Abstract
A hallmark of most neurodegenerative diseases, including those caused by polyglutamine expansion, is the formation of ubiquitin (Ub)-positive protein aggregates in affected neurons. This finding suggests that the Ub system may be involved in common mechanisms underlying these otherwise unrelated diseases. Here we report the finding of ataxin-3 (Atx-3), whose mutation is implicated in the neurodegenerative disease spinocerebellar ataxia type 3, in a bioinformatics search of the human genome for components of the Ub system. We show that wild-type Atx-3 is a Ub-binding protein and that the interaction of Atx-3 with Ub is mediated by motifs homologous to those found in a proteasome subunit. Both wild-type Atx-3 and the otherwise unrelated Ub-binding protein p62/Sequestosome-1 have been shown to be sequestered into aggregates in affected neurons in several neurodegenerative diseases, but the mechanism for this recruitment has remained unclear. In this article, we show that functional Ub-binding motifs in Atx-3 and p62 proteins are required for the localization of both proteins into aggregates in a cell-based assay that recapitulates several features of polyglutamine disease. We propose that the Ub-mediated sequestration of essential Ub-binding protein(s) into aggregates may be a common mechanism contributing to the pathogenesis of neurodegenerative diseases.
Ubiquitin (Ub) is an 8-kDa protein that is covalently attached to lysine residues in specific substrates (and to Ub itself, to form polymeric chains) through the action of Ub ligases (1, 2). The best known function of this posttranslational modification (ubiquitylation) is to promote the recognition of substrates by the proteasome degradation machinery (1). It has recently become clear that Ub also serves as a signal for other processes, such as membrane trafficking. Ub-elicited processes can be mediated by proteins that interact with it in a noncovalent manner and that contain one of a growing list of Ub-binding motifs, such as the Ub-associated (UBA) domain (3), the Cue1p-homologous (CUE) domain (4–6), and the Ub-interaction motif/polyUb-binding motif (UIM and PUB motif, respectively) (refs. 7–14 and this article). In addition to specifying the fate of Ub-modified proteins, proteins that bind to Ub noncovalently may also, for example, assist in Ub chain synthesis (15, 16) and could perform other, yet undiscovered functions in normal or pathological processes.
Cells respond to protein misfolding [caused by, for example, expansion of polyglutamine (polyQ) tracts] by various mechanisms, including by attempting to refold the protein or by promoting its ubiquitylation for proteasomal degradation. Failure in the clearance of misfolded proteins can lead to their accumulation in aggregates, which are rich in Ub (17–19). In addition to containing the ubiquitylated mutant or misfolded protein, aggregates in disease brains recruit normal proteins, including chaperones, proteasomes, and others. Although such aggregates are a hallmark of neurodegenerative disorders, their precise role in the pathological process remains elusive. In this article, we used a cell-based assay that recapitulates several features of polyQ disease to show that Ub-binding motifs mediate the localization of normal Ub-binding proteins into polyQ aggregates. We propose that the Ub-mediated sequestration of essential Ub-binding protein(s) into aggregates may be a common mechanism contributing to the pathogenesis of neurodegenerative diseases.
Materials and Methods
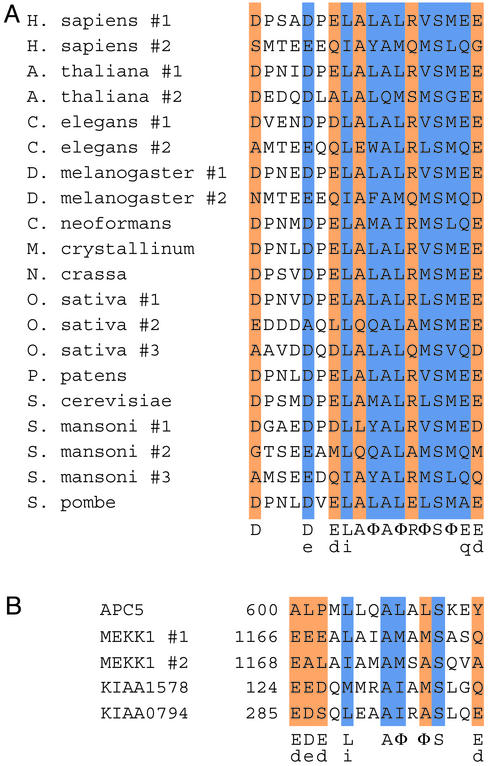
Bioinformatics Analyses. Multihit local hidden Markov models (HMMs) were built by using alignments of PUB motifs of the Rpn10p/S5a family (Fig. 1A) and were calibrated with hmmer 2.1, which can be accessed at http://hmmer.wustl.edu. Public and private sequence databases were searched with these HMMs to identify candidate PUB homologues. Because the PUB motif is short and important residues for Ub binding have been verified solely in the context of the Rpn10p/S5a family, we used one or more of the following criteria for assigning hits as PUB homologues: (i) statistical significance estimates; (ii) presence of more than one hit in the same protein [as in the case of human S5a (Fig. 1 A)]; (iii) greater conservation, among orthologues, of amino acids within the motif when compared with surrounding sequences; and (iv) a known relationship of the protein to the Ub system. Putative homologues were used to construct multiple alignments and iterated HMMs. In some cases, to reduce redundancy and subfamily bias within sequences, only one sequence from closely related proteins was used for the alignment and reiteration of the database search procedure.
Fig. 1.
