryotic cells from various organisms, including humans, yeast, and plants (2). The intracellular migration of DNA from mitochondria to the nucleus has been detected experimentally in Saccharomyces cerevisiae strains that contained nuclear genetic markers inserted into their mitochondrial genomes (3, 4). In contrast to the widely affirmed transfer of genetic information from mitochondria to the nucleus, only limited evidence exists for gene transfer from the nuclear to the mitochondrial genome. So far, the only experiments involving mitochondrial expression of an endogenous nuclear gene involved a recoded ARG8 auxotrophic marker, called ARG8m, flanked by the mitochondrial untranslated sequences originating from the cox2, cox3, or var1 genes. The product of ARG8m has been used as a reporter/passenger to study the role of mRNA-specific translational activators in mitochondrial gene regulation and the topogenesis, insertion, and export of membrane proteins (5–9).
In this study we report the successful expression from the mitochondrial genetic system of the nuclear RIP1 gene whose product, the Rieske FeS protein, is essential for respiration and shows a remarkable evolutionary conservation. The Rieske FeS protein is the only ubiquitous essential protein identified in cytochrome bc1 complexes from bacteria and mitochondria and in the very similar cytochrome b6f complexes from chloroplasts (10–12). In S. cerevisiae and other eukaryotes, the Rieske FeS protein is encoded by the nuclear gene RIP1, synthesized on cytoplasmic ribosomes, and targeted to the mitochondria by its amino-terminal leader sequence. A yeast strain bearing the RIP1 deletion is viable, but respiratory-deficient (13, 14).
In this study we demonstrate that a mitochondrially encoded copy of the gene (RIP1m) can be stably integrated into the mtDNA and expressed within the organelle. Although the RIP1m integration creates a substantial duplication of the regions carrying the mitochondrial signals that originally ensure the expression of cox1 gene, the mitochondrial chromosome with extended gene content is fully functional. We show that in vivo the Rieske FeS protein produced in either homoplasmic or heteroplasmic mitochondria provides the cell with a sufficient amount of functional protein to complement the deletion of the nuclear gene.
Materials and Methods
Yeast Strains, Genetics Methods, and Media. The Δrip1 rho+ RGLT1 strain carrying 777-3A mtDNA was obtained by sporulation of diploids issued from the cross between the wild-type strain and the Δrip1 strain JPJ1 (13). All other Δrip1 rho+ strains with various mtDNAs (listed in Table 1) were obtained by a series of cytoductions using a rho0 derivative of RGLT1 (MATα, ura3, his3, ade2, leu2, trp1, rip1::LEU2) as the final recipient strain. Diploids homozygous for the deletion of the nuclear copy of RIP1 were generated by mating Δrip1 rho+ strains to synthetic Δrip1 rho– (RIP1m) strains. The progeny of each cross was spread onto fermentable glucose medium, and the respiring diploids were identified by replica-plating on nonfermentable glycerol-containing medium. To measure the mitotic stability of the mitochondrial genotypes, several individual glycerol-growing colonies were grown overnight in liquid glucose medium, and diluted aliquots were plated on glucose medium and replicated onto glycerol medium to score respiring colonies. Up to three successive passages in fermentable medium were made. All of the media used to grow yeast cells were as described (15).
Table 1. Complementation tests and mitotic stability of respiring diploids.
|
Cross
|
mtDNA of ρ+ strains
|
Mitotic stability of diploids
|
|||||
|---|---|---|---|---|---|---|---|
| ρ+Δrip1 × ρ- | RIP1m | Origin | Genotype | Name | I | II | III |
| RGLT1 | PG09 | 777-3A | int+, “long” ENS2nf | DR3 | 4 | 0 | |
| RPG2 | PG09 | S. capensis | int+, ENS2- | DR4 | 3 | 0 | |
| RGLT1 | PG10 | 777-3A | int+, “long” ENS2nf | RR1 | 70 | 71 | 81 |
| RPG2 | PG10 | S. capensis | int+, ENS2- | RR2 | 83 | 92 | 91 |
| RPG1 | PG10 | D273-10B | int+, “short” ENS2nf | RR3 | 32 | 12 | 5 |
| RJL2 | PG10 | WF3/5-2 | ai2+, ENS2nf | RR4 | 34 | 33 | 27 |
| RJL3 | PG10 | AF/32 | bi+, ENS2nf | RR5 | 30 | 20 | 23 |
| RJL6 | PG10 | KL14-4A | int+, “long”, ENS2nf | NO | |||
| RJL1 | PG10 | KF4/3-4 | ai1+, ENS2+ | NO | |||
| RJL4 | PG10 | CKYL1 | int-, ENS2+ | NO | |||
| RJL5 | PG10 | S. douglasii | int+, ENS2- | NO | |||
Two Δrip1 (rho- RIP1m) strains were crossed to a serie of Δrip1 rho+ strains (mtDNA origin and genotype are indicated) to test their ability to complement the deletion of the nuclear gene. int+ or int- indicates that mitochondrial introns are present or absent, respectively; long and short designate the various number of introns present in the cox1 (7 or 5) and cyt b (5 or 2) genes; ai1+ or ai2+ indicates that only the designated intron of the cox1 gene is present; bi+ indicates that only all introns (long version) of the cyt b gene are present. ENS2: + gene active; nf, gene nonfunctional; -, gene absent. n.o., not observed. Mitotic stability of respiring diploids was determined after three successive passages (I, II, III) on nonselective medium (see Materials and Methods). The numbers correspond to the percentage of respiring diploids in the population analyzed.
