Abstract
ErbB2 is a receptor tyrosine kinase whose activity in normal cells depends on dimerization with another ligand-binding ErbB receptor. In contrast, amplification of c-erbB2 in tumors results in dramatic overexpression and constitutive activation of the receptor. Breast cancer cells overexpressing ErbB2 depend on its activity for proliferation, because treatment of these cells with ErbB2-specific antagonistic antibodies or kinase inhibitors blocks tumor cells in the G1 phase of the cell cycle. Intriguingly, loss of ErbB2 signaling is accompanied by a decrease in the phosphotyrosine content of ErbB3. On the basis of these results, it has been proposed that ErbB3 might be a partner for ErbB2 in promoting cellular transformation. To test this hypothesis and directly examine the role of the “kinase dead” ErbB3, we specifically ablated its expression with a designer transcription factor (E3). By infection of ErbB2-overexpressing breast cancer cells with a retrovirus expressing E3, we show that ErbB3 is an essential partner in the transformation process. Loss of functional ErbB2 or ErbB3 has similar effects on cell proliferation and cell cycle regulators. Furthermore, expression of constitutively active protein kinase B rescues the proliferative block induced as a consequence of loss of ErbB2 or ErbB3 signaling. These results demonstrate that ErbB2 overexpression and activity alone are insufficient to promote breast tumor cell division. Furthermore, we identify ErbB3's role, which is to couple active ErbB2 to the phosphatidylinositol 3-kinase/protein kinase B pathway. Thus, the ErbB2/ErbB3 dimer functions as an oncogenic unit to drive breast tumor cell proliferation.
The family of ErbB receptor tyrosine kinases includes four members: epidermal growth factor (EGF) receptor/ErbB1, ErbB2, ErbB3, and ErbB4. Binding of peptides of the EGF-related growth factor family to the extracellular domain of ErbB receptors results in the formation of homo- and heterodimers. Ligand binding induces the intrinsic receptor kinase activity, ultimately leading to stimulation of intracellular signaling cascades (1, 2). The physiological role of ErbB2, in the context of ErbB ligand signaling, is to serve as a coreceptor (3, 4). In fact, ErbB2 appears to be the preferred partner of the other ligand-bound ErbBs (5, 6). The importance of heterodimer-mediated signaling in normal development is obvious from studies in genetically modified mice. This is particularly true for ErbB2/ErbB3 and ErbB2/ErbB4 heterodimers. Loss of ErbB2 or ErbB3 has a similar impact on neuronal development (7), whereas loss of ErbB2 or ErbB4 has major effects on heart development (8, 9).
A wealth of clinical data has demonstrated that ErbB receptor tyrosine kinases, in particular ErbB1 and ErbB2, have roles in human cancer development, thus making them attractive targets for cancer therapies (10–13). ErbB2 overexpression, generally attributable to gene amplification, occurs in 25–30% of breast cancer and correlates with shorter time to relapse and lower overall survival (14). Overexpressed ErbB2 is constitutively phosphorylated in breast cancer cell lines and in human tumors (15, 16). It has been observed that targeting overexpressed active ErbB2 results in efficient inhibition of breast cancer cell proliferation, which proceeds via inhibition of intracellular signaling pathways and directly targets various members of the cell cycle machinery (17–20).
Interestingly, expression of ErbB3 is seen in many tumors that express ErbB2, including breast (21), bladder (22), and others. Furthermore, in many ErbB2-overexpressing breast tumors, ErbB3 has elevated levels of phosphotyrosine (15). ErbB3 itself has impaired tyrosine kinase activity (23) and needs a dimerization partner to become phosphorylated and acquire signaling potential (24). Indeed, we and others have shown that inactivation of ErbB2 leads to decreased ErbB3 tyrosine phosphorylation (17, 18, 25, 26). ErbB3, which contains six docking sites for the p85 adaptor subunit of phosphatidylinositol 3-kinase (PI3K), efficiently couples to this pathway (27, 28). Interestingly, it has been observed that a major consequence of targeting overexpressed ErbB2 is decreased PI3K/protein kinase B (PKB) activity (17, 18, 26), suggesting a role for ErbB3 in stimulation of this pathway downstream of the active ErbB2.