The PUB motifs of the proteasome subunit Rpn10p/S5a and PUBH motifs found in otherwise unrelated proteins. (A) Alignment of the PUB motif found in Rpn10p homologues. Conserved amino acids or side chain features are indicated and colored as follows: blue, very high conservation (>90%); orange, high conservation (65–90%); φ, conserved hydrophobicity. Lowercase letters indicate less frequently occurring residues that nonetheless preserve a side chain feature. (B) Alignment of PUBH motifs from selected human proteins. Numbers to the left indicate the position of the first amino acid in the sequence. Legend is as in A. APC5 is a subunit of the Ub ligase APC. MEKK1 is a protein serine/threonine kinase that also exhibits Ub ligase activity. KIAA1578 and KIAA0794 have other domains that are associated with the Ub system (see text and ref. 9). Conserved PUBH/UIM residues are indicated as in ref 9.
Pull-Down Assays. Protein expression and purification were done essentially as described (21), except that cells were lysed in 50 mM Tris·HCl (pH 8)/500 mM NaCl/5 mM 2-mercaptoethanol/5 mM imidazole/10% glycerol and protease inhibitors. For pull-down assays, 200 pmol of His-tagged protein was incubated with 0.5 μg of a mixture of K48-linked Ub chains containing mostly 2–7 Ub units (Ub2–7; Affiniti, Exeter, U.K.) for 1.5–4 h at 4°C in 1 ml of binding buffer [20 mM Tris·HCl (pH 7.5)/100 mM NaCl/0.1% Nonidet P-40/25 μg/ml BSA], and then with 30 μl of Talon cobalt beads (CLONTECH) for 45 min. Proteins pelleted with beads were washed twice in binding buffer without BSA, eluted in SDS sample buffer, subjected to SDS/PAGE in 4–20% acrylamide Tris-glycine gel (Invitrogen), and immunoblotted with anti-Ub mAb (Zymed).
DNA Constructs. Ataxin-3 (Atx-3) and p62 cDNAs were cloned by PCR from a human brain library (GIBCO/BRL). We used the Atx-3 isoform MJD1-1 (MJD, Machado–Joseph's disease) (ref. 22; GenBank accession no. NM_004993). Amino acid numbers in this text correspond to that sequence, but with a polyQ tract of 26 residues (which is within the normal range of 13–36 residues). Vectors used were pZsGreen1-C1 (CLONTECH), pHis8, and pFLAG. Mutations were introduced as follows: in a 50-μl volume, 100 ng of template was combined with 12.5 pmol of each primer, 0.2 mM dNTPs, and 2 μl of Pfu (Stratagene) in Pfu buffer. After thermocycling (16 cycles of 95°C for 30 sec, 54°C for 1 min, and 68°C for 13 min), reactions were incubated with DpnI and transformed into Escherichia coli. When necessary, multiple rounds of mutagenesis were performed (e.g., S236A/S256A/S359A). cDNA encoding truncated proteins was generated either by PCR with internal oligonucleotides or by mutagenesis. Complete coding sequences were verified by sequencing. Cyan fluorescent protein (CFP)-Q78 was generated by PCR amplification of an expanded polyQ tract from Atx-3 (isoform MJD1a) (22) using MJDtr-Q78 (23) as template and cloning into pCMX-CFP.
Immunoprecipitation. HEK293 cells were transfected with FLAG plasmids by using Fugene 6 (Roche Diagnostics). Two days after transfection, cells were lysed for 20 min at 4°C in either 50 mM Tris·HCl (pH 7.5)/120 mM NaCl/1 mM EDTA/0.1% Nonidet P-40 (Nonidet P-40 buffer), or 50 mM Tris (pH 8.0)/150 mM NaCl/1% Triton X-100/0.5% sodium deoxycholate/0.1% SDS (RIPA buffer), both containing protease inhibitors. Cleared lysates were mixed with beads covalently coupled to the FLAG mAb M2 (Sigma), or with uncoupled M2 Ab for 2 h at 4°C, followed by pull-down with protein A Sepharose beads. Pelleted beads were washed four times with lysis buffer. Bound proteins were eluted with 125 mM Tris·HCl (pH 7.0)/4% SDS and 20% glycerol, separated on SDS/polyacrylamide gels and subjected to immunoblotting with polyclonal Ab to Ub (DAKO) or mAb M2 to FLAG (Sigma).
PolyQ Aggregate Colocalization Assay. HEK293 cells were transfected with Fugene 6, plated onto poly-l-lysine-coated chamber slides (Becton Dickinson) the following day, fixed 48 h later by addition of equal volume of 8% formaldehyde in PBS directly to culture media, and mounted in Prolong Antifade media (Molecular Probes). Cells were analyzed by fluorescence microscopy with an Olympus fluorescence microscope, and images were captured with an Orca 100 charge-coupled device camera (Hamamatsu, Middlesex, NJ). Filter sets were Chroma Technology (Brattleboro, VT) cyan GFP (excitation 436/20 nm, emission 480/40 nm) and Chroma 41001 (excitation 489/16 nm, emission 535/50 nm). CFP and GFP filter sets were deliberately selected such that excitation wavelengths were narrow and completely nonoverlapping. Confocal analyses were done with an Olympus Fluoview 500 laser scanning confocal on an Olympus IX61 upright microscope. The GFP was imaged with the 488-nm line of the argon laser, and the emission filter was a 505IF or long pass filter. The CFP was acquired sequentially with the 442-nm line of a helium–cadmium laser and with a BA465–495 emission filter (band-pass).
Results
The finding that insoluble Ub-rich protein aggregates are a hallmark of most neurodegenerative disorders (17–19) has generated interest in understanding whether the Ub system plays a common role in these otherwise unrelated pathologies. We noticed that different factors that are sequestered into such protein aggregates are also Ub-binding proteins (Table 1), and we reasoned that a possible pathogenic mechanism common to neurodegenerative disorders could be the sequestration of normal cellular proteins through interaction with Ub chains attached to aggregated proteins. Proteasomes are one such Ub-binding factor recruited to protein aggregates in many types of neurodegenerative disorders (refs. 24 and 25 and references therein), presumably mediated by its Ub-binding subunits, including S5a.