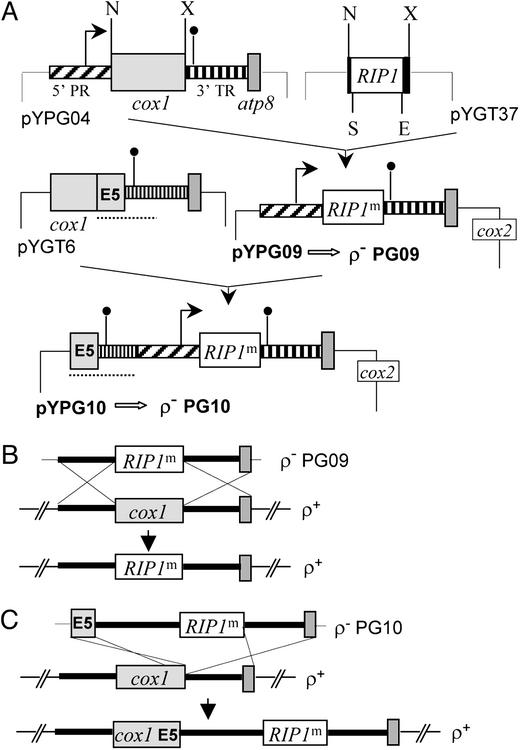
Construction of an Engineered RIP1m Gene. To construct the engineered form of the RIP1 gene, a synthetic fragment containing 14 bp of RIP1 upstream sequence including the endogenous Sau3A site, fused to the endogenous EcoRI site and its downstream 32 bp including the stop codon (Fig. 1A, pYGT37, filled boxes), was cloned into pUC13. In this fragment, an NdeI site was introduced at the start codon (CAT added before ATG), the ATA at the fourth codon of RIP1 was replaced by ATT, the TAG stop codon was replaced by TAA, and an XbaI site was introduced immediately downstream from the stop codon. The RIP1m ORF was assembled by cloning the internal Sau3A–EcoRI fragment (602 bp) excised from plasmid PISP2 (provided by J. P. di Rago, Institut de Biochimie et Génétique Cellulaire, Bordeaux, France) into the corresponding sites of the synthetic fragment to yield plasmid pYGT37. To ensure the expression of RIP1m within the organelle an intermediate plasmid, pYPG04 (Fig. 1 A), was constructed. It carries the flanking upstream (5′ PR) and downstream (3′ TR) regions of the intronless cox1 gene to which the NdeI and XbaI sites were introduced at the initiation and at the termination codons, respectively. The RIP1m ORF (NdeI–XbaI fragment) was then excised from pYGT37 and used to replace the NdeI–XbaI cox1 fragment in pYPG04. Finally, the sequences around the start and stop codons were reverted to wild type, and the insert was cloned into the mitochondrial transformation vector pJM2 (16) to give plasmid pYPG09 (Fig. 1 A).
Fig. 1.
Construction of rho– mtDNA carrying RIP1m and its integration into the rho+ mtDNA. (A) Diagram of plasmids constructed for the expression of RIP1m in mitochondria. The mitochondrial genes, cox1 and atp8, are designated by light gray and dark gray boxes, respectively. The cox1 gene upstream promoter region (hatched boxes, 5′ PR) and downstream terminator region (vertical stripes, 3′ TR) are represented. The limits of the cox1 transcript are indicated by arrows (promoter) and filled circles (3′ end). The filled boxes designate synthetic sequences of the RIP1m gene carrying some modifications compared with the nuclear version of the gene. The locations of the introduced restriction sites for NdeI (N), XbaI (X), Sau3A (S), and EcoRI (E) are shown. Two final constructs (pYPG09 and pYPG10) were introduced by biolistic transformation into rho0 mitochondria to give the synthetic rho– (ρ–) PG09 and PG10, respectively. See the text for details. (B and C) Recombinational integration of RIP1m into the rho+ mtDNA. The homologous recombination events are depicted by dotted lines. Recombination between rho+ mtDNA and rho– PG09 leads to a nonfunctional mitochondrial chromosome, lacking the cox1 gene (B), and recombination between rho+ mtDNA and rho– PG10 leads to a functional mitochondrial chromosome with extended gene content (C). For simplicity, the flanking intergenic sequences are denoted only by thick lines.
To obtain the larger plasmid pYPG10, the 868-bp KpnI–ClaI fragment of mtDNA from Saccharomyces douglasii was added at the 5′ end of the insert present in pYPG09. This fragment, containing the 144 codons of the last exon (E5) of the cox1 gene and 436 bp of its 3′ flanking region (dotted underlined segment in pYGT6, Fig. 1 A), was PCR amplified by using pYGT6 as template. The PCR product was digested with KpnI and ClaI and inserted into the same sites of pYPG09 to yield pYPG10 (Fig. 1 A).