Here we have investigated whether ErbB3 tyrosine phosphorylation is a consequence of ErbB2 signaling or a necessary step to activate the PI3K pathway, thus contributing directly to proliferation of breast cancer cells. Signaling originating from either ErbB2 or ErbB3 was specifically down-regulated. ErbB2 was functionally inactivated by intracellular expression of a single-chain antibody [single-chain variable region fragment (scFv-5R)], which retains the receptor in the endoplasmic reticulum and prevents its translocation to the plasma membrane (29, 30). ErbB3 was targeted by a transcription factor designed to bind a specific region in the 5′ UTR of c-erbB3 and down-regulate its expression (31, 32). We conclude that in breast cancer cells, constitutive tyrosine phosphorylation on ErbB3 depends on the activity of overexpressed ErbB2. Furthermore, activity of the PI3K/PKB pathway depends fully on p85's recruitment to phospho-ErbB3. Importantly, inactivation of ErbB3 blocks proliferation of breast cancer cells as efficiently as impeding ErbB2 signaling. Finally, expression of constitutively active PKB rescues the proliferative block induced as a consequence of loss of ErbB2 or ErbB3 signaling. These results demonstrate that the ErbB2/ErbB3 dimer functions as an oncogenic unit to drive tumor cell proliferation.
Materials and Methods
Antibodies and Reagents. Antibodies used for surface staining, Western blotting, and immunoprecipitation were: ErbB3 -SGP1 (NeoMarkers, Fremont, CA), C-17 (Santa Cruz Biotechnology); ErbB2-21N (18); phospho-ErbB2 -Y1248 (Upstate Biotechnology, Lake Placid, NY); scFv-5R (17); p85 PI3K and antiphosphotyrosine-G10 (both from Upstate Biotechnology); phospho-pRb (Ser-795); PKB (Akt), phospho-PKB (Ser-473), extracellular-signal regulated kinase (ERK)1+2, phospho-ERK1+2 (p44/42; Thr-202/Tyr-204) (all from Cell Signaling, Beverly, MA); cyclin D3-C-16; cyclin E-HE12; cyclin B1-H-433; HA-Y-11 (all from Santa Cruz Biotechnology); cyclin A was kindly supplied by W. Krek (FMI, Basel). PKI116 was provided by P. Traxler (Novartis Pharma AG, Basel).
Expression Vectors. pMD.G, encoding the vesicular stomatitis virus-G envelope protein (33), and the pCLMFG (MFG) packaging vector (34) were obtained from I. Verma (Salk Institute, San Diego). E3, scFv-5R, and EGFP were cloned into MFG by standard cloning procedures. MFG–internal ribosome entry site (IRES)-EGFP was made by excision of the IRES-EGFP fragment from pAdloxCMV-IRES-EGFP (provided by U. Muller, FMI) and cloned into MFG NotI/BamHI. myristoylated PKB (MyrPKB) was blunt-cloned into the XhoI site of MFG-IRES-EGFP. The MyrPKB construct was obtained from B. Hemmings (FMI) (35). All expression constructs were verified by sequence analysis.
Cell Culture, Transfections, and Retroviral Infections. SKBR3, MDA-MB-361 (MB361), BT474, and T47D cells were obtained from American Type Culture Collection. The 293-polgag packaging cell line was obtained from I. Verma. Cells were cultured at 37°C/5% CO2 in RPMI 1640 (BT474) or DMEM (all others) supplemented with 10% FCS. SKBR3 cells were transfected with pcDNA3.1 expressing ErbB3 or with empty vector by using the FuGENE6 (Roche Diagnostics) reagent according to the manufacturer's protocol. Stable pools of transfectants were selected in medium containing 1 mg/ml G418 (Sigma). For retroviral infections, pMD.G and MFG plasmids were cotransfected into 293-polgag cells using Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol. After 16 h medium was replaced, and 1 day later culture supernatants were collected, passed through 0.45-μm filters, and transferred to target cells. For MB361 and T47D cells, the procedure was repeated 1 day later.
Flow Cytometry and Proliferation Assay. To analyze cell cycle profiles, trypsinized cells were stained with propidium iodide staining and analyzed on a Becton Dickinson FACScan flow cytometer, as described (17). Surface expression of ErbB3 was determined as described (31). To analyze cell cycle profiles of EGFP expressing cells, trypsinized cells were washed in cold PBS, then fixed and permeabilized in PBS/4% paraformaldehyde/0.1% saponin for 10 min at room temperature. After two washes with PBS/1% BSA/0.1% saponin, DNA was stained with 7-aminoactinomycin (Sigma; 5 μg/ml in PBS) for 30 min on ice, then subjected to flow cytometry analysis. For proliferation assays, equal numbers of SKBR3 cells were plated in triplicate 2 days after infection; 3 days later, cells were trypsinized and counted in a hemocytometer.
Cell Lysate Preparation, Immunoblots, and Northern Blotting. Cell extracts were prepared as described (17). Fifteen to 50 μg of protein lysates were resolved by SDS/PAGE (7.5–14%), transferred to poly(vinylidene difluoride) membranes, probed with specific antibodies and detected by enhanced chemiluminescence (Amersham Pharmacia Biotech). Northern blotting was done as described (17).