Table 1. Normal proteins or protein complexes that contain Ub-interacting motifs and are recruited to protein aggregates in neurodegenerative diseases.
| Protein (Ub-binding motif) | Neurodegenerative disease | Ref(s). |
|---|---|---|
| p62/Sequestosome (UBA) | AD, PiD, PD, MSA, DLB | 39 |
| AD, PD | 40 | |
| Ataxin-3 (PUBH/UIM) | SCA-1, SCA-2, DRPLA | 34 |
| NIHID | 35 | |
| S5a (PUB) | SCA-3 | 24 |
| Proteasome (PUB, others) | SCA-1, SCA-3, SCA-7, AD, HD, PD | References in 24; 25 |
| A1Up (UBA) | Cell-based assay for ataxin-1 aggregates | 46 |
SCA, spinocerebellar ataxia; DRPLA, dentatorubral-pallidoluysian atrophy; NIHID, neuronal intranuclear hyaline inclusion disease; AD, Alzheimer's disease; HD, Huntington's disease; PD, Parkinson's disease; PiD, Pick's disease; MSA, multiple system atrophy; DLB, dementia with Lewy bodies.
Rpn10p/S5a orthologues contain motifs that mediate noncovalent PUB motifs (Fig. 1 A) (7, 26). Using a bioinformatics approach, we found a number of proteins that play varied cellular roles and contain PUB-homologous (PUBH) motifs (Figs. 1B and 2 A and data not shown). Using related HMMs, Hofmann and Falquet (9) found putative UIMs in a set of proteins that is overlapping but nonidentical to those we found, because of presumably small differences in the way the HMMs for PUBH and UIM were built, and differences in the criteria used for selection of the hits (see Materials and Methods). In fact, we have found significant hits that do not conform well to the UIM consensus. For example, anaphase-promoting complex (APC)-5 lacks the conserved negatively charged residues in the N terminus of the UIM consensus (Fig. 1B). We nonetheless assigned a PUBH motif to APC5 because this protein is a subunit of a Ub ligase, the APC (27). Other hits unique to our analyses included MEKK1, a protein serine/threonine kinase that is also a Ub ligase (ref. 28; the two PUBH motifs of MEKK1 overlap); KIAA1578, which also has a UBA domain and a WWE domain; and KIAA0794, which also has a UAS and a UBX domain (the latter four types of domain are associated with the Ub system; Fig. 1B). Hits common to both analyses include the endocytic regulators Eps15, epsins, and STAM (C.A.P.J., K.A.C., and S.B., unpublished observations; ref 9).
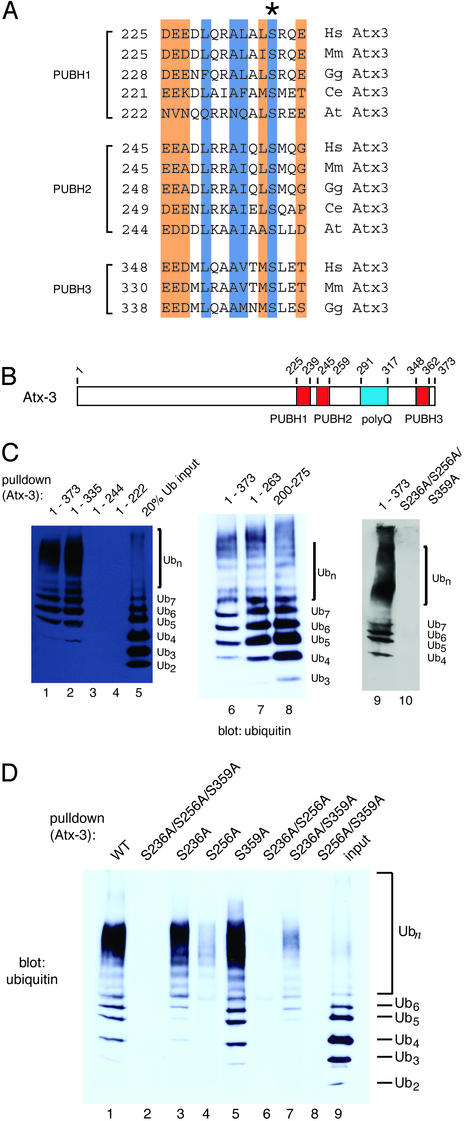
Fig. 2.
UIM/PUBH motifs mediate noncovalent binding of Atx-3 to Ub chains in vitro.(A) Alignment of UIM/PUBH motifs present in Atx-3 orthologues. Numbers indicate the position of the first amino acid. Conserved amino acids or side chain features are indicated as in Fig. 1. *, The Ser residue that is conserved in PUBH motifs and that was mutated in this study (below). (B) Domain structure of Atx-3. Red boxes represent PUBH motifs and the polyQ tract is blue. Numbers indicate amino acid positions. (C) Atx-3 binding to Ub chains. Effect of truncations and of PUBH motif mutations. N-terminal His8-tagged Atx-3 protein, its fragments, or a triple PUBH mutant (S236A/S256A/S359A) were used to pull down Ub chains. The equivalent of 20% of Ub chain input is shown in lane 5. Pull-down products were revealed by immunoblot with Abs against Ub. The number of Ub units in the chain mixture is indicated by the subscript in Ubn.(D) Atx-3 PUBH motifs required for interaction with Ub chains. His-tagged wild-type or mutant Atx-3 (Upper) were used. Legend is as in C.