Construction of rho– RIP1m Strains. The plasmids pYPG09 or pYPG10 were introduced by cotransformation with the nuclear selectable URA3 plasmid YEp352 into the rho0 strain W303-1B/A/50 (MATa, ura3, his3, leu2, ade2, trp1) by microprojectile bombardment using a Biolistic PDS-1000/He Particle Delivery System (Bio-Rad) as described (15). Mitochondrial transformants were identified among the URA3+ nuclear transformants by their ability to produce respiring diploids when mated to the nonrespiring tester strain (TF145, MATα, ade2, ura3) bearing a cox2 deletion mutant (16). Mitochondria of two stable rho– clones carrying the pYPG09 and pYPG10 plasmids were transferred into a rho0 kar1-1 strain and cytoducted into a rho0 derivative of JPJ1 (MATa, ura3, his3, ade2, leu2, trp1, rip1::LEU2) (13). These Δrip1 rho– clones, PG09 (pYPG09) and PG10 (pYPG10), were used for complementation tests by crossing to a Δrip1 rho+ haploid strain.
DNA and RNA Manipulations. Total mitochondrial RNA extraction, purification, and Northern blot analysis were carried out as described (15). mtDNA was purified as described (17), and restriction and Southern blot analyses were carried out according to ref. 18. Two probes specific to the cox1 gene were used: pYJL17, which carries a fragment of exon 4 (19), served in the Northern blot experiments and the synthetic oligonucleotide (5′-GCGTCGACTATGTATTATCAATGGGT-3′), corresponding to part of exon 5 sequences, was used in the Southern blot experiments. The RIP1 probe was pYGT37 (described above). The atp8/atp6 probe was pYGT7, which contains the 3′ part of atp8, the intergenic region, and the atp6 sequences cloned in pUC13.
Plasmid DNAs were sequenced with the Sequenase version 2.0 kit (Amersham Pharmacia) by using either double-stranded or single-stranded templates. For sequencing of the last exon of the cox1 gene from Saccharomyces capensis and from recombined mtDNAs, 3–5 μg of mtDNA was cycle-sequenced in the Perkin–Elmer GeneAmp 2400 thermocycler by using the Thermo-Sequenase kit (Amersham Pharmacia) and the primer (5′-ATGTATTATCAATGGGTGCT-3′) labeled at the 5′ end with [γ-32P]dATP by using T4 polynucleotide kinase.
Immunodetection and bc1 Complex Activities. Mitochondrial extracts made from purified mitochondria of RGLT1, RR1, and isogenic wild-type strains were run on SDS-15% polyacrylamide gels and the proteins were transferred to an Optitran BA-S83 membrane as described (20). Immunoblotting was performed by using a Bio-Rad Western processor with the following antibody dilutions: Homo sapiens polyclonal anti-Rieske FeS protein (a gift from C. Godinot, Centre de Génétique Moléculaire et Cellulaire, Lyon, France), 1:50,000; anti-rabbit secondary antibody, 1:20,000; monoclonal anti-outer membrane porin, 1:20,000; and anti-mouse secondary antibody, 1:10,000; and the signals were revealed by using the Pierce Super Signal Pico detection system. The antimycin-sensitive bc1 complex activity was measured according to ref. 21.
Results
A Synthetic Mitochondrial Version of the Nuclear Gene RIP1 Is Functional in Heteroplasmons. To express a nuclear gene within mitochondria, two basic requirements should be fulfilled. (i) The sequence of the nuclear gene should be modified to adjust for the differences in the genetic code between the two cellular compartments (22). (ii) The recoded gene should be placed under the control of mitochondrial signals necessary for its expression within the organelle. The appropriate construction cloned in a plasmid can then be introduced into yeast mitochondria lacking mtDNA (rho0) by biolistic transformation, and maintained in trans with or integrated into a complete mitochondrial chromosome (rho+) by crosses between synthetic rho– and recipient rho+ strains.
The 216-codon RIP1 gene sequence contains four CTN leucine codons that specify threonine in yeast mitochondria, one ATA isoleucine codon that specifies methionine in mitochondria, and a TAG termination codon that is rarely used in mitochondrial genes. To construct RIP1m, a full-length mitochondrial version of the RIP1 ORF able to encode the precursor form of Rip1p, we replaced the fourth codon by ATT and changed the termination codon to TAA (see Materials and Methods). Among the four CTN leucine codons, two (L22 and L28) are located in the presequence, one (L65) in the transmembrane domain, and the last one (L156) in the catalytic domain (23). The comparison of the Rieske protein sequences from a variety of organisms (13) shows that these four leucines are not conserved; we therefore hypothesized that their substitution by threonine would not dramatically affect the function of the protein.
To ensure the expression of RIP1m in the organelle, its ORF was precisely flanked with the sequences originally surrounding the S. cerevisiae cox1 ORF to yield pYPG09. The upstream cox1 region used (0.97 kb, 5′ PR, Fig. 1 A) carried the conserved nonanucleotide motif considered as a yeast mitochondrial promoter from which the synthesis of the cox1 mRNA is initiated (24). The downstream cox1 region used (1.1 kb, 3′ TR, Fig. 1 A) contained, in addition to the atp8 gene, the conserved dodecamer sequence, located 78 bp downstream from the stop codon of the cox1 gene, which determines a site of processing of the 3′ end of the cox1 mature mRNA (25). Thus, plasmid pYPG09 contains all of the sequences required for the mitochondrial expression of RIP1m in trans. This construct could not be used for the stable integration of RIP1m into a rho+ mtDNA, because recombination would simultaneously eliminate the cox1 ORF (Fig. 1B), but it allowed us to test the complementation of RIP1 deletion by coexpression of RIP1m and cox1 in an heteroplasmon.