Results
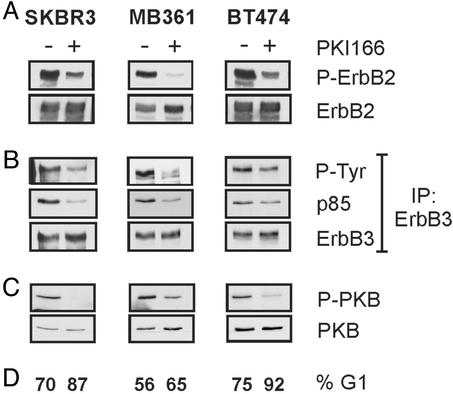
Sensitivity of ErbB2-Overexpressing Breast Tumor Cells to PKI166: Inhibition of ErbB2 Blocks ErbB3 Signaling. The kinase-impaired ErbB3 acquires signaling activity, i.e., phosphotyrosine, only after associating with another active ErbB family member. In breast tumor cells with overexpressed ErbB2, ErbB3 has relatively high amounts of phosphotyrosine, which might result from an interaction with ErbB2. To examine this experimentally, we treated three ErbB2-overexpressing breast tumor cell lines, SKBR3, BT474, and MB361, with PKI166, an ErbB-selective kinase inhibitor (36). Short-term treatment of each tumor cell line with PKI166 resulted in a strong decrease in ErbB2's phosphotyrosine content (Fig. 1A). After 24 h of treatment, each cell line showed an accumulation in the G1 phase of the cell cycle (Fig. 1D). Importantly, the phosphotyrosine content of ErbB3 was reduced in parallel to that of ErbB2 (Fig. 1B), showing the significance of ErbB2 in maintaining ErbB3's activity. Of the four ErbB receptors, ErbB3 couples best to the PI3K pathway (27, 28). In the PKI166-treated breast tumor cells, we observed a decrease in the level of the ErbB3/p85 complex (Fig. 1B) and the activity of PKB, as measured by a phosphospecific antibody against Ser-473, was down-regulated (Fig. 1C). These results suggest the signaling activity of ErbB3 to the downstream PI3K/PKB pathway depends on constitutively active ErbB2.
Fig. 1.
An ErbB2 kinase inhibitor blocks ErbB3 signaling and activity of the PI3K pathway. SKBR3, MB361, and BT474 breast cancer cells were treated for 30 min (A–C) or 24 h (D) with medium containing 5 μM PKI166 (+) or DMSO vehicle control (–). Protein lysates were probed for phospho-ErbB2 (P-ErbB2) (A) or phospho-PKB (P-PKB) (C). Membranes were stripped and reprobed to control for ErbB2 and PKB levels. (B) ErbB3 was immunoprecipitated from 500 μg of protein lysates, immunoprecipitates were resolved by SDS/PAGE, transferred onto a poly(vinylidene difluoride) membrane, and probed for phosphotyrosine (P-Tyr) or p85PI3K. Membranes were stripped and reprobed to control for ErbB3 levels. (D) Cells were harvested by trypsinization, nuclei were stained with propidium iodide, and a flow cytometric analysis was performed. The percentage of cells in the G1 phase of the cell cycle is indicated.
Specific Down-Regulation of ErbB2 and ErbB3. The previous results raise the intriguing possibility that ErbB3 is an essential partner for ErbB2, and that the ErbB2/ErbB3 heterodimer functions as a signaling unit. To address this possibility, we used the transcription factor E3 (31) to down-regulate ErbB3 and the single-chain antibody scFv-5R (29, 30) to functionally inactivate ErbB2.
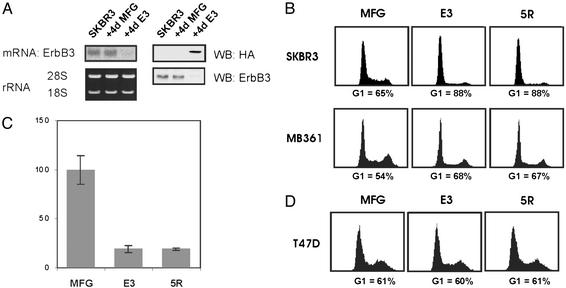
E3, an artificial transcription factor that blocks ErbB3 expression, is composed of a polydactyl zinc-finger domain designed to specifically recognize an 18-bp stretch in the 5′ UTR of c-erbB3. This domain is fused to a Krüppel-associated box (a transcriptional repressor domain), a nuclear localization signal, and a hemagglutinin tag (31). E3 was expressed in target cells through retroviral infection. The control retrovirus (MFG) had no effect on ErbB3 expression (Fig. 2A). E3 infection efficiency, often close to 100%, was routinely monitored by ErbB3 surface staining (not shown). Four days after infection, there was no detectable ErbB3 mRNA or protein (Fig. 2 A).