Interestingly, among the PUBH/UIM-containing hits, a protein that had already been implicated in neurodegeneration was found, Atx-3 (Fig. 2A). Expansion of a polyQ tract in Atx-3 leads to aggregate formation in the dominantly inherited SCA-3 or MJD (18, 29–32).
Normal Atx-3 is a predominantly cytoplasmic, widely expressed protein of unknown function (18, 30, 33). In addition to the role of mutant Atx-3 in a specific neurodegenerative disease (SCA-3), wild-type Atx-3 is recruited to aggregates in brains of SCA types 1 and 2, dentatorubral-pallidoluysian atrophy (DRPLA), and neuronal intranuclear hyaline inclusion disease (NIHID), as well as to Marinesco bodies, which are aggregates of unclear relationship to disease (Table 1). Like proteasomes, cytoplasmic Atx-3 can become depleted in neurons affected by these diseases (31, 34–36). These observations, together with the presence of PUBH motifs in Atx-3, raised the possibility that Atx-3 is recruited to aggregates through interaction with Ub.
We first tested the possibility that Atx-3 is a Ub-binding protein and that its interaction with Ub is mediated by PUBH motifs. E. coli-purified full-length Atx-3 (1–373) was used in pull-down assays with a mixture of lysine 48 (K48)-linked Ub chains, containing mostly 2–7 Ub units (Fig. 2C, lane 5). The results show that, like Rpn10p/S5a and the 26S proteasome (7, 26), Atx-3 prefers to bind to chains of 4 or more Ub units (Fig. 2C, lanes 1, 6, and 9). Atx-3 contains three PUBH motifs (Fig. 2 A and B), the C-terminal-most one being absent in some splicing variants (e.g., MJD1a) (22) and in Atx-3 orthologues (Fig. 2 A). Deletion of the C-terminal 26 residues, including PUBH3 (Atx-3 1–335), or of the polyQ tract and amino acids C-terminal to it (Atx-3 1–263) was not detrimental for Ub binding (Fig. 2C, lanes 2 and 7, respectively). (Although we have not examined the consequence of polyQ expansion on Ub binding by Atx-3 with a biochemical approach, it is expected that the aggregation and altered subcellular localization of the expanded protein is likely to cause the loss of its normal cellular functions in SCA-3.) Further C-terminal truncation leaving only PUBH1 (Atx-3 1–244) or no PUBH motifs (Atx-3 1–222) impaired binding to Ub chains (Fig. 2C, lanes 3 and 4, respectively). Conversely, Atx-3 200–275, containing only PUBH1 and PUBH2 and no other recognizable motifs, sufficed for binding to Ub chains (Fig. 2C, lane 8). We next investigated whether the interaction between the C-terminal half of Atx-3 and Ub was mediated by PUBH motifs. Rpn10p S235 is the only residue absolutely conserved among PUB and PUBH motifs (Fig. 1) and is required for binding of Rpn10p to polyUb chains (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). The equivalent Ser residue was replaced with Ala in all three PUBH motifs of Atx-3 (Fig. 2 A). The results in Fig. 2C show that the triple mutant was defective in binding to Ub (lane 10).
The PUBH motifs were also mutated individually in the context of the full-length Atx-3 to determine their relative contribution to Ub binding (Fig. 2D). Mutation of PUBH2 (S256A, lane 4) was the most deleterious to Ub binding, suggesting that the remaining functional PUBH1 and PUBH3 together cannot efficiently interact with Ub. On the other hand, mutation of PUBH3, leaving both PUBH1 and PUBH2 intact (S359A, lane 5), did not affect binding to Ub compared with the wild-type protein. To a lesser extent, PUBH2 and PUBH3 together also bound to Ub (S236A, lane 3). Among the double mutants, only Atx-3 S236A/S359A, which has only a functional PUBH2, bound to Ub, but did so very poorly (lane 7). We conclude that at least two PUBH motifs working together, preferably PUBH1 and PUBH2, are required for optimal binding to Ub chains.
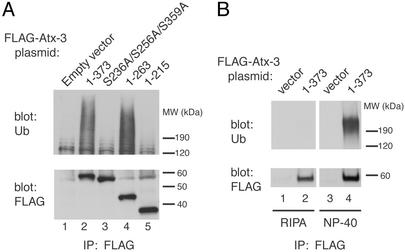
PUBH motifs mediate the association of Atx-3 with Ub chains in vivo as well. FLAG-tagged Atx-3-expressing cells were lysed in Nonidet P-40-containing buffer and subjected to immunoprecipitation with an Ab to the FLAG epitope. Proteins recognized by an Ab against Ub were coimmunoprecipitated along with Atx-3 (Fig. 3A, lane 2, Upper) in a PUBH motif-dependent manner [lane 3; mutation of PUBH motifs did not affect Atx-3 protein levels (Lower, lanes 2 and 3)]. As expected from the results in Fig. 2C, the Atx-3 polyQ tract and amino acids C-terminal to it were not required for Ub coimmunoprecipitation (lane 4, Upper), and the conserved N-terminal half of Atx-3 did not coimmunoprecipitate with Ub (lane 5). The coimmunoprecipitated Ub signal was not due to the covalent attachment of Ub chains to Atx-3 itself, because the Atx-3–Ub coimmunoprecipitation was sensitive to a more stringent, SDS-containing lysis buffer (RIPA; Fig. 3B Upper). The simplest interpretation of these results is that Atx-3 noncovalently binds to polyubiquitylated proteins in vivo through PUBH motif–Ub chain interactions.
Fig. 3.