A synthetic rho– strain carrying pYPG09 RIP1m in its mitochondria was obtained by biolistic transformation of a rho0 recipient, and these rho– mitochondria were transferred into a Δrip1 nuclear background (13) by cytoduction. The resulting rho– strain, PG09 (Fig. 1 A, Table 1), was used as a donor of RIP1m in crosses with several Δrip1 rho+ isonuclear strains carrying differently organized mitochondrial genomes (Table 1), to determine whether differences in mtDNA structures or gene content could influence the mitochondrial stability and expression of a foreign gene. The mtDNAs differed mainly by the intron content and presence of various alleles of the ENS2 gene, the active product of which encodes the mitochondrial subunit of the Endo.SceI endonuclease involved in mtDNA cleavage and recombination (26).
Among nine rho+ recipient strains crossed with the synthetic rho– PG09, only two, carrying 777-3A (27) or S. capensis (28) mtDNAs, respectively, generated respiring diploids (DR3 and DR4, Fig. 2A). The respiratory-proficient phenotype of these two diploids was highly unstable on growth under nonselective conditions with fermentable medium, showing a rate of mitotic segregation typical of heteroplasmons (Table 1). Southern blot analysis of the DR3 diploid mtDNA with various probes gave the signals expected for the superposition of the rho+ and rho– mtDNAs, with nonstoichiometric proportions of different restriction fragments characteristic of the heteroplasmic state (Fig. 3 B–E).
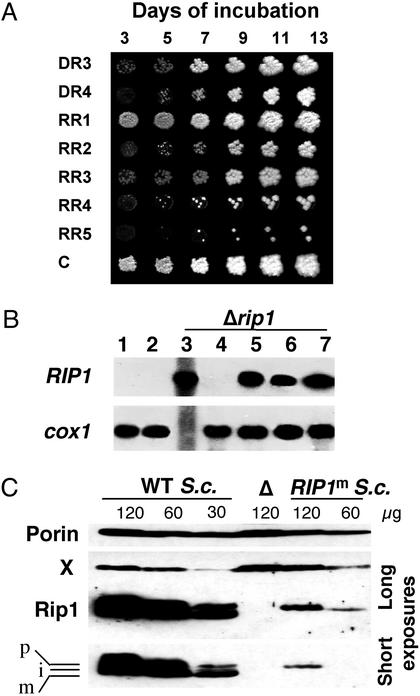
Fig. 2.
RIP1m complements the deletion of the nuclear gene and is expressed inside the organelle. (A) Glycerol growth of diploid strains. The names of diploids are on the left (see Table 1). C is the result of a control cross between the rho– (PG10) and a tester strain. Drops of diploids were replica plating onto selective medium and incubated at 28°C. (B) Northern blot analysis. Total mitochondrial RNAs were purified from isolated mitochondria, separated on a 1.1% denaturing agarose gel, transferred to Hybond C filter, and hybridized with a RIP1 probe and a cox1 exon 4 probe. Mitochondrial RNAs were from wild-type S. cerevisiae (lane 1) and wild-type S. capensis (lane 2) strains used as controls, and Δrip1 strains PG10 (lane 3), RGLT1 (lane 4), RR1 (lane 5), RR2 (lane 6), and DR3 (lane 7). (C) Immunodetection of the Rip1 protein. Mitochondrial proteins from S. cerevisiae (S.c.) wild-type, Δrip1 (Δ), or RIP1m (RR1) strains with 777-3A mtDNA were analyzed by Western blotting and hybridized with anti-human Rieske FeS protein or anti-outer membrane porin antibodies. Two different exposures are given for Rip1p detection. The porin signal as well as an unknown protein (X) cross-reacting with the anti-FeS protein antibody reflect the different quantities loaded on each lane (120, 60, or 30 μg). p, i, and m indicate the precursor, intermediate, and mature forms of Rip1p, respectively.
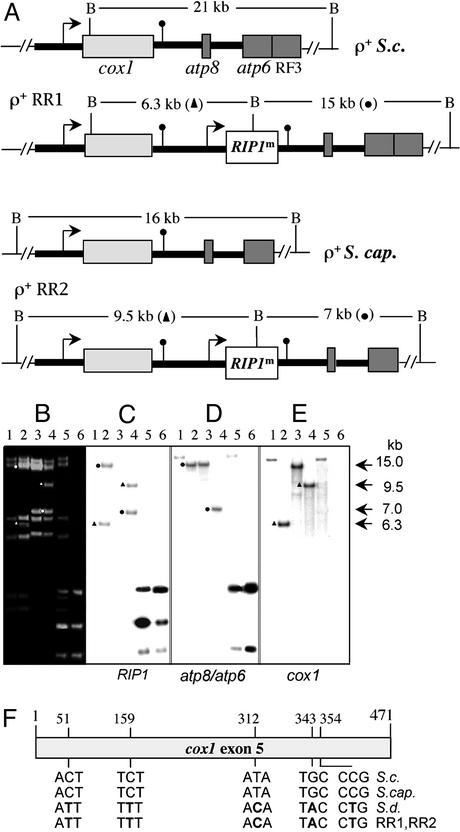
Fig. 3.
