Abstract
Ig-like transcript 4 (ILT4) (also known as leukocyte Ig-like receptor 2, CD85d, and LILRB2) is a cell surface receptor expressed mainly on myelomonocytic cells, whereas ILT2 (also known as leukocyte Ig-like receptor 1, CD85j, and LILRB1) is expressed on a wider range of immune cells including subsets of natural killer and T cells. Both ILTs contain immunoreceptor tyrosine-based inhibitory receptor motifs in their cytoplasmic tails that inhibit cellular responses by recruiting phosphatases such as SHP-1 (Src homology 2 domain containing tyrosine phosphatase 1). Although these ILTs have been shown to recognize a broad range of classical and nonclassical human MHC class I molecules (MHCIs), their precise binding properties remain controversial. We have used surface plasmon resonance to analyze the interaction of soluble forms of ILT4 and ILT2 with several MHCIs. Although the range of affinities measured was quite broad (Kd = 2–45 μM), some interesting differences were observed. ILT2 generally bound with a 2- to 3-fold higher affinity than ILT4 to the same MHCI. Furthermore, ILT2 and ILT4 bound to HLA-G with a 3- to 4-fold higher affinity than to classical MHCIs, suggesting that ILT/HLA-G recognition may play a dominant role in the regulation of natural killer, T, and myelomonocytic cell activation. Finally, we show that ILT2 and ILT4 effectively compete with CD8 for MHCI binding, raising the possibility that ILT2 modulates CD8+ T cell activation by blocking the CD8 binding as well as by recruiting inhibitory molecules through its immunoreceptor tyrosine-based inhibitory receptor motif.
Keywords: leukocyte Ig-like receptors, major histocompatibility complex, surface plasmon resonance, natural killer cell, coreceptor
Ig-like transcripts (ILTs) (also called leukocyte Ig-like receptors, CD85, or LILRB) are encoded by a family of immunoreceptor genes located at human chromosome 19q13.4. This locus is called the leukocyte receptor complex and includes, in addition to ILT genes, the genes encoding killer cell Ig-like receptors (KIRs), leukocyte-associated Ig-like receptors, NKp46, and the Fcα receptor (1). Although ILT2 is broadly expressed on monocytes, B cells, dendritic cells, and subsets of natural killer (NK) and T cells, ILT4 expression is largely confined to the myelomonocytic lineage (2–8). Both ILT2 and ILT4 have four tandem Ig-like extracellular domains and four and three immunoreceptor tyrosine-based inhibitory receptor motifs, respectively, in their cytoplasmic tails. Immunoreceptor tyrosine-based inhibitory receptor motifs recruit the protein tyrosine phosphatase SHP-1 (Src homology 2 domain containing phosphatase 1), which is thought to inhibit early signaling events triggered by stimulatory receptors. Indeed engagement of ILT2 on T cells has been shown to inhibit T cell antigen receptor (TCR) signaling and downstream events such as actin reorganization (9). Studies on CD8+ cells suggest that ILT2 is expressed early on in contrast to KIRs, which are expressed primarily on the subset of stimulated CD8+ cells that become long-term memory cells (10). In addition, ILT2 is a major inhibitory receptor on NK cells for the nonclassical HLA-G ligand. On the other hand, ILT4 may be involved in regulating the activation of inflammatory cells in a range of systems: (i) CD68+ macrophages and neutrophils in synovium from rheumatoid arthritis patients (11); (ii) myelomonocytic cells in materno-fetal tolerance (12); and (iii) tolerogenic antigen-presenting cells induced by regulatory T cells (13).
Based on killing and cell-binding assays, ILT2 and ILT4 have been shown to bind to classical (HLA-A and -B) and nonclassical (HLA-G1, -E, and -F) MHC class I molecules (MHCIs). In these cellular assays, minimal or no binding to HLA-C was detected (3–5, 7, 8, 12, 14–16). Cellular assays also indicate that ILT2 binds to the MHCI homolog from human cytomegalovirus UL18, whereas ILT4 does not. Using purified proteins, however, Chapman et al. (17) detected ILT2 binding to HLA-C alleles and ILT4 binding to UL18, with affinities that are within the range typical of cell–cell recognition interactions.
Domain deletion and mutational analyses of the ILT2 ectodomain have revealed that the N-terminal domain 1 (D1) is the main MHCI-binding region (17, 18). Conversely, domain-swapping experiments have shown that ILTs bind to the α3 domain of MHCIs. This is distinct from the KIR-binding site, which lies within the α1–α2 region (17, 19, 20). Because the α3 domain is relatively more conserved among the MHCI alleles than the polymorphic α1–α2 peptide-binding region, these data account for the broader binding specificity of ILT2 compared with KIRs. The crystal structure of the D1 and D2 region of ILT2 (ILT2D1D2) has been solved (18), demonstrating that ILT2D1D2 had two Ig-like domains in tandem related by an acute elbow angle, similar to KIRs. However, the MHC-binding sites are distinct. The binding site on ILT2 is confined to the GFC β-sheet surface of the D1 domain [Ig domains possess two β-sheets, made up of ABE(D) or GFC(C′C″) β-strands, respectively], whereas the corresponding site on KIRs involves the interdomain region between domains 1 and 2.