UIM/PUBH motifs mediate noncovalent binding of Atx-3 to Ub chains in vivo.(A) PUBH motif-dependent interaction of Atx-3 and Ub chains in vivo. Cells were transfected with the indicated plasmids encoding N-terminal (FLAG)3-tagged Atx-3 proteins. Lysates were subjected to immunoprecipitation with Ab to the FLAG epitope. Immunoprecipitated proteins were blotted and reacted with Ab to Ub (Upper) or to the FLAG epitope (Lower). (B) Noncovalent interaction of Atx-3 and Ub chains in vivo. Cells were transfected with empty vector or vector encoding FLAG-tagged wild-type Atx-3. Lysates made in Nonidet P-40 or RIPA buffers, as indicated, were subjected to immunoprecipitation with Ab to the FLAG epitope. Immunoprecipitated proteins were blotted and reacted with Ab to Ub (Upper) or to the FLAG epitope (Lower).
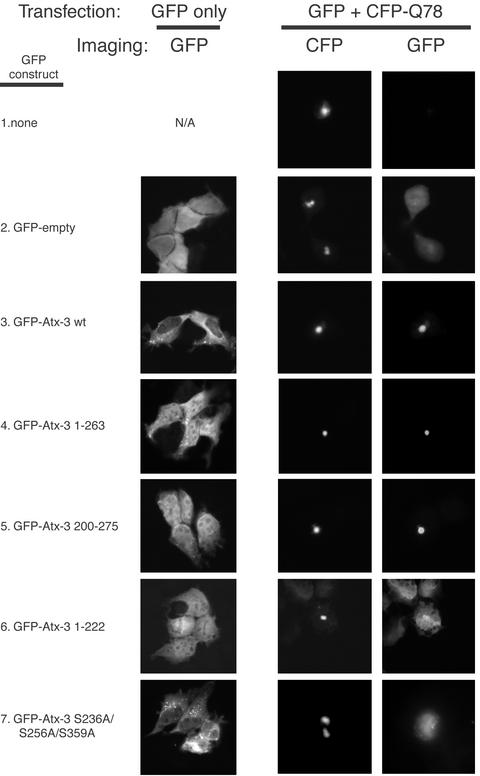
Wild-type Atx-3 is recruited to Ub-rich aggregates in various neurodegenerative disorders (Table 1). It has been argued that the recruitment of normal polyQ tract-containing proteins to polyQ aggregates, such as CBP/p300, might occur through polyQ–polyQ interactions (37). However, in an assay that recapitulates many features that expanded polyQ proteins exhibit in vivo (18, 31, 32), normal Atx-3 was recruited to aggregates formed by a transiently expressed expanded polyQ tract independent of its own polyQ tract (31, 32). Our findings that Atx-3 interacts with Ub prompted the examination of whether this recruitment might be mediated by the Atx-3 PUBH motifs (Fig. 4). For these analyses, we used the cell-based aggregate formation assay mentioned above (18, 31, 32). An expanded polyQ tract flanked, at the N terminus and C terminus, by 12 and 43 aa, respectively, of the SCA-3-derived Atx-3 isoform MJD1a (22) was fused to CFP-Q78 and formed readily visible aggregates in transfected cells (Fig. 4 Center). These aggregates stained positive for Ub (see Fig. 7, which is published as supporting information on the PNAS web site). In the absence of CFP-Q78, full-length Atx-3 fused to GFP and containing or not functional PUBH motifs [wild-type (wt) and S236A/S256A/S359A, respectively] localized mostly to the cytoplasm (Fig. 4 Left). For unknown reasons, truncated proteins (1–222, 1–263, and 200–275) localized equally in the nucleus and cytoplasm (Fig. 4). All proteins were expressed at comparable levels (see Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 4.
PUBH motif-dependent recruitment of Atx-3 to polyQ aggregates. HEK293 cells were transfected with constructs encoding GFP-Atx-3 fusions and/or a CFP fusion with expanded polyQ (CFP-Q78), as indicated. The GFP-Atx-3 S236A/S256A/S359A mutant is in the context of the full-length protein (1–373). Imaging was done with filters for GFP (Left and Right) or CFP (Center).
As previously reported, coexpression of an expanded polyQ tract with normal Atx-3 led to recruitment of the latter to aggregates (Fig. 4 Right, row 3; ref 31). The results in Fig. 4 also confirm that the normal-length polyQ tract in Atx-3 is not required for the recruitment of Atx-3 into expanded polyQ aggregates (31), because recruitment can happen with both Atx-3 1–263 and Atx-3 200–275 proteins, which lack the polyQ tract (Right, rows 4 and 5, respectively). Furthermore, even on overexpression, the normal-length polyQ tract that is present in Atx-3 S236A/S256A/S359A does not mediate efficient sequestration of this protein in aggregates (Right, row 7; the difference in the distribution of Atx-3 S236A/S256A/S359A in cells expressing or not the expanded polyQ tract presumably reflects morphological changes caused by the latter and seems also to depend on Atx-3 sequence). Strikingly, CFP-Q78 aggregates efficiently recruited only those Atx-3 proteins bearing functional PUBH motifs, namely, the Atx-3 wild-type (wt; full-length), Atx-3 1–263, and Atx-3 200–275 (Right, rows 3, 4, and 5, respectively), but not those without (Atx-3 S236A/S256A/S359A and Atx-3 1–222; Right, rows 6 and 7, respectively). Moreover, Atx-3 200–275 contains only two PUBH motifs and no other recognizable motif. Thus, functional PUBH motifs are required and sufficient for recruitment of Atx-3 to polyQ aggregates.