Molecular analysis of heteroplasmic and recombinant mtDNAs from the respiratory-competent diploids. (A) Diagram of the polygenic transcription unit from rho+ (ρ+) S. cerevisiae (S. c.), S. capensis (S. cap.) RR1 and RR2 strains. The location of BspHI (B) sites and the lengths of wild-type and new recombined fragments (triangles and circles) are indicated. Thick lines represent intergenic sequence; designation of boxes and transcriptional signals are as given in the legend to Fig. 1. (B) Ethidium bromide-stained agarose gel with BspHI fragments of mtDNA from the S. cerevisiae wild-type strain 777-3A (lane 1), RR1 (lane 2), S. capensis wild-type strain (lane 3), RR2 (lane 4), DR3 (lane 5), and PG10 (lane 6). (C–E) Autoradiograms of blots hybridized with the indicated probes. Sizes of new recombined fragments are indicated. (F) Sequence of the last exon of the cox1 gene in recombinant mtDNAs from RR1 and RR2. S.c., S.cap., S.d., the wild-type sequences of S. cerevisiae (GenBank accession no. V00694), S. capensis (this work), and S. douglasii (GenBank accession no. M97514), respectively. Variable nucleotides at indicated positions are in bold.
RIP1m Can Be Stably Integrated in a New Form of mtDNA with Extended Gene Content. Because the RIP1m gene appeared to be functional at least in some mtDNA backgrounds, a second recombinant plasmid pYPG10 (Fig. 1 A) was constructed to direct stable integration of RIP1m into rho+ mtDNA. We reasoned that the best way to ensure the expression of RIP1m was to insert it into the largest multigenic transcriptional unit of yeast mtDNA containing three (cox1, atp8, atp6) or four (plus RF3) genes, depending on the strain (24, 29, 30; Fig. 3A). To allow homologous recombination between the new construct and rho+ mtDNA, the last S. douglasii cox1 exon and part of its terminator region were cloned upstream of the cox1::RIP1m gene. This large additional region homologous to the 3′ part of the cox1 gene (886 bp) should promote integration of RIP1m between the cox1 and atp8 genes in rho+ mtDNA (Fig. 1C). S. douglasii rather than bona fide S. cerevisiae cox1 sequences were used, because repeated sequences in S. cerevisiae mtDNA are known to be highly unstable. Because the portion of S. douglasii sequence displays several polymorphic changes compared with the S. cerevisiae and S. capensis relevant sequences (Fig. 3F), we reasoned that this would lower the excision of the RIP1m gene by recombination and should allow us to discriminate wild-type and recombinant molecules.
A synthetic rho– strain, PG10, carrying this new pYPG10 RIP1m construct in a Δrip1 nuclear background was crossed to the same Δrip1 rho+ strains as before, selecting for respiring colonies on nonfermentable medium. As for PG09, respiring diploid colonies were obtained from crosses with strains carrying 777-3A and S. capensis mtDNA, but also with three other recipient strains (Fig. 2 A and Table 1). Respiratory growth of the best-complemented strain, RR1 carrying 777-3A mtDNA, was a little slower than that of wild type (3 days instead of 2 for growth on glycerol medium after replica-plating). However, the respiratory deficiency of four of the Δrip1 rho+ strains tested was not complemented (Table 1).
Analysis of the mitotic stability showed that the 777-3A- and S. capensis-derived diploids RR1 and RR2 were highly stable (Table 1), suggesting that they were homoplasmic strains. On the contrary, the three other respiring diploids issued from D273-10B (31), WF3/5 (30), or AF/32 mitochondrial backgrounds displayed an intermediate stability; the D273-10B-derived strain was the most unstable (Table 1). Another unusual feature of the WF3/5- and AF/32-derived diploids, RR4 and RR5, is their extremely low frequency of appearance (Fig. 2 A). Preliminary analysis of their mtDNAs (not shown) has shown that a predominant rho– mtDNA much larger than the original rho– mtDNA coexists with the rho+ mtDNA present in a minute amount. In contrast, Southern blot analyses of the RR1 and RR2 stable diploids revealed in all cases the bands expected new from a precise stable integration of RIP1m between the cox1 and atp8 genes (Fig. 3 A–E). The restriction patterns of the RR1 and RR2 mtDNAs using BspHI (Fig. 3B) or HaeIII (not shown) restriction endonucleases were characteristic of a single type of mtDNA. Furthermore, sequencing of the recombined molecules (Fig. 3F) clearly indicated that the last exon of cox1 gene in both RR1 and RR2 mtDNAs exhibited a sequence typical of S. douglasii and therefore originated from the pYPG10 construct.