Fig. 2.
ErbB3 expression is required for ErbB2-dependent proliferation. (A) Four days after infection of SKBR3 cells, total RNA or protein was prepared from noninfected cells (SKBR3) or cells infected with the MFG control virus (MFG) or the E3 virus (E3). (Left) An ErbB3 Northern analysis was carried out on 3 μg of total RNA; ribosomal RNA levels are shown below. (Right) Fifty micrograms of protein lysates were probed for HA or ErbB3. (B and D) Cultures were infected with the indicated viruses and collected 4 days later. Cells were harvested by trypsinization, and the nuclei were stained with propidium iodide. The percentage of cells in G1 is indicated. (C) Proliferation assay of SKBR3 cells was performed as described in Materials and Methods. The bars indicate standard deviations.
We have previously shown that intracellular expression of the endoplasmic reticulum (ER)-targeted scFv-5R causes retention of ErbB2 in this compartment, leading to loss of receptor function (29, 30). ER retention of ErbB2 had no effect on proliferation of cells with low levels of ErbB2 (29); however, it blocked growth of ErbB2-overexpressing tumor cells (17). To directly compare the effects of E3 and scFv-5R, the latter was cloned into the MFG vector to allow production of vesicular stomatitis virus-G pseudotyped virus (5R). In individual experiments, we have consistently observed that transient virus preparations have comparable infection efficiencies, which has allowed us to directly compare the effects of E3 and 5R.
ErbB2-Overexpressing Breast Tumor Cells Require ErbB3 to Proliferate. In the next experiments, we compared the sensitivity of the breast tumor cells lines SKBR3, MB361, and T47D to down-regulation of ErbB3 or functional inactivation of ErbB2. Four days after infection with the MFG, the E3, or the 5R virus, we examined cell proliferation using flow cytometry. The proliferation of the low ErbB2-expressing T47D cells was unaffected by either E3 or 5R expression (Fig. 2D). In contrast, both ErbB2-overexpressing lines were severely affected by E3 or 5R expression (Fig. 2B). SKBR3 cells went from a G1 population of 65% to 88% as a consequence of ErbB3 or ErbB2 loss (Fig. 2B Upper). Similarly, MB361 cells went from 54% to 68% and 67%, respectively (Fig. 2B Lower). Furthermore, after 5 days, there were 80% fewer cells in the E3- and 5R-expressing SKBR3 cultures vs. control cultures (Fig. 2C). Thus, in the ErbB2-overexpressing cell lines, expression of E3 or 5R had comparable effects, suggesting that both receptors are required for proliferation. Furthermore, down-regulation of ErbB3 expression has antiproliferative effects similar to those of the ErbB kinase inhibitor PKI166 (Fig. 1). We conclude that overexpression and activity of ErbB2 alone are not sufficient to drive proliferation of breast cancer cells; ErbB3 appears to be an essential partner.
The Antiproliferative Effect of E3 Is Due to Loss of ErbB3 Expression. To prove that E3's effect was a consequence of ErbB3 down-regulation, we tested it in cells ectopically expressing ErbB3 driven from a cytomegalovirus promoter and lacking the E3 target sequence. SKBR3-ErbB3 cells have elevated levels of the receptor, in comparison to the SKBR3-neo control cells (Fig. 3A Top). To test for E3's specificity, SKBR3-ErbB3 and SKBR3-neo cells were infected with the MFG control, the E3 and the 5R viruses. MFG-infected SKBR3-neo and SKBR3-ErbB3 cells had essentially the same G1 population (Fig. 3B, 67% vs. 65%, respectively), showing that ErbB3 overexpression had no effect on proliferation. Although transfected cells are not as efficiently infected as naive cells (T.H., unpublished observations), the viruses infected the SKBR3-neo and -ErbB3 cells with similar efficiency. This is attested to by the fact that E3 was expressed at the same level in both cell lines; the same holds true for 5R (Fig. 3A, hemagglutinin and scFv-5R, respectively). Thus, it is possible to directly compare the two cell lines for effects of ErbB receptor loss. The antiproliferative effects of E3 and 5R were equivalent in control SKBR3-neo (Fig. 3B Upper, 74% and 75% in G1 vs. 67% in MGF control). Turning to the SKBR3-ErbB3 cells, 5R-mediated down-regulation of ErbB2 had the same effect as in SKBR3-neo control cells (Fig. 3B Lower Right; G1 = 76%). These results show that high ErbB3 expression does not rescue the cells from the effects of 5R, i.e., ErbB3 still needs to be transactivated by ErbB2. In striking contrast to the results with 5R, SKBR3-ErbB3 cells were unaffected by E3 expression (Fig. 3B Lower,G1 = 65% in MFG- and E3-infected cells). These results demonstrate that loss of ErbB3 is directly responsible for the antiproliferative effect of the E3 transcription factor.