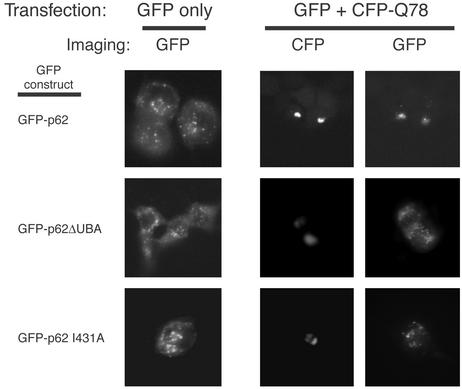
Because the Ub-binding motifs of Atx-3 are required for its sequestration into aggregates, we next investigated whether the recruitment of an unrelated Ub-binding protein to aggregates was mediated by a similar mechanism. p62/Sequestosome-1 interacts directly with Ub through a UBA domain (38). p62 is also recruited to Ub-rich aggregates formed in several neurodegenerative diseases, including Alzheimer's, Pick's, and Parkinson's (refs. 39 and 40; Table 1). The different GFP-p62 fusion proteins exhibited mostly a punctate cytoplasmic staining pattern when expressed in the absence of CFP-Q78 (Fig. 5, Left). GFP-p62 I431A has a mutation in a conserved UBA domain residue whose equivalent in human Rad23, L355, is important for binding to Ub (41). The coexpression of GFP-p62 fusion proteins with CFP-Q78 (Right) was accompanied by the selective sequestration of GFP-p62 wild-type to aggregates, but not GFP-p62 lacking the UBA domain (amino acids 393–440; p62 ΔUBA) or GFP-p62 I431A. Thus, similarly to Atx-3 requiring functional PUBH motifs, p62 requires an intact UBA domain to be sequestered into aggregates.
Fig. 5.
Ub-binding-motif-dependent recruitment of p62/Sequestosome-1 to polyQ aggregates. HEK293 cells were transfected with constructs encoding GFP-p62 fusions and/or a CFP fusion with expanded polyQ (CFP-Q78), as indicated. Imaging was done with filters for GFP (Left and Right) or CFP (Center).
Discussion
Motifs that are homologous to the PUB motifs found in the proteasome subunit Rpn10p/S5a (Figs. 1 and 2 A; ref. 9) are conserved among the worm, plant, and vertebrate orthologues of Atx-3 (Fig. 2 A), suggesting that these motifs are important for the normal function of the protein. Like the PUB motifs in Rpn10p/S5a, the PUBH motifs in Atx-3 mediate binding to Ub chains in vivo (Fig. 3 A and B) and in vitro (Fig. 2 C and D), indicating that wild-type Atx-3 functions as a player in the Ub system. This finding is important because, whereas mutant Atx-3 with an expanded polyQ tract is known to cause SCA-3, the function of the normal protein has remained largely unknown.
There has been great interest in understanding the molecular mechanisms underlying neurodegenerative diseases. Molecular links have already been established between the Ub system and neurodegenerative disorders. Mutations in parkin, a Ub ligase, and in UCH-L1, a deubiquitylating enzyme, are found in hereditary parkinsonism (42). Mutations in other genes encoding a deubiquitylating enzyme (43), a Ub ligase (44), and Ub itself (45) have also been associated with neurodegeneration. In this article we reveal yet another link between the Ub system and neurodegeneration: Ub-interacting motifs in wild-type Atx-3 and p62, two known factors sequestered into aggregates in several forms of neurodegenerative disease, are essential for the localization of both proteins into polyQ aggregates. Another UIM/PUBH motif-containing protein, Eps15, binds to Ub (10) and is also recruited to polyQ aggregates in this assay (W.L. and C.A.P.J., unpublished observations). The UBA domain-containing protein A1Up is similarly recruited to aggregates formed in cultured cells by an expanded Atx-1 (46), but in this case it is possible that other interactions are also involved. Whether Eps15 and A1Up are recruited to aggregates in disease is not known.
There is evidence to suggest that depletion of Ub-binding proteins by sequestration in disease aggregates should have deleterious consequences. The proteasome is one such Ub-binding factor that becomes redistributed to aggregates in the affected neurons of many neurodegenerative diseases (Table 1). The relevance of this observation is underscored by the findings that aggregate sequestration leads to defective proteasomal degradation of otherwise unstable proteins (47) and that defective proteasomal function can cause apoptosis of neuronal cells (e.g., ref. 48). Antisense oligonucleotide-mediated p62 depletion has been used to mimic the loss of p62 caused by aggregate sequestration and has indicated that a reduction in p62 levels can lead to neuronal dysfunction (20). Affected neurons in certain neurodegenerative disorders can also become depleted of cytoplasmic Atx-3 (31, 34–36). Whether, like p62 and proteasomes, Atx-3 is also essential for neuronal function and/or survival remains to be determined.
The conclusion that Ub-rich aggregates actively recruit normal Ub-binding proteins provides a significant conceptual advance toward understanding the etiology of neurodegeneration: although neurodegenerative disorders are very distinct from each other, one aspect that a majority of these diseases have in common is the presence of Ub-rich protein aggregates in affected neurons. Thus, the depletion of cellular Ub-binding proteins caused by their sequestration into aggregates may be a common element contributing to neurodegenerative disorders.
Supplementary Material
Acknowledgments
We thank F. Supek, D. Sherman, M. C. Sogayar, E. P. Geiduschek, T. Hunter, and C. Masuda for comments on the manuscript; J. Min and J. Chen for technical assistance; H.-k. Huang and T. Hunter for the pFLAG vector; M. Verdecia and J. Noel for pHis8; N. Bonini for UAS-MJDtr-Q78; Y. Trottier for 1H9 mAb; E. Gardiner and K. Spencer for extraordinary help with fluorescence microscopy; T. Orth for the brain cDNA library; and R. Evans, S. Kay, G. Hampton, and P. Schultz for support. This work was funded by the Novartis Research Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SCA, spinocerebellar ataxia; Atx-3, ataxin-3; Ub, ubiquitin; PUB, polyUb-binding; PUBH, PUB-homologous; polyQ, polyglutamine; UIM, Ub-interaction motif; UBA, Ub-associated; HMM, hidden Markov model; MJD, Machado–Joseph's disease; APC, anaphase-promoting complex; CFP, cyan fluorescent protein.