Altogether, our results showed that the expression and stability of the relocated gene seems to depend on the mtDNA of the recipient strain, and that the RIP1m gene could be stably inserted into either S. cerevisiae (777-3A) or S. capensis rho+ mtDNAs. Moreover, sporulation and tetrad analysis of both diploids generated only respiratory competent spores, as expected for a mitochondrially inherited RIP1m gene (not shown).
RIP1m Is Expressed Within the Organelle and Restores Cytochrome bc1 Complex Function. To confirm that expression of RIP1m in the respiring diploids indeed occurred in the mitochondrial compartment we analyzed the mitochondrial transcripts from the heteroplasmic DR3 and the homoplasmic RR1 and RR2 diploids compared with wild-type and rho– controls (Fig. 2B). By using the RIP1 probe, the mRNA of RIP1 was detected in mitochondrial RNAs from the rho– and from the respiring diploids. The same blot was hybridized to the cox1 probe and, as expected, the mature cox1 mRNA was present in all of the rho+ strains, both diploid and haploid, and absent from the synthetic rho– strain, showing that the introduction of RIP1m did not affect transcription of cox1.
Western blot analysis was performed on purified mitochondria from the wild-type, Δrip1, and RIP1m strains carrying 777-3A mtDNA by using an anti-human Rieske FeS protein antibody known to react with the yeast Rip1p (21). Whereas no signal was detected in 120 μg of Δrip1mitochondrial proteins, the three typical (precursor/intermediate/mature) bands were already strongly detected in 30 μg of wild-type proteins (Fig. 2C). In the extract from the RIP1m strain, Rip1p could be detected only in 120 to 60 μg of mitochondrial proteins and apparently corresponded to the precursor form of Rip1p. Thus, the RIP1m gene seems to produce a low amount of unprocessed, but nevertheless functional Rip1p. However the possibility that a very low amount of mature Rip1p might be present but undetectable under our conditions cannot be excluded. The bc1 complex activities were recorded from the same mitochondria and showed that the RIP1m strain activity (expressed as nanomoles of cytochrome c reduced per minute and per milligram of mitochondrial proteins) was 9% (236 ± 66) of that of the wild-type cells (2,609 ± 244), compared with the complete lack of activity found in the Δrip1 strain. Analysis of cytochrome spectra showed that the RR1 [RIP1m] strain exhibited substantial reduction in cytochrome c1 and c (not shown).
These data show that in an appropriate mitochondrial genome, the RIP1m gene can promote significant growth on glycerol medium (Fig. 2 A) despite the low level of protein produced and the low level of bc1 complex activity.
Discussion
Relocation of a nuclear gene into mitochondria is not trivial because the genetic codes as well as the transcriptional and translational signals are different in the two compartments. An earlier relocated gene, the auxotrophic marker TRP1, was inserted into the mtDNA upstream of the cox2 gene without affecting the integrity of residing genes, but was not phenotypically expressed because it served as a mitochondrially inert DNA escape marker (4). Two recoded reporter genes, ARG8m and GFPm, have since been constructed (5, 32). However, in all of the cases where ARG8m was inserted, it was in place of, or in fusion with the cox2, cox3, or var1 coding sequences without duplicating any sequences, and most of the time its insertion led to respiratory deficiency (6–9).
We show that the mitochondrial copy of the relocated RIP1 gene can be stably integrated into the mtDNA and is expressed, and it complements the deficiency in its nuclear counterpart. Whereas it is known that the repeated sequences in yeast mtDNA are usually highly unstable, we show that large mtDNA duplications can be maintained in the cox1 region. The introduction and expression of RIP1m involved the creation of substantial (≈1 kb) tandem repeats of the cox1 flanking regions, while keeping the mitochondrial genes fully functional in a stable mtDNA. However, the fact that the repeated 3′ cox1 sequences were slightly divergent in the recombined molecules (S. douglasii versus S. cerevisiae) probably helped to stabilize such large duplications.
The mtDNA region where a foreign gene is inserted is likely to be important for expression of the transgene, because the S. cerevisiae mtDNA is expressed as multigenic transcriptional units. RNA synthesis of polygenic transcriptional units is initiated at a mitochondrial promoter, upstream of the first gene of the transcriptional unit (in this case cox1), and the following genes lacking this motif, are cotranscribed (24, 33). The duplication of the cox1 promoter in the larger transcriptional unit that we generated raises the question of how such a giant precursor RNA, at least 20 kb long, would be processed. If the duplicated promoter upstream of RIP1m is ignored by the RNA polymerase, the transcription could be initiated at the original promoter upstream of the cox1 gene, and thus the resulting polygenic primary transcript (including cox1, RIP1m, atp8, atp6, and RF3) would undergo multiple processing steps to yield mature mRNAs. Alternatively, if both initiation sites are recognized, the mature mRNA of cox1 could be transcribed alone and processed in a single step, and the RNA synthesis of the polycistronic primary transcript would then start at the promoter upstream of the RIP1m gene. In this case, a first endonucleolytic cleavage at the site of processing (24) downstream of RIP1m would yield the 3′ terminus of the RIP1m mRNA and the 5′ terminus of the RNA coding for atp8, atp6, and RF3, which would be later processed to produce mature mRNAs.