In contrast to ILT2, the binding studies on recombinant ILT4 have been limited to the ILT4–UL18 interaction (18). Here we report an investigation of the binding of ILT4 to a wide range of classical and nonclassical MHCIs, together with a thorough analysis of ILT2 binding. ILT4 and ILT2 bound with low affinities to all MHCIs tested. Unexpectedly, ILT4 and ILT2 bound to the nonclassical MHCI HLA-G with 3- to 4-fold higher affinity than to classical MHCIs. Because the HLA-G molecule is uniquely expressed on the immunologically relevant sites such as trophoblasts in placenta, thymic epithelial cells, and some tumors (21–25) including glioma cells (26), this result suggests an important in vivo role for ILT–HLA-G interactions. Finally, competition assays showed that ILTs compete with CD8 for binding to MHCI. We discuss the implications of these findings for the function of ILTs on CD8+ T cells and NK cells.
Materials and Methods
Production of ILT2 and ILT4 Ectodomains. DNA encoding the first two extracellular domains (residues 1–197) of ILT4 was amplified from cDNA (6) by using 5′-G GAA CAT ATG GGG ACC ATC CCC AAG CCC-3′ as forward primer and 5′-CC CAA GCT TAC TAT GGG ACC AGG AAG CTC CAG G-3′ as reverse primer. The resultant fragments were digested with the restriction enzymes NdeI and HindIII and ligated into the pGMT7 vector (27) (designated pGMILT4D1D2). Escherichia coli strain BL21(DE3)pLysS cells (Novagen) harboring pGMILT4D1D2 produced ILT4D1D2 inclusion bodies. They were isolated from cell pellet by sonication and washed repeatedly with wash solution including 0.5% Triton X-100. The DNA encoding ILT2D1D2 region (residues 1–197) designed by using E. coli-favored codons was constructed by PCR with 10 chemically synthesized DNAs (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). The fragment was inserted into the pGMT7 vector (designated pGMILT2D1D2), and inclusion bodies containing ILT2D1D2 were obtained by using the same method as described above.
The purified ILT2D1D2 inclusion bodies were solubilized in denaturant solution including 6 M guanidine hydrochloride. By using the refolding buffer (0.1 M Tris·HCl, pH 8.0/0.4 M l-arginine/2 mM EDTA/5 mM reduced glutathione/0.5 mM oxidized glutathione/0.1 mM PMSF), the solubilized protein solution was diluted slowly to the final protein concentration of 1–2 μM and stirred for 48 h at 4°C. Then the refolding mixture of ILT2D1D2 or ILT4D1D2 was concentrated with a VIVAFLOW50 system (Sartorius). ILT2D1D2 was purified by gel filtration with Superdex 75 (Amersham Pharmacia). In the case of ILT4D1D2, after concentrating to 5 μM, the buffer was exchanged gradually to 50 mM phosphate buffer, pH 6.0, with the VIVAFLOW50 system. ILT4D1D2 was purified by anion-exchange chromatography (SP Sepharose, Amersham Pharmacia) followed by gel filtration (Superdex 75).
The whole ectodomain of ILT2 was obtained from transfected J558L cells producing chimeric ILT2 molecule fused with the Fc portion of human IgG1 (6). On the other hand, for the whole ectodomain of ILT4, an expression system producing soluble extracellular domains of ILT4 tagged at the C terminus with a c-Myc tag and poly-His tail was constructed.
Production of Soluble and Biotinylated MHC Molecules. Soluble biotinylated HLA-A*1101 (with peptide AIFQSSMTK), HLA-B*3501 (with peptide IPLTEEAEL), HLA-Cw*0401 (with peptide QYDDAVYKL), and HLA-G1 (with peptide RIIPRHLQL) with C-terminal biotin ligase (BirA) recognition sequence (GSLHHILDAQKMVWNHR) were prepared as described (27, 28). Each MHCI was biotinylated with 50 mM d-biotin/100 mM ATP/15 μM BirA for 15 μM MHCI in the reaction buffer. After biotinylation, MHCIs were separated from the reaction mixture by gel filtration (Superdex 75). Biotinylated HLA-Cw*0702 was refolded with peptide [DS11 (RYRPGTVAL) or DS12 (NKADVILKY)] and chemically biotinylated β2-microglobulin (β2m) in the same way as other MHCI complexes. Purification was accomplished by using Superdex 200 and MonoQ columns.
Surface Plasmon Resonance (SPR). SPR experiments were performed by using a BIAcore2000 (BIAcore, St. Albans, U.K.). The biotinylated MHCIs and control protein (biotinylated OX48 antibody or BSA) were immobilized on the research-grade CM5 chip (BIAcore), onto which streptavidin was covalently coupled. ILT2D1D2 and ILT4D1D2, after buffer exchange to HBS-EP (10 mM Hepes, pH7.4/150 mM NaCl/3.4 mM EDTA/0.005% Surfactant P20), were injected over the immobilized MHCIs. The binding response at each concentration was calculated by subtracting the equilibrium response measured in the control flow cell from the response in the MHCIs flow cell. Kinetic constants were derived by using the curve-fitting facility of the biaevaluation 3.0 program (BIAcore) to fit rate equations derived from the simple 1:1 Langmuir binding model (A + B ↔ AB). Other curve fitting was performed by origin 3 (Microcal Software, Northampton, MA). Affinity constants (Kd) were derived by Scatchard analysis or nonlinear curve fitting of the standard Langmuir binding isotherm.
The whole ectodomain of ILT2 fused with Fc was indirectly coupled to the SPR sensor surface by using an anti-human-Fc mAb. Sensor flow-cell surfaces coated with mAb alone served as paired controls. With regards to ILT4, the whole ectodomain was coupled indirectly to the SPR sensor chip by using the anti-ILT 40H2 mAb. MHCIs were injected over the immobilized ILTs in HBS-EP.