Fig. 3.
The antiproliferative effect of E3 is due to loss of ErbB3. Stable pools of SKBR3 cells transfected with empty pcDNA3 vector (SKBR3-neo) or pcDNA3-ErbB3 (SKBR3-ErbB3) were infected with control (MFG), E3 or 5R viruses. Four days after infection parallel cultures were used to prepare cell lysates (A) or analyzed for cell cycle distribution (B). (A) Fifty micrograms of protein lysates were probed with the indicated antibodies. Reprobing with an ERK1+2 antiserum was used to control for loading. (B) Cells were harvested by trypsinization, and nuclei were stained with propidium iodide. The percentage of cells in G1 is indicated.
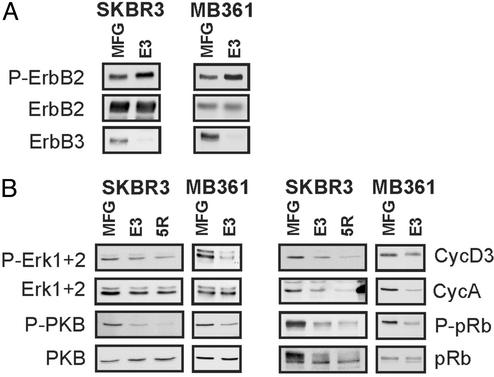
Effects of ErbB3 Down-Regulation on Cytoplasmic and Nuclear-Signaling Molecules. The mitogen-activated protein kinase and PI3K pathways are major signaling cascades downstream of activated ErbB receptors (1). Antisera specific for the active phosphorylated forms of ERK1+2 and PKB, the major kinases on the respective pathways, were used to probe for their activity in E3- and 5R-expressing cells. As observed in many tumor cells, these pathways are also constitutively active in SKBR3 and MB361 cells, as demonstrated by the high basal levels of P-ERK1+2 and P-PKB (Fig. 4B, MFG). Loss of ErbB3 had no major effect on the level of P-ERK1+2 in SKBR3 tumor cells but led to a strong decrease in P-PKB content in both cell lines (Fig. 4B Left). These results are intriguing considering that in both E3-expressing tumor cell lines, overexpressed ErbB2 was still highly phosphorylated on tyrosine (Fig. 4A Top) and thus has full signaling potential.
Fig. 4.
E3 down-regulates cytoplasmic and nuclear signaling molecules but not ErbB2 activity in breast tumor cells. Cultures of SKBR3 or MB361 cells were infected with the indicated viruses, and 4 days later, protein lysates were prepared. Fifty micrograms of protein lysates were resolved by SDS/PAGE and transferred onto poly(vinylidene difluoride) membranes. (A) The same membrane was sequentially probed for phospho-ErbB2 (P-ErbB2), ErbB2, and ErbB3. (B) Membranes were probed for phospho-ERK1+2, phospho-PKB, cyclin D3, cyclin A, phospho-pRb, pRb, ERK1+2, and PKB.
Turning to nuclear cell cycle regulators, we observed lower levels of cyclin A in 5R- and E3-expressing SKBR3 tumor cells (Fig. 4B), which likely reflects the decreased number of S-phase cells after loss of functional ErbB receptors. In E3-expressing SKBR3 and MB361 tumor cells, there was also a strong decrease in phosphorylation of pRb on Ser-795, a Cdk4/cyclin D site, which might be attributed to the drop in cyclin D3 levels evident in these cells (Fig. 4B Right). Interestingly, the level of cyclin D3 closely reflects proliferation. In Fig. 3, SKBR3-ErbB3 cells, which were rescued from the antiproliferative effect of E3, had high levels of cyclin D3, in comparison to the E3-blocked SKBR3-neo cells and the 5R-blocked cultures, which all showed reduced cyclin D3 expression, a result also confirmed by analysis of pRb Ser-795 phosphorylation (Fig. 3A). In summary, the loss of ErbB3 expression had major effects on nuclear cell cycle regulators and ErbB2 activity alone was not sufficient to maintain strong activation of the signaling pathways, despite the fact that the receptor was heavily phosphorylated in the E3-expressing tumor cells. These results support our hypothesis that ErbB2/ErbB3 functions as an oncogenic unit.