Although the RIP1m mRNA was efficiently transcribed and processed in the most stable complemented RIP1m strain, RR1, the steady-state amount of Rip1p and bc1 complex activity were rather low. The respiratory competent phenotype of this strain supports once again the idea that respiratory complex subunits are far more abundant than necessary to sustain normal respiratory growth under laboratory conditions. This low protein level could be due to inefficient translation, because the overall codon bias of the RIP1m ORF is not optimal. Alternatively, Rip1p produced inside the organelle might be less efficiently inserted/assembled into the bc1 complex, leading to instability of the RIP1m product. In this sense, it might be important that the RIP1m mRNA contains the 5′ untranslated leader from cox1, which directs its usual product to the inner mitochondrial membrane, and that the first transmembrane segment of Cox1p presents the same topology (N-terminal inside) as Rip1p, enhancing membrane insertion of Rip1p and ensuring subsequent assembly into the bc1 complex. However, because the Rieske protein is the last member of complex III to be added to a partially assembled but stable complex (34), we cannot draw more general conclusions about how an allotopically expressed protein in the mitochondrion can integrate into the assembly pathway of a multisubunit complex. Still, we can propose that the unusual route of production and insertion of the RIP1m product might be responsible for the inefficient processing of Rip1p in this strain. Indeed, cotranslational insertion might circumvent the proteases that normally process the protein during posttranslational import. Alternatively, the substitution of leucines 22 and 28 by threonines in the precursor protein might also affect processing, because they are located in the presequence near the two proteolytic cleavage sites. Although we cannot exclude it, we do not favor this hypothesis because these substitutions at positions 22 and 28 do not affect the recognition consensus RX↓(F/L/I)XX(T/S/G)XXXX↓, which corresponds in wild-type S. cerevisiae Rip1p to RL↓ISQSLLAS↓ (35), and in the RIP1m product to RT↓ISQSLTAS↓. Also, it has been shown that the presence of various aberrant processing sites or uncleaved presequence does not dramatically disturb complex bc1 assembly or functionality (35, 36). Two remaining CTN leucines, 65 and 156, were translated as threonines in the RIP1m product. The threonine at position 65 is included in the long helical transmembrane domain of Rip1p that does not appear to interact directly with any other subunit. This T65 should not disrupt the helical structure of this segment and is thus unlikely to affect the assembly of Rip1p into the complex (37). The substitution of L156 by threonine is located in a β-sheet fold of the extrinsic catalytic domain and should not disturb the overall folding of the domain. This region does not directly interact with either the FeS cluster or other subunits of the complex. Thus, according to the crystal structure of the yeast bc1 complex (23), these mutations should not drastically affect the assembly of Rip1p into the complex.
Finally, that the mitochondrial background clearly plays a role in the stability and/or expression of the transgene was an unexpected observation. Nine isonuclear strains carrying differently organized mitochondrial genomes were tested in complementation assays and gave very different results. Based on what is known about the structure of the rho+ mtDNAs used, it seems that the known polymorphic features like the optional presence of introns or reading frames outside the genes RF1, RF2, and RF3 (ENS2 if active) do not influence the stability and/or expression of RIP1m. The unequal ability to express the transgene observed between different recipient strains might rather be caused by allelic variations of mitochondrial genes encoding components of the mitochondrial translation apparatus, like ribosomal RNAs and the Var1p protein.
The results of this study also have an evolutionary impact. According to current theories, genes related to mitochondrial biogenesis and function have moved from the genome of the endosymbiotic mitochondrial ancestor into the nucleus in the process of reductive genome evolution (1, 38). The protein products of such genes are still imported and functional in mitochondria. Our demonstration that a “present-day” nuclearly encoded gene can be “returned” to and expressed in mitochondria, its original location, is in some way an artificial reversal of evolutionary events, albeit on a very limited scale. Nevertheless, it opens possibilities in the research on the unsolved problems concerning the evolutionary origin of organelles.
Acknowledgments
We thank G. L. Tian, B. Meunier, T. D. Fox, B. L. Trumpower, J. P. di Rago, and C. Godinot for providing yeast strains, plasmids, and antibodies, C. J. Herbert for critical reading and looking over the English, and P. P. Stepien for his interest. We are deeply grateful to P. P. Slonimski for helpful discussions and encouragement. P.G. was the recipient of a postdoctoral fellowship from the Centre National de la Recherche Scientifique within the framework of the Centre Franco-Polonais. N.B. is supported by the Association Française contre les Myopathies. Y.S.-G. is the recipient of a Fondation pour la Recherche Médicale fellowship.