Competitive Binding Assays. The ectodomain of KIR2DL1 (residues 1–224) and CD8α homodimer (CD8αα) (residues 1–120) were produced as described (27, 29). ILT2D1D2 and ILT4D1D2 with or without a fixed amount of KIR2DL1(38 μM), which was almost saturated, were flowed over the immobilized HLA-Cw*0401. ILT2D1D2 and ILT4D1D2, with or without a fixed concentration of CD8αα (92 μM), were flowed over the immobilized HLA-B*3501, -Cw*0401, and -G1.
Results and Discussion
Production of Soluble ILTs and MHCIs. We initially attempted to express three forms of the ILT4 ectodomain: domain 1 alone (D1, residues 1–98); domains 1 and 2 (D1D2, residues 1–197); and the entire extracellular domains (D1–D4, residues 1–435). These constructs were expressed in E. coli as inclusion bodies and subsequently refolded in vitro. ILT4D1 and ILT4D1–D4 were difficult to refold and aggregated readily, making it impossible to produce enough material for further study. However, ILT4D1D2 was successfully refolded and purified as described in Materials and Methods. Because stored ILT4D1D2 tended to aggregate, we prepared freshly refolded and purified ILT4D1D2 for each experiment. The equivalent ILT2 fragment (ILT2D1D2) was prepared by using a very similar method (see Materials and Methods). The far-UV circular dichroism spectrum of ILT4D1D2 indicated that it had mainly β-sheet secondary structure, similar to ILT2D1D2 and as expected for a protein with two Ig-like domains (ref. 18 and data not shown). For comparison, soluble fragments comprising the entire ectodomain of ILT2 and ILT4 (ILT4D1–D4 and ILT2D1–D4) were produced by using the eukaryotic expression systems (see Materials and Methods).
All the MHCIs were expressed in bacterial inclusion bodies, refolded in vitro with bacterially expressed β2m and appropriate peptides, and purified as described in Material and Methods. With the exception of HLA-Cw*0702, all MHC heavy chains incorporated a C-terminal biotinylation tag (LHHILDAQKMVWNHR). After refolding and purification they were biotinylated with biotin ligase, which specifically biotinylates a lysine in the tag (underlined). HLA-Cw*0702 was refolded with chemically biotinylated β2m. Biotin-HLA-Cw*0702 produced in this way has been used successfully for KIR2DL3-binding studies (30).
Affinities of ILT2 and ILT4 Binding Toward MHCIs. Affinity measurements were performed by using SPR as implemented in the BIAcore2000 instrument. Soluble ILT4D1D2 or ILT2D1D2 was injected over sensor surfaces to which biotinylated MHCI complexes or (as negative controls) biotinylated OX48 antibody or BSA had been immobilized. The affinities of ILT4D1D2 and ILT2D1D2 binding to MHCIs were measured by equilibrium binding analysis. A range of concentrations of ILT4D1D2 or ILT2D1D2 was injected through flow cells with MHCIs immobilized.
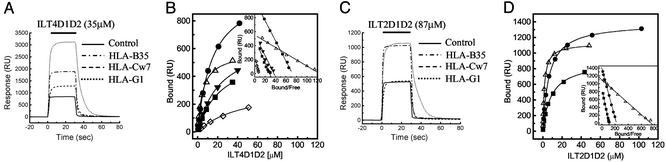
Representative data for equilibrium binding of ILT4D1D2 to various MHCIs are shown in Fig. 1A. ILT4D1D2 binding reached equilibrium and dissociated rapidly, indicating that it bound MHCI with the fast kinetics that are typical of interactions mediating cell–cell recognition [for example, see Maenaka et al. (30, 31)]. ILT2D1D2 binding also displayed fast kinetics (Fig. 1C and see below). Fitting of conventional and Scatchard plots of ILT4D1D2 (Fig. 1B and Inset) and ILT2D1D2 (Fig. 1D and Inset) binding data indicated that binding conformed to the simple 1:1 (Langmuir) binding model.
Fig. 1.
Equilibrium binding of ILT2 and ILT4 to MHCIs. (A and C) ILT4D1D2 (35 μM) (A) and ILT2D1D2 (87 μM) (C) were injected for 30 s through flow cell 1 with control (BSA, solid line), flow cell 2 with HLA-B35 (broken and dotted line), flow cell 3 with HLA-Cw7 refolded by using chemically biotinylated β2m (broken line), and flow cell 4 with HLA-G1 (dotted line). Biotinylated BSA was used as a control. (B and D) Plots of the equilibrium binding responses of ILT4D1D2 (B) and ILT2D1D2 (D) versus concentration. Diamonds, HLA-A11; squares, HLA-B35; circles, HLA-Cw4; downward triangles, HLA-Cw7; upward triangles, HLA-G1. The solid lines represent direct nonlinear fits of the 1:1 Langmuir binding isoform to the data. (Insets) Scatchard plots of the same data are shown. The solid lines are linear fits. RU, response units.