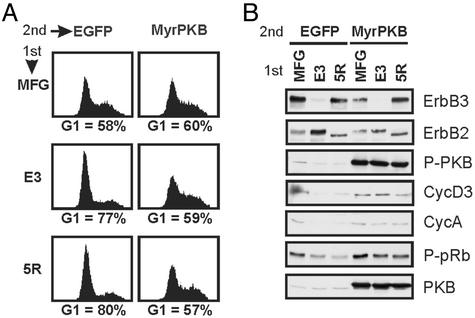
Constitutively Active PKB Reverts the Antiproliferative Effects of E3 and 5R. Considering ErbB3's potential to interact with p85 and the fact that the PI3K/PKB pathway was consistently down-regulated in response to loss of ErbB3, we examined whether restoration of PKB activity could rescue the E3 phenotype. To accomplish this, a membrane-targeted constitutively active form of PKB (35) was introduced into cells via infection with a vesicular stomatitis virus-G pseudotyped, bicistronic virus expressing MyrPKB and an IRES-driven EGFP. Experimentally, SKBR3 cells were first infected with MFG, E3, or 5R viruses. Then, 36 h later, these cultures were divided and infected with either EGFP-expressing control virus or MyrPKB virus. Sixty hours later, EGFP-positive cells were analyzed for their DNA content by flow cytometry (Fig. 5A), and parallel cultures were used to analyze specific proteins (Fig. 5B).
Fig. 5.
Expression of constitutively active PKB rescues the G1 block of SKBR3 cells lacking functional ErbB2 or ErbB3. SKBR3 cells were infected at day 0 with either MFG, E3, or 5R viruses (1st). Thirty-six hours later, cultures were infected with a second virus (2nd) expressing either EGFP or myristoylated PKB-IRES-EGFP (MyrPKB). Parallel cultures were collected 4 days after the first infection and stained for DNA content (A) or used to prepare protein lysates (B). In A, cells were harvested by trypsinization and fixed, and the cell cycle distribution of EGFP-positive cells was determined. The percentage of cells in the G1 phase of the cycle is indicated. In B, 15 μg of protein lysates was resolved by SDS/PAGE, transferred onto poly(vinylidene difluoride) membranes, and probed for the indicated proteins.
There was little or no ErbB3 in the E3-infected cultures, and essentially all ErbB2 was in the ER in the 5R-infected cells (Fig. 5B Top, ER retention causes ErbB2 to electrophorese more rapidly). After the second round of infection, the MyrPKB-infected cultures displayed higher levels of the kinase, in comparison to control cultures (Fig. 5B, PKB). Importantly, MyrPKB was active, as shown by high levels of Ser-473 phosphorylation (Fig. 5B, P-PKB). Expression of MyrPKB had essentially no effect on basal SKBR3 proliferation (Fig. 5A, G1 = 58% and 60% in EGFP- and MyrPKB-infected cells, respectively). Furthermore, as expected, E3- and 5R-expressing cells infected with the EGFP virus still accumulated in G1 (Fig. 5A Left, 77% and 80%, respectively, vs. 58% in control MFG cultures). In striking contrast, expression of MyrPKB completely rescued the antiproliferative effects mediated by loss of both ErbB receptor (Fig. 5A Right). E3- and 5R-expressing cells concomitantly expressing MyrPKB had approximately the same G1 population, 59% and 57%, respectively, as MFG-infected cultures (60%). Turning to the cell cycle regulators, E3- and 5R-expressing cells rescued by MyrPKB had the same level of cyclin D3, cyclin A, and P-pRb as control cells (Fig. 5B).
In conclusion, the results suggest that activity of the PI3K/PKB pathway is essential for proliferation of these tumor cells. Furthermore, fact that MyrPKB rescues the 5R-expressing cells suggests that ErbB2's proliferative effect funnels mainly through ErbB3 and the downstream PI3K/PKB pathway.
Discussion
The results presented here demonstrate that ErbB3 functions as an indispensable ErbB2 dimerization partner and is required for proliferation of ErbB2-overexpressing tumor cells. We and others have previously shown that blocking ErbB2 had antiproliferative effects that were consistently accompanied by a decrease in ErbB3's signaling ability (17, 18, 25, 26). The availability of E3 allowed us to directly test the role of endogenous ErbB3 in breast cancer cells without affecting ErbB2 signaling activity and dimerization potential. Importantly, by rescuing E3-blocked cells with ectopic ErbB3, the possibility that other unspecific effects cause the G1 arrest can be ruled out. The combination of the binding specificity and the ease with which such transcription factors can be assembled make them attractive and versatile tools (32).
Although in normal cells ErbB2 signaling is controlled by the EGF-related ligands through obligate formation of heterodimers with other ErbBs (2, 5, 6), in tumor cells overexpressing ErbB2, two possibilities for maintaining constitutive ligand-independent stimulation of signaling pathways can be considered. On the one hand, overexpressed active ErbB2 might function on its own; on the other hand, ErbB2 might still need to dimerize with another ErbB receptor. On the basis of the results presented here, we propose that in cancer cells, ErbB2 cannot act alone but requires ErbB3 for its full signaling potential.