References
- 1.Gray, M. W., Burger, G. & Lang, F. B. (1999) Science 283, 1476–1481. [DOI] [PubMed] [Google Scholar]
- 2.Thorsness, P. E. & Weber, E. R. (1996) Int. Rev. Cytol. 165, 207–234. [DOI] [PubMed] [Google Scholar]
- 3.Thorsness, P. E. & Fox, T. D. (1990) Nature 346, 376–379. [DOI] [PubMed] [Google Scholar]
- 4.Thorsness, P. E. & Fox, T. D. (1993) Genetics 134, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steele, D. F., Butler, C. A. & Fox, T. D. (1996) Proc. Natl. Acad. Sci. USA 93, 5253–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchirico, M. E., Fox, T. D. & Mason, T. L. (1998) EMBO J. 17, 5796–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnefoy, N., Bsat, N. & Fox, T. D. (2001) Mol. Cell. Biol. 21, 2359–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green-Willms, N. S., Butler, C. A., Dunstan, H. M. & Fox, T. D. (2001) J. Biol. Chem. 276, 6392–6397. [DOI] [PubMed] [Google Scholar]
- 9.Saracco, S. A. & Fox, T. D. (2002) Mol. Biol. Cell 13, 1122–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trumpower, B. L. (1990) Microbiol. Rev. 54, 101–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabellini, N. (1988) J. Bioenerg. Biomembr. 20, 59–83. [DOI] [PubMed] [Google Scholar]
- 12.Kurowski, B. & Ludwig, B. (1987) J. Biol. Chem. 262, 13805–13811. [PubMed] [Google Scholar]
- 13.Beckmann, J. D., Ljungdahl, P. O. & Trumpower, B. L. (1989) J. Biol. Chem. 264, 3713–3722. [PubMed] [Google Scholar]
- 14.Beckmann, J. D., Ljungdahl, P. O., Lopez, J. L. & Trumpower, B. L. (1987) J. Biol. Chem. 262, 8901–8909. [PubMed] [Google Scholar]
- 15.Szczepanek, T. & Lazowska, J. (1996) EMBO J. 15, 3758–3767. [PMC free article] [PubMed] [Google Scholar]
- 16.Mulero, J. J. & Fox, T. D. (1993) Mol. Biol. Cell 4, 1327–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gargouri, A. (1989) Curr. Genet. 15, 235–237. [Google Scholar]
- 18.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 19.Tian, G. L., Macadre, C., Kruszewska, A., Szczesniak, B., Ragnini, A., Grisanti, P., Rinaldi, T., Palleschi, C., Frontali, L., Slonimski, P. P. & Lazowska, J. (1991) J. Mol. Biol. 218, 735–746. [DOI] [PubMed] [Google Scholar]
- 20.Lemaire, C., Hamel, P., Velours, J. & Dujardin, G. (2000) J. Biol. Chem. 275, 23471–23475. [DOI] [PubMed] [Google Scholar]
- 21.Saint-Georges, Y., Bonnefoy, N., di Rago, J. P., Chiron, S. & Dujardin, G. (2002) J. Biol. Chem. 277, 49397–49402. [DOI] [PubMed] [Google Scholar]
- 22.Fox, T. D. (1987) Annu. Rev. Genet. 21, 67–91. [DOI] [PubMed] [Google Scholar]
- 23.Hunte, C., Koepke, J., Lange, C., Rossmanith, T. & Michel, H. (2000) Struct. Fold. Des. 8, 669–684. [DOI] [PubMed] [Google Scholar]
- 24.Osinga, K. A., De Vries, E., Van der Horst, G. T. J. & Tabak, H. F. (1984) Nucleic Acids Res. 12, 1889–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osinga, K. A., De Vries, E., Van der Horst, G. T. J. & Tabak, H. F. (1984) EMBO J. 3, 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawa, K., Morishima, N. & Shibata, T. (1992) EMBO J. 11, 2702–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazowska, J., Jacq, C. & Slonimski, P. P. (1980) Cell 22, 333–348. [DOI] [PubMed] [Google Scholar]
- 28.Lazowska, J., Szczepanek, T., Macadre, C. & Dokowa, M. (1992) C. R. Acad. Sci. Paris 315, 37–41. [PubMed] [Google Scholar]
- 29.Simon, M. & Faye, G. (1984) Mol. Gen. Genet. 196, 266–274. [DOI] [PubMed] [Google Scholar]
- 30.Lazowska, J., Meunier, B. & Macadre, C. (1994) EMBO J. 13, 4963–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grivell, L. A. (1989) Eur. J. Biochem. 182, 477–493. [DOI] [PubMed] [Google Scholar]
- 32.Cohen, J. S. & Fox, T. D. (2001) Mitochondrion 1, 181–189. [DOI] [PubMed] [Google Scholar]
- 33.Christianson, T. & Rabinowitz, M. (1983) J. Biol. Chem. 258, 14025–14033. [PubMed] [Google Scholar]
- 34.Crivellone, M. D., Wu, M. A. & Tzagoloff, A. (1988) J. Biol. Chem. 263, 14323–14333. [PubMed] [Google Scholar]
- 35.Nett, J. H., Schagger, H. & Trumpower, B. L. (1998) J. Biol. Chem. 273, 8652–8658. [DOI] [PubMed] [Google Scholar]
- 36.Nett, J. H., Denke, E. & Trumpower, B. L. (1997) J. Biol. Chem. 272, 2212–2217. [DOI] [PubMed] [Google Scholar]
- 37.Beattie, D. S., Wang, Y. & Obungu, V. H. (1999) J. Bioenerg. Biomembr. 3, 215–224. [DOI] [PubMed] [Google Scholar]
- 38.Kurland, C. G. & Andersson, S. G. E. (2000) Microbiol. Mol. Biol. Rev. 64, 786–820. [DOI] [PMC free article] [PubMed] [Google Scholar]