The results of multiple independent measurements for both ILT4D1D2 and ILT2D1D2 are summarized in Table 1 and Figs. 4 and 5, which are published as supporting information on the PNAS web site. ILT4D1D2 binds to a broad range of classical MHCIs (HLA-A*1101, -B*3501, -Cw*0401, and -Cw*0702) with affinities (Kd) ranging from ≈14 to ≈45 μM (Fig. 5 and Table 1). Interestingly, ILT4D1D2 binds with an even higher affinity (Kd ≈ 5 μM) to the nonclassical MHCI HLA-G1. A previous cellular binding study (7) reported that ILT4 binds to HLA-A2, -A3, -B8, -B27, -B35, and -G1 but not to HLA-Cw3 or -Cw5. This result is surprising given the small differences (one or two residue changes) in the relevant (α3) portions of HLA-Cw4, which we show binds ILT4 and HLA-Cw3 and -Cw5 (see Fig. 5C). However, there have been previous reports of discrepancies between cell-based and direct binding studies when measuring ILT2–HLA-C interactions (6–8). The reason for these discrepancies remains unclear, but it may be related to avidity effects (cell-based assays rely on multivalent binding) and/or the low level of cell surface expression of HLA-C when compared with other MHC alleles.
Table 1. Summary of affinity constants of the interactions of ILT2D1D2 and ILT4D1D2 to MHCIs.
|
Injected, Kd at 25°C, μM
|
|||
|---|---|---|---|
| Immobilized | sILT2D1D2 | sILT4D1D2 | CD8αα |
| HLA-A11 | ND | 45 ± 17 (3) | ND |
| HLA-B35 | 8.8 ± 0.2 (5) | 26 ± 4.6 (8) | 126 ± 3 (4) |
| HLA-Cw4 | 6.5 ± 0.5 (5) | 14 ± 2.0 (4) | 210 ± 10 (2) |
| HLA-G1 | 2.0 ± 0.7 (11) | 4.8 ± 1.4 (10) | 72 ± 1.4 (4) |
| HLA-Cw7/DS11* | NB | 26 ± 6.0 (8) | ND |
| HLA-Cw7/DS12* | NB | 23 ± 6.2 (4) | ND |
Shown is the mean ± standard deviation. The number of measurements is shown in parentheses. The immobilized levels of MHCs are from 800 to 3,000 response units. NB, no binding observed at the ILT2D1D2 concentration of 87 μM; ND, not determined.
Chemically biotinylated β2m was used.
ILT2D1D2 binds to HLA-B35 and HLA-Cw4 with affinities (Kd ≈ 8.8 and 6.5 μM, respectively) similar to those measured for KIR–MHCI interactions (Kd ≈ 7–10 μM) (30, 32). As found with ILT4D1D2, the ILT2D1D2 bound with the highest affinity to HLA-G1 (Kd ≈ 2.0 μM). As with ILT4, these results are consistent with the previous cellular-based assays with the exception of HLA-Cs. HLA-C transfectants do not bind efficiently to ILT2 Fc fusion protein or inhibit the killing activity of ILT2+ NK cells (6, 15).
The higher affinity of ILT2 and ILT4 for HLA-G1 versus other MHCIs (HLA-A2, -A68, -B8, and -E) was confirmed by using entire ectodomains (D1–D4) of both ILT2 and ILT4 produced by eukaryotic cells (data not shown). In contrast, Chapman et al. (17) reported that ILT2 binds HLA-G1 with a lower affinity (Kd ≈ 100 μM) than it binds other MHCIs. The reason for this discrepancy is uncertain, but it may be the result of differences in the way MHCIs were immobilized. In our study we typically used the site-specific biotinylated tag at the C terminus of the MHCI heavy chain, whereas Chapman et al. (17) randomly coupled the MHCI via amines. The former method presents MHCI in a manner optimal for ligand binding, whereas it is possible that the random coupling of MHC disrupts or interferes with ILT binding.
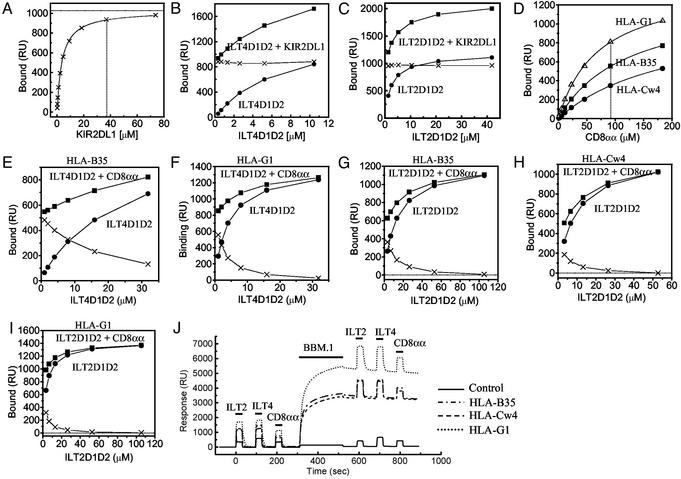
ILTs Compete with CD8 for Binding to MHCIs. Although there is evidence from domain-swapping experiments that ILT2 binds the α3 domain of MHCI heavy chains (17), it is not known whether ILT binding interferes with the binding of other MHCI ligands. We therefore used SPR to examine whether ILT binding to MHCI influenced the binding of the MHCI ligands KIR2DL1 and CD8 and the mAb BBM.1, which binds β2m. KIR2DL1 binds the region adjacent to the peptide C terminus on the peptide-binding platform of HLA-Cw4 and related alleles [Fig. 2A and Fan et al. (20)].
Fig. 2.