References
- 1.Glickman, M. H. & Ciechanover, A. (2002) Physiol. Rev. 82, 373–428. [DOI] [PubMed] [Google Scholar]
- 2.Joazeiro, C. A. & Weissman, A. M. (2000) Cell 102, 549–552. [DOI] [PubMed] [Google Scholar]
- 3.Madura, K. (2002) Cell Cycle 1, 235–244. [PubMed] [Google Scholar]
- 4.Donaldson, K. M., Yin, H., Gekakis, N., Supek, F. & Joazeiro, C. (2003) Curr. Biol. 13, 258–262. [DOI] [PubMed] [Google Scholar]
- 5.Davies, B. A., Topp, J. D., Sfeir, A. J., Katzmann, D. J., Carney, D. S., Tall, G. G., Friedberg, A. S., Deng, L., Chen, Z. & Horazdovsky, B. F. (2003) J. Biol. Chem. 278, 19826–19833. [DOI] [PubMed] [Google Scholar]
- 6.Shih, S. C., Prag, G., Francis, S. A., Sutanto, M. A., Hurley, J. H. & Hicke, L. (2003) EMBO J. 22, 1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young, P., Deveraux, Q., Beal, R. E., Pickart, C. M. & Rechsteiner, M. (1998) J. Biol. Chem. 273, 5461–5467. [DOI] [PubMed] [Google Scholar]
- 8.van Nocker, S., Sadis, S., Rubin, D. M., Glickman, M., Fu, H., Coux, O., Wefes, I., Finley, D. & Vierstra, R. D. (1996) Mol. Cell. Biol. 16, 6020–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmann, K. & Falquet, L. (2001) Trends Biochem. Sci. 26, 347–350. [DOI] [PubMed] [Google Scholar]
- 10.Polo, S., Sigismund, S., Faretta, M., Guidi, M., Capua, M. R., Bossi, G., Chen, H., De Camilli, P. & Di Fiore, P. P. (2002) Nature 416, 451–455. [DOI] [PubMed] [Google Scholar]
- 11.Bilodeau, P., Urbanowski, J. L., Winistorfer, S. C. & Piper, R. C. (2002) Nat. Cell Biol. 4, 534–539. [DOI] [PubMed] [Google Scholar]
- 12.Oldham, C. E., Mohney, R. P., Miller, S. L., Hanes, R. N. & O'Bryan, J. P. (2002) Curr. Biol. 12, 1112–1116. [DOI] [PubMed] [Google Scholar]
- 13.Raiborg, C., Bache, K. G., Gillooly, D. J., Madshus, I. H., Stang, E. & Stenmark, H. (2002) Nat. Cell Biol. 4, 394–398. [DOI] [PubMed] [Google Scholar]
- 14.Shih, S. C., Katzmann, D. J., Schnell, J. D., Sutanto, M., Emr, S. D. & Hicke, L. (2002) Nat. Cell Biol. 4, 389–393. [DOI] [PubMed] [Google Scholar]
- 15.McKenna, S., Spyracopoulos, L., Moraes, T., Pastushok, L., Ptak, C., Xiao, W. & Ellison, M. J. (2001) J. Biol. Chem. 276, 40120–40126. [DOI] [PubMed] [Google Scholar]
- 16.VanDemark, A. P., Hofmann, R. M., Tsui, C., Pickart, C. M. & Wolberger, C. (2001) Cell 105, 711–720. [DOI] [PubMed] [Google Scholar]
- 17.Alves-Rodrigues, A., Gregori, L. & Figueiredo-Pereira, M. E. (1998) Trends Neurosci. 21, 516–520. [DOI] [PubMed] [Google Scholar]
- 18.Zoghbi, H. Y. & Orr, H. T. (2000) Annu. Rev. Neurosci. 23, 217–247. [DOI] [PubMed] [Google Scholar]
- 19.Lowe, J., Mayer, J., Landon, M. & Layfield, R. (2001) Adv. Exp. Med. Biol. 487, 169–187. [DOI] [PubMed] [Google Scholar]
- 20.Samuels, I. S., Seibenhener, M. L., Neidigh, K. B. & Wooten, M. W. (2001) J. Cell. Biochem. 82, 452–466. [DOI] [PubMed] [Google Scholar]
- 21.Joazeiro, C. A., Wing, S. S., Huang, H., Leverson, J. D., Hunter, T. & Liu, Y. C. (1999) Science 286, 309–312. [DOI] [PubMed] [Google Scholar]
- 22.Ichikawa, Y., Goto, J., Hattori, M., Toyoda, A., Ishii, K., Jeong, S. Y., Hashida, H., Masuda, N., Ogata, K., Kasai, F., et al. (2001) J. Hum. Genet. 46, 413–422. [DOI] [PubMed] [Google Scholar]
- 23.Warrick, J. M., Paulson, H. L., Gray-Board, G. L., Bui, Q. T., Fischbeck, K. H., Pittman, R. N. & Bonini, N. M. (1998) Cell 93, 939–949. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt, T., Lindenberg, K. S., Krebs, A., Schols, L., Laccone, F., Herms, J., Rechsteiner, M., Riess, O. & Landwehrmeyer, G. B. (2002) Ann. Neurol. 51, 302–310. [DOI] [PubMed] [Google Scholar]
- 25.Chai, Y., Koppenhafer, S. L., Shoesmith, S. J., Perez, M. K. & Paulson, H. L. (1999) Hum. Mol. Genet. 8, 673–682. [DOI] [PubMed] [Google Scholar]
- 26.Fu, H., Sadis, S., Rubin, D. M., Glickman, M., van Nocker, S., Finley, D. & Vierstra, R. D. (1998) J. Biol. Chem. 273, 1970–1981. [DOI] [PubMed] [Google Scholar]