Loss of ErbB3 was strongly antiproliferative in the three ErbB2-overexpressing breast tumor cell lines we examined: SKBR3, MB361, and BT474 (shown here and T.H., unpublished data). Each one also expresses ErbB1; in addition, BT474 cells contain ErbB4. Thus, to drive proliferation, neither ErbB1 nor ErbB4 could replace ErbB3 as ErbB2's partner. In the same vein, ErbB4, was not able to rescue loss of ErbB3 when introduced into SKBR3 cells (T.H., unpublished data). These results suggest that ErbB3 is the biologically relevant partner for overexpressed ErbB2. A hint that ErbB2/ErbB3 dimers are also important in vivo comes from results with a transgenic mammary tumor model induced by expression of activated forms of ErbB2. Here it was observed that expression of ErbB3, but not ErbB1 or ErbB4, was increased in the tumors. Furthermore, ErbB3 was tyrosine phosphorylated, suggesting that during ErbB2-driven tumor development ErbB3 activity might be important (37).
Previous publications suggested a role for ErbB3, in the context of an active ErbB2/ErbB3 heterodimer, in maintaining constitutive activity of the PI3K/PKB pathway (18, 20, 25, 38). Here we unambiguously show that without ErbB3, the activity of this pathway is severely compromised. The observation that tumor cells lacking ErbB3 maintain the same high level of phosphorylated ErbB2 as do control cells yet are proliferatively blocked is a striking result, highlighting the essential role of ErbB3. That ERK1+2 activity was nearly unaffected by E3 expression in SKBR3 cells (Fig. 4B) and that treatment of SKBR3 cells with the kinase inhibitor PKI166 decreased the activity of the mitogen-activated protein kinase (MAPK) pathway (not shown) prove that signaling pathways originating from ErbB2 are still active in these cells. Nevertheless, PKI166 was no more effective in blocking proliferation of SKBR3 cells than E3-mediated ErbB3 down-regulation (Fig. 1). These results demonstrate the importance of the PI3K/PKB pathway to the in vitro proliferation of these tumor cells. However, it is very likely that in vivo this as well as other pathways, e.g., the MAPK pathway, play important roles in other aspects of tumorigenesis.
Further proof of the importance of the PI3K/PKB pathway in breast cancer comes from clinical studies. Phosphatase and tensin homolog deleted on chromosome 10 (PTEN), a negative regulator of the PI3K/PKB pathway, has been increasingly implicated in breast carcinogenesis. Cowden's syndrome, which, in affected family members, is associated with an increased risk of breast cancer, is due to germ-line mutations in PTEN (39). Although PTEN mutations are rare in spontaneous breast cancers (40), a significant number of breast tumors have low PTEN levels (41). Alterations of other proteins in the PI3K pathway have also been described. Overexpression of PKB has been reported in various types of human cancer (reviewed in ref. 42) and amplification of the gene encoding PKBβ has been found in some primary breast tumors (43). Furthermore, amplification of PIK3CA, which encodes the catalytic subunit of PI3K, has been described in primary ovarian tumors and cell lines (44). Intriguingly, some of these ovarian cancer cell lines overexpress ErbB2 but lack ErbB3 (45), raising the interesting possibility that overexpression of PI3K substitutes for ErbB3 in some ovarian tumors. That expression of a constitutively active PKB completely rescues loss of ErbB2/ErbB3 heterodimer signaling confirms the importance of the PI3K/PKB pathway in breast cancer cell proliferation.
The results presented here might have important clinical implications. We have shown that in ErbB2-overexpressing breast tumor cell lines there is a correlation between ErbB3 expression and sensitivity to ErbB2-directed inhibitors. Attesting to this correlation, the MKN7 tumor cell line, which has high ErbB2 levels but lacks ErbB3, is insensitive to ErbB2-directed inhibitors (18, 20). We propose that tumors with active ErbB2/ErbB3 dimers might be particularly sensitive to ErbB2-targeted therapeutics, in comparison to tumors that have other molecular alterations leading to activation of the PI3K/PKB pathway. We suggest that ErbB3 might function as a diagnostic marker for such tumors and that screening for ErbB3 and components of the PI3K pathway might be useful to design the best cancer treatment. Finally, the results presented here suggest that targeting ErbB2-driven tumors with agents disrupting heterodimer formation should be a feasible alternative to targeting ErbB2 activity alone. In this respect, it was recently shown that mAb 2C4, which blocks ErbB2's ability to heterodimerize with other ErbBs, efficiently inhibits growth of BT474 tumor cells in a xenograft model (46). By demonstrating ErbB3's essential role as an ErbB2 dimerization partner, coupling it to the PI3K/PKB pathway and driving proliferation of tumor cells, we provide a model that could explain why preventing ErbB2 heterodimerization can be a powerful tool in tumors where activation of the receptor is achieved by its overexpression.