The effect of KIR2DL1, anti-β2m mAb (BBM.1), or CD8αα on the binding of a soluble ILT2 and ILT4 to MHCIs. (A) Equilibrium binding analysis of KIR2DL1 against HLA-Cw*0401 (Kd ≈ 3.3 μM) on the same sensor chip used in the experiments shown in B and C. The estimated saturation level of KIR2DL1 was calculated by nonlinear curve fitting [1,026 response units (RU)]. (B and C) Binding of ILT4D1D2 (B) and ILT2D1D2 (C) (filled circles) alone or mixed with KIR2DL1 (filled squares). The concentration of KIR2DL1 was 38 μM (A, dotted line). The difference in the binding seen with or without KIR2DL1 was plotted (crosses). (D) Equilibrium binding analysis of CD8αα against HLA-B*3501 (squares), HLA-Cw*0401 (circles), and HLA-G1 (upward triangles) on the same sensor chip used in the experiments shown in E–I. The Kd values are listed in Table 1. (E and F) Binding of ILT4D1D2 with or without CD8αα to HLA-B*3501 and HLA-G1, respectively. The concentration of CD8αα was 92 μM(D, dotted line). The difference in the binding seen with or without CD8αα was plotted (crosses) in the experiments shown in E–I.(G–I) Binding of ILT2D1D2 with or without CD8αα to HLA-B*3501 (G), HLA-Cw*0401 (H), and HLA-G1 (I). The concentration of CD8αα was 92 μM. (J) Binding of ILT2D1D2 (105 μM), ILT4D1D2 (33 μM), and CD8αα (92 μM) before and after injection of anti-β2m BBM.1 mAb to saturation level.
Having established that KIR2DL1 binds HLA-Cw4 with an affinity of Kd ≈ 3.3 μM (Fig. 2A), we examined the binding response when increasing concentrations of ILT4 (Fig. 2B) or ILT2 (Fig. 2C) were injected over HLA-Cw4 with (squares) or without (circles) a fixed, high concentration (38 μM) of KIR2DL1. The difference between the responses remained the same (crosses) whatever the concentration of ILTs, indicating that the binding of ILTs and KIR2DL1 to HLA-Cw4 was purely additive. This demonstrates that the binding of ILTs does not affect the binding of KIR2DL1 to the same molecule, consistent with previous data showing that they bind to different regions of the MHCI.
We next examined whether ILTs interfered with CD8 binding to MHCIs (Fig. 2 E–I). Having showed that soluble recombinant CD8αα bound to HLA-B35, -Cw4, and -G1 with affinities of Kd ≈ 126, 210, and 72 μM, respectively (Fig. 2D and Table 1), we injected increasing concentrations of ILT2 and ILT4 in the presence (squares) or absence (circles) of CD8αα (92 μM) over different MHCIs (Fig. 2 E–I). The difference between the responses with or without CD8αα (crosses) decreased as the concentration of ILTs increased, indicating that binding was not additive and that ILTs inhibited CD8αα binding to these MHCIs. In assays in which the concentration of ILT used was saturating, this inhibition was complete (Fig. 2 F–I), which is typical of competitive inhibition and indicates that the ILT and CD8αα-binding sites on MHCI overlap or are close enough to each other to block the binding. This is consistent with the demonstration that ILT binding requires the α3 domain of MHCIs (17).
We next examined the effect of the anti-β2m mAb BBM.1 on the binding of ILTs and CD8αα (Fig. 2J). The binding responses observed for ILT2, ILT4, and CD8αα to three different MHCIs were the same as before and after saturating the MHCI with BBM.1 mAb (Fig. 2J), indicating that the BBM.1-binding site does not overlap with either the CD8αα- or ILT-binding sites. There is some controversy as to the exact position of the BBM.1-binding site on β2m; one putative site overlaps with the CD8αα site (residues 58–63 and 91–95; shown in cyan on β2m in Fig. 3C) (33), whereas the other proposed site (residues 38, 44, and 45; blue surfaces in Fig. 3C) does not (34). Our results suggest that the latter site is the BBM.1 epitope.
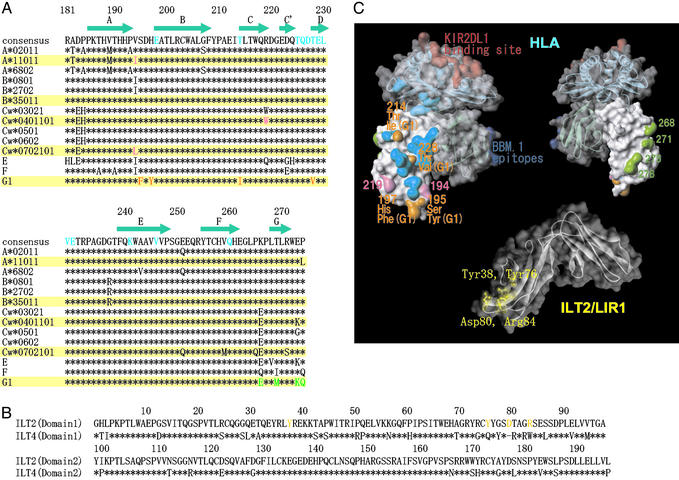
Fig. 3.
The putative ILT-binding site of MHCI. (A) Amino acid sequence alignment of α3 domains of HLA class I alleles (183–276). (B) Amino acid sequence alignment of the D1 and D2 domains of ILT2 and ILT4. (C Upper Left) Surface and ribbon diagram of HLA class I molecule with solid (α3 domain) and transparent surface. The putative residues playing an important role in strong binding of ILT2 and ILT4 to HLA-G1 (see Results and Discussion) are shown in orange. The residues of the α3 domain involved in MHCI–CD8αα binding are shown in cyan (35). The putative residues playing a part in the differentiation of ILT binding to different MHCI alleles are shown in pink. The residues interacting with KIR2DL1 are shown in red. The important epitopes of anti-β2m antibody BBM.1 (residues 38, 44, and 45) are shown in blue (34). (Upper Right) The reverse side view of that shown in the Upper Left. The residues of HLA-G1, which differ from those of the other MHCI alleles on the G strand, are shown in green. (Lower) Surface and ribbon diagram of ILT2D1D2. The residues playing an important role in binding to UL18 are shown in yellow (18). The colors used in this figure correspond to those of alignment shown in A and B. The diagrams in C were created with weblab viewer lite (Accelrys, San Diego).