- 27.Zachariae, W., Shevchenko, A., Andrews, P. D., Ciosk, R., Galova, M., Stark, M. J., Mann, M. & Nasmyth, K. (1998) Science 279, 1216–1219. [DOI] [PubMed] [Google Scholar]
- 28.Lu, J., Xu, S., Joazeiro, C., Cobb, M. H. & Hunter, T. (2002) Mol. Cell 9, 945–956. [DOI] [PubMed] [Google Scholar]
- 29.Kawaguchi, Y., Okamoto, T., Taniwaki, M., Aizawa, M., Inoue, M., Katayama, S., Kawakami, H., Nakamura, S., Nishimura, M. & Akiguchi, I. (1994) Nat. Genet. 8, 221–228. [DOI] [PubMed] [Google Scholar]
- 30.Paulson, H. L., Das, S. S., Crino, P. B., Perez, M. K., Patel, S. C., Gotsdiner, D., Fischbeck, K. H. & Pitman, R. N. (1997) Ann. Neurol. 41, 453–462. [DOI] [PubMed] [Google Scholar]
- 31.Perez, M. K., Paulson, H. L., Pendse, S. J., Saionz, S. J., Bonini, N. M. & Pittman, R. N. (1998) J. Cell Biol. 143, 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paulson, H. L., Perez, M. K., Trottier, Y., Trojanowski, J. Q., Subramony, S. H., Das, S. S., Vaig, P., Mandel, J. L., Fischbeck, K. H. & Pittman, R. N. (1997) Neuron 19, 333–344. [DOI] [PubMed] [Google Scholar]
- 33.Trottier, Y., Cancel, G., An-Gourfinkel, I., Lutz, Y., Weber, C., Brice, A., Hirsch, E. & Mandel, J.-L. (1998) Neurobiol. Dis. 5, 335–347. [DOI] [PubMed] [Google Scholar]
- 34.Uchihara, T., Fujigasaki, H., Koyano, S., Nakamura, A., Yagishita, S. & Iwabuchi, K. (2001) Acta Neuropathol. 102, 149–152. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi, J., Tanaka, J., Arai, K., Funata, N., Hattori, T., Fukuda, T., Fujigasaki, H. & Uchihara, T. (2001) J. Neuropathol. Exp. Neurol. 60, 369–373. [DOI] [PubMed] [Google Scholar]
- 36.Fujigasaki, H., Uchihara, T., Takahashi, J., Matsushita, H., Nakamura, A., Koyano, S., Iwabuchi, K., Hirai, M. & Mizusawa, H. (2001) J. Neurol. Neurosurg. Psychiatry 71, 518–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nucifora, F. C., Jr., Sasaki, M., Peters, M. F., Huang, H., Cooper, J. K., Yamada, M., Takahashi, H., Tsuji, S., Troncoso, J., Dawson, V. L., et al. (2001) Science 291, 2423–2428. [DOI] [PubMed] [Google Scholar]
- 38.Geetha, T. & Wooten, M. W. (2002) FEBS Lett. 512, 19–24. [DOI] [PubMed] [Google Scholar]
- 39.Kuusisto, E., Salminen, A. & Alafuzoff, I. (2001) NeuroReport 12, 2085–2090. [DOI] [PubMed] [Google Scholar]
- 40.Zatloukal, K., Stumptner, C., Fuchsbichler, A., Held, H., Schnoelzer, M., Kenner, L., Kleinert, R., Prinz, M., Aguzzi, A. & Denk, H. (2002) Am. J. Pathol. 160, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertolaet, B. L., Clarke, D. J., Wolff, M., Watson, M. H., Henze, M., Divita, G. & Reed, S. I. (2001) Nat. Struct. Biol. 8, 417–422. [DOI] [PubMed] [Google Scholar]
- 42.Giasson, B. I. & Lee, V. M. (2001) Neuron 31, 885–888. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, S. M., Bhattacharyya, B., Rachel, R. A., Coppola, V., Tessarollo, L., Householder, D. B., Fletcher, C. F., Miller, R. J., Copeland, N. G. & Jenkins, N. A. (2002) Nat. Genet. 32, 420–425. [DOI] [PubMed] [Google Scholar]
- 44.Cummings, C. J., Reinstein, E., Sun, Y., Antalffy, B., Jiang, Y., Ciechanover, A., Orr, H. T., Beaudet, A. L. & Zoghbi, H. Y. (1999) Neuron 24, 879–892. [DOI] [PubMed] [Google Scholar]
- 45.van Leeuwen, F. W., de Kleijn, D. P., van den Hurk, H. H., Neubauer, A., Sonnemans, M. A., Sluijs, J. A., Koycu, S., Ramdjielal, R. D., Salehi, A., Martens, G. J., et al. (1998) Science 279, 242–247. [DOI] [PubMed] [Google Scholar]
- 46.Davidson, J. D., Riley, B., Burright, E. N., Duvick, L. A., Zoghbi, H. Y. & Orr, H. T. (2000) Hum. Mol. Genet. 9, 2305–2312. [DOI] [PubMed] [Google Scholar]
- 47.Bence, N. F., Sampat, R. M. & Kopito, R. R. (2001) Science 292, 1467–1468. [DOI] [PubMed] [Google Scholar]
- 48.Pasquini, L. A., Besio Moreno, M., Adamo, A. M., Pasquini, J. M. & Soto, E. F. (2000) J. Neurosci. Res. 59, 601–611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.