The binding sites of KIR2DL1 (20), BBM.1 mAb (34), and CD8αα (35), as defined by mutagenesis and structural studies, are shown in Fig. 3C. Our results show that the ILT-binding site overlaps the CD8αα-binding site (cyan surfaces in Fig. 3C). A previous domain-swapping study demonstrated that the α3 domain is required for ILT binding (17). Taken together these data suggest that the ILT-binding site overlaps with the portion of the CD8αα-binding site located on the α3 domain. Consistent with this observation, ILT4 binding to HLA-Cw7 and -G1 was not affected by changing the peptide in the peptide groove (data not shown).
To localize the ILT-binding site on MHCIs further, we took advantage of our finding that ILTs bind to HLA-G1 with a significantly higher affinity than to classical MHCIs and the fact that these differences are likely to result from a relatively small number of sequence differences (Fig. 3A). The regions on the surface of MHCIs that differ between HLA-G1 and the classical MHCIs are shown in Fig. 3C (orange and green surfaces on Upper Left and Upper Right, respectively). They cluster in two sites: (i) the G strand residues 268, 271, 275, and 276 (green in Fig. 3C Upper Right) and (ii) the CC′E β-strand residues 195, 197, 214, and 228 (orange in Fig. 3C Upper Left). Given that the CD8αα- and ILT-binding sites overlap, it is unlikely that site 1 residues (green) contribute to ILT binding. However, site 2 residues (orange) are well positioned to contribute to ILT binding (Fig. 3C).
There is some evidence that the ILT2- and ILT4-binding sites are not identical. First, as shown in Table 1, HLA-Cw7 produced by using the chemically biotinylated β2m could bind to ILT4 but not to ILT2. Second, Allen et al. (36) reported that the free form of HLA-B27 lacking β2m bound ILT4-transfected but not ILT2-transfected cells. These results suggest that ILT2 binding depends more on β2m than ILT4 binding. Perhaps the ILT2-binding site incorporates the region around the α3–β2m interface.
Our results indicate that, although both ILT2 and ILT4 bind to a range of MHCIs, there are significant variations in the binding affinities. On the one hand, ILTs bind HLA-G with a higher affinity than they bind classic MHCIs. One notable difference in the putative ILT-binding site is the relatively hydrophobic patch formed by F195/Y197 on HLA-G, which is absent in other MHCs where the corresponding residues are S195/H197. On the other hand, ILT2 has a higher affinity and narrower specificity than ILT4. What might be the structural basis for this difference? Comparison of sequences in the putative MHCI-binding site of ILT molecules indicates notable differences (Fig. 3B). For example the putative binding region on ILT2D1D2 (18) includes residues (Y76, D80, and R84) that are not conserved in ILT4 (Q76, R80, and R84) (Fig. 3C). Furthermore, there is evidence, noted above, that ILT2 but not ILT4 binding to MHCI depends on β2m, suggesting a difference in the ILT2-versus ILT4-binding sites on MHCI. Thus there is significant variation in the ILT–MHCI binding interfaces that could easily account for the observed variation in binding affinities. Whether these differences are physiologically important remains to be shown.
Functional Implications. Our finding that ILT2 and ILT4 bind with a higher affinity to HLA-G than to classical MHCIs raises the possibility that both the ILTs may contribute to functional interactions between leukocytes expressing ILTs (T, NK, and myelomonocytic cells) and cells expressing HLA-G. The latter include thymic epithelial cells, fetal trophoblast tissue, some cancers, and the cells infected by human cytomegalovirus (37).
ILT2 may influence thymocyte development by interacting with HLA-G and classical MHCIs on thymic epithelial cells, thereby modulating the threshold of TCR triggering. It is noteworthy that the binding affinity of the ILT2–HLA-G interaction is at the high end of the range of affinities measured for TCR–MHCs interactions (Kd = 1–50 μM).
We also show that ILTs compete directly with CD8αα for binding to MHCIs. Whereas intraepithelial T cells express CD8αα, most T cells express CD8αβ. However, our results are likely to be relevant for all T cells, because CD8αα and CD8αβ have comparable affinities for MHC, and these affinities are considerably lower than the affinity of ILT2 binding to MHCI. The higher affinity of ILT versus CD8 binding suggest that ILTs may effectively block CD8 binding at the cell–cell interface. Interestingly, Dietrich et al. (9) showed that ILT2 and TCR colocalize at the immunological synapse formed between T cells and antigen-presenting cells expressing ligands for ILT2 (HLA-B27) and TCR (superantigen) on their surface. These data suggested that ILT2 could potentially function as an “inhibitory” coreceptor, first by blocking binding of CD8 and second by bringing immunoreceptor tyrosine-based inhibitory receptor motifs with their associated tyrosine phosphatases into proximity with the TCR engaging the same peptide-MHCI. There is at present no direct evidence for a “competitive” inhibitory effect, and the fact that CD8+ T cells that express ILT2 can be activated by recognition of MHCIs suggests that this competition may be modulated in some way. One possibility is that CD8 is effectively recruited to the TCR–CD3 complex, thereby enhancing its ability to engage MHCIs.
The relatively high affinity of ILT2 binding to HLA-G is consistent with previous observations that the effects of ILT2-mediated inhibition on peripheral blood NK cells can be largely attributed to HLA-G1 recognition (38). Similarly, the ILT2–HLA-G interaction may also effectively inhibit NK cell recognition of trophoblast and HLA-G-expressing tumor cells, thereby contributing to materno-fetal stolerance and escape of tumor cells.
Although the affinity of ILT4 binding to HLA-G is slightly lower than ILT2, ILT4 shows a much stronger preference for HLA-G versus classical MHCIs than does ILT2, which suggests that the ILT4–HLA-G interaction may be of considerable significance for regulating the maturation and/or function of cells in the myelomonocytic lineage. Indeed, it has been suggested that ILT4 also may regulate the threshold of myelomonocytic activation in materno-fetal tolerance (12), inflammatory responses (11), and tolerogenic antigen-presenting cells induced by regulatory T cells (13).
In conclusion, we report here that ILT2 and ILT4 bind more strongly to HLA-G than to classical MHCIs, that ILT2 binds with a higher affinity than ILT4, and that ILTs compete with CD8 for binding to MHCIs. These observations provide insights into the possible role of ILT–MHCI interactions in regulating immunological recognition.
Supplementary Material
Acknowledgments
We thank D. I. Stuart and G. F. Gao for discussion; L. Lanier for the 40H2 mAb; and E. Davies for assistance. We also thank L. M. Thomas and P. J. Bjorkman for giving us the unpublished information of the ILT–MHC complex structure. K.M. was supported in part by the Ministry of Education, Science, Sports, Culture, and Technology of Japan, the 2000th year Joint Research Project (Soken/K01-4) of Sokendai, and a research grant from the Nakajima foundation. E.Y.J. is supported by Cancer Research U.K. and the Medical Research Council.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ILT, Ig-like transcript; KIR, killer cell Ig-like receptor; NK, natural killer; TCR, T cell antigen receptor; MHCI, MHC class I molecule; Dn, N-terminal domain n; β2m, β2-microglobulin; SPR, surface plasmon resonance.
Note Added in Proof. The conclusion that site 2 residues are well positioned to contribute to ILT binding is supported by a recently determined crystal structure of LIR-1 (ILT2) bound to a classical MHCI (B.W., L. M. Thomas, and P. J. Bjorkman, unpublished data).
References
- 1.Martin, A. M., Kulski, J. K., Witt, C., Pontarotti, P. & Christiansen, F. T. (2002) Trends Immunol. 23, 81–88. [DOI] [PubMed] [Google Scholar]
- 2.Samaridis, J. & Collona, M. (1997) Eur. J. Immunol. 27, 660–665. [DOI] [PubMed] [Google Scholar]
- 3.Cosman, D., Fanger, N., Borges, L., Kubin, M., Chin, W., Peterson, L. & Hsu, M.-L. (1997) Immunity 7, 273–282. [DOI] [PubMed] [Google Scholar]
- 4.Borges, L., Hsu, M.-L., Fanger, N., Kubin, M. & Cosman, D. (1997) J. Immunol. 159, 5192–5196. [PubMed] [Google Scholar]
- 5.Wagtmann, N., Rojo, S., Eichler, E., Mohrenweiser, H. & Long, O. E. (1997) Curr. Biol. 7, 615–618. [DOI] [PubMed] [Google Scholar]
- 6.Colonna, M., Navarro, F., Bellón, T., Llano, M., García, P., Samaridis, J., Angman, L., Cella, M. & López-Botet, M. (1997) J. Exp. Med. 186, 1809–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colonna, M., Samaridis, J., Cella, M., Angman, L., Allen, R. L., O'Callaghan, C. A., Dunber, R., Ogg, G. S., Cerundolo, V. & Rolink, A. (1998) J. Immunol. 160, 3096–3100. [PubMed] [Google Scholar]
- 8.Fanger, N. A., Cosman, D., Peterson, L., Braddy, S. C., Maliszewski, C. R. & Borges, L. (1998) Eur. J. Immunol. 28, 3423–3434. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich, J., Cella, M. & Colonna, M. (2001) J. Immunol. 166, 2514–2521. [DOI] [PubMed] [Google Scholar]
- 10.Young, N. T., Uhrberg, M., Phillips, J. H., Lanier, L. L. & Parham, P. (2001) J. Immunol. 166, 3933–3941. [DOI] [PubMed] [Google Scholar]
- 11.Tedla, N., Gibson, K., McNeil, H. P., Cosman, D., Borges, L. & Arm, J. P. (2002) Am. J. Pathol. 160, 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allan, D. S. J., Colonna, M., Lanier, L. L., Churakova, T. D., Abrams, J. S., Ellis, S. A., McMichael, A. J. & Braud, V. M. (1999) J. Exp. Med. 189, 1149–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, C. C., Ciubotariu, R., Manavalan, J. S., Yuan, J., Colovai, A. I., Piazza, F., Lederman, S., Colonna, M., Cortesini, R., Dalla-Favera, R. & Suciu-Foca, N. (2002) Nat. Immunol. 3, 237–243. [DOI] [PubMed] [Google Scholar]
- 14.Navarro, F., Liano, M., Bellon, T., Collona, M., Geraghty, D. E. & Lopez-Botet, M. (1999) Eur. J. Immunol. 29, 277–283. [DOI] [PubMed] [Google Scholar]
- 15.Vitale, M., Castriconi, R., Parolini, S., Pende, D., Hsu, M.-L., Moretta, L., Cosman, D. & Moretta, A. (1999) Int. Immunol. 11, 29–35. [DOI] [PubMed] [Google Scholar]
- 16.Lupin, E. J. M., Bastin, J. M., Allan, D. S. J., Roncador, G., Braud, V. M., Mason, D. Y., van der Merwe, P. A., McMichael, A. J., Bell, J. I., Powis, S. H. & O'Callaghan, C. A. (2000) Eur. J. Immunol. 30, 3552–3561. [DOI] [PubMed] [Google Scholar]
- 17.Chapman, T. L., Heikema, A. P. & Bjorkman, P. J. (1999) Immunity 11, 603–613. [DOI] [PubMed] [Google Scholar]
- 18.Chapman, T. L., Heikema, A. P., West, A. P., Jr., & Bjorkman, P. J. (2000) Immunity 13, 727–736. [DOI] [PubMed] [Google Scholar]
- 19.Boyington, J. C., Motyka, S. A., Schuck, P., Brooks, A. G. & Sun, P. D. (2000) Nature 405, 537–543. [DOI] [PubMed] [Google Scholar]
- 20.Fan, Q. R., Long, E. O. & Wiley, D. C. (2001) Nat. Immunol. 2, 452–460. [DOI] [PubMed] [Google Scholar]
- 21.Kovats, S., Main, E. K., Librach, C., Stubblebine, M., Fisher, S. J. & DeMars, R. (1990) Science 248, 220–223. [DOI] [PubMed] [Google Scholar]
- 22.Crisa, L., McMaster, M. T., Ishii, J. K., Fisher, S. J. & Salomon, D. R. (1997) J. Exp. Med. 186, 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul, P., Rouas-Freiss, N., Khalil-Daher, I., Moreau, P., Riteau, B., Le Gal, F. A., Avril, M. F., Dausset, J., Guillet, J. G. & Carosella, E. D. (1998) Proc. Natl. Acad. Sci. USA 95, 4510–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibrahim, E. C., Guerra, N., Lacombe, M.-J. T., Angevin, E., Chouaib, S., Carocella, E. D., Caignard, A. & Paul, P. (2001) Cancer Res. 61, 6838–6845. [PubMed] [Google Scholar]
- 25.Lefebvre, S., Antoine, M., Uzan, S., McMaster, M., Dausset, J., Carosella, E. D. & Paul, P. (2002) J. Pathol. 196, 266–274. [DOI] [PubMed] [Google Scholar]
- 26.Wiendl, H., Mitsdoerffer, M., Hofmeister, V., Wischhusen, J., Bornemann, A., Meyermann, R., Weiss, E. H., Melms, A. & Weller, M. (2002) J. Immunol. 168, 4772–4780. [DOI] [PubMed] [Google Scholar]
- 27.Reid, S. W., Smith, K. J., Jakobsen, B. K., O'Callaghan, C. A., Reyburn, H., Harlos, K., Stuart, D. I., McMichael, A. J., Bell, J. I. & Jones, E. Y. (1996) FEBS Lett. 383, 119–123. [DOI] [PubMed] [Google Scholar]
- 28.Garboczi, D. N., Hung, D. T. & Wiley, D. C. (1992) Proc. Natl. Acad. Sci. USA 89, 3429–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao, G. F., Gerth, U. C., Wyer, J. R., Willcox, B. E., O'Callaghan, C. A., Zhang, Z., Jones, E. Y., Bell, J. I. & Jakobsen, B. K. (1998) Protein Sci. 7, 1245–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maenaka, K., Juji, T., Nakayama, T., Wyer, J. R., Gao, G. F., Maenaka, T., Zaccai, N. R., Kikuchi, A., Yabe, T., Tokunaga, K., et al. (1999) J. Biol. Chem. 274, 28329–28334. [DOI] [PubMed] [Google Scholar]
- 31.Maenaka, K., van der Merwe, P. A., Stuart, D. I., Jones, E. Y. & Sondermann, P. (2001) J. Biol. Chem. 276, 44898–44904. [DOI] [PubMed] [Google Scholar]
- 32.Valez-Gomez, M., Reyburn, H. T., Mandelboim, M. & Strominger, J. L. (1998) Immunity 9, 337–344. [DOI] [PubMed] [Google Scholar]
- 33.Williams, R. C. & Malone, C. C. (1993) J. Lab. Clin. Med. 121, 805–820. [PubMed] [Google Scholar]
- 34.Trymbulak, W. P., Jr., & Zeff, R. A. (1997) Transplantation 64, 640–645. [DOI] [PubMed] [Google Scholar]
- 35.Gao, G. F., Tormo, J., Gerth, U. C., Wyer, J. R., McMichael, A. J., Stuart, D. I., Bell, J. I., Jones, E. Y. & Jakobsen, B. K. (1998) Nature 387, 630–634. [DOI] [PubMed] [Google Scholar]
- 36.Allen, R. L., Raine, T., Haude, A., Trowsdale, J. & Wilson, M. J. (2001) J. Immunol. 167, 5543–5547. [DOI] [PubMed] [Google Scholar]
- 37.Onno, M., Pangault, C., Friec, G. L., Guilloux, V., Andre, P. & Fauchet, R. (2000) J. Immunol. 164, 6426–6434. [DOI] [PubMed] [Google Scholar]
- 38.Riteau, B., Menier, C., Khalil-Daher, I., Martinozzi, S., Pla, M., Dausset, J., Carosella, E. D. & Rouas-Freiss, N. (2001) Int. Immunol. 13, 193–201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.