Acknowledgments
We thank Drs. U. Muller, B. Hemmings, W. Krek, and I. Verma for providing plasmids, cells, and antibodies. Thanks to the Hynes lab members, in particular I. Boschke and A. Badache, for their helpful discussions. The laboratory of N.E.H. was supported by Novartis Forschungsstiftung Zweigniederlassung Friedrich Miescher Institute for Biomedical Research. T.H. was partially supported by grants from the Novartis Stiftung für Medizinisch-Biologische Forschung and the Krebsliga Beider Basel. The laboratory of C.F.B. was supported by National Institutes of Health Grants CA86258 and DK61803.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: scFv, single-chain variable region fragment; IRES, internal ribosome entry site; ER, endoplasmic reticulum; PKB, protein kinase B; MyrPKB, myristoylated PKB; PI3K, phosphatidylinositol 3-kinase; ERK, extracellular-signal regulated kinase; EGF, epidermal growth factor; EGFP, EGF protein.
References
- 1.Olayioye, M. A., Neve, R. M., Lane, H. A. & Hynes, N. E. (2000) EMBO J. 19, 3159–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yarden, Y. & Sliwkowski, M. X. (2001) Nat. Rev. Mol. Cell Biol. 2, 127–137. [DOI] [PubMed] [Google Scholar]
- 3.Pinkas-Kramarski, R., Soussan, L., Waterman, H., Levkowitz, G., Alroy, I., Klapper, L., Lavi, S., Seger, R., Ratzkin, B. J., Sela, M., et al. (1996) EMBO J. 15, 2452–2467. [PMC free article] [PubMed] [Google Scholar]
- 4.Riese, D. J., van Raaij, T. M., Plowman, G. D., Andrews, G. C. & Stern, D. F. (1995) Mol. Cell. Biol. 15, 5770–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graus-Porta, D., Beerli, R. R., Daly, J. M. & Hynes, N. E. (1997) EMBO J. 16, 1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzahar, E., Waterman, H., Chen, X., Levkowitz, G., Karunagaran, D., Lavi, S., Ratzkin, B. J. & Yarden, Y. (1996) Mol. Cell. Biol. 16, 5276–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britsch, S., Li, L., Kirchhoff, S., Theuring, F., Brinkmann, V., Birchmeier, C. & Riethmacher, D. (1998) Genes Dev. 12, 1825–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee, K. F., Simon, H., Chen, H., Bates, B., Hung, M. C. & Hauser, C. (1995) Nature 378, 394–398. [DOI] [PubMed] [Google Scholar]
- 9.Gassmann, M., Casagranda, F., Orioli, D., Simon, H., Lai, C., Klein, R. & Lemke, G. (1995) Nature 378, 390–394. [DOI] [PubMed] [Google Scholar]
- 10.Shawver, L. K., Slamon, D. & Ullrich, A. (2002) Cancer Cell 1, 117–123. [DOI] [PubMed] [Google Scholar]
- 11.Mendelsohn, J. & Baselga, J. (2000) Oncogene 19, 6550–6565. [DOI] [PubMed] [Google Scholar]
- 12.Salomon, D. S., Brandt, R., Ciardiello, F. & Normanno, N. (1995) Crit. Rev. Oncol. Hematol. 19, 183–232. [DOI] [PubMed] [Google Scholar]
- 13.Hynes, N. E. & Stern, D. F. (1994) Biochim. Biophys. Acta 1198, 165–184. [DOI] [PubMed] [Google Scholar]
- 14.Slamon, D. J., Clark, G. M., Wong, S. G., Levin, W. J., Ullrich, A. & McGuire, W. L. (1987) Science 235, 177–182. [DOI] [PubMed] [Google Scholar]
- 15.Alimandi, M., Romano, A., Curia, M. C., Muraro, R., Fedi, P., Aaronson, S. A., Di Fiore, P. P. & Kraus, M. H. (1995) Oncogene 10, 1813–1821. [PubMed] [Google Scholar]
- 16.DiGiovanna, M. P., Chu, P., Davison, T. L., Howe, C. L., Carter, D., Claus, E. B. & Stern, D. F. (2002) Cancer Res. 62, 6667–6673. [PubMed] [Google Scholar]
- 17.Neve, R. M., Sutterluty, H., Pullen, N., Lane, H. A., Daly, J. M., Krek, W. & Hynes, N. E. (2000) Oncogene 19, 1647–1656. [DOI] [PubMed] [Google Scholar]
- 18.Lane, H. A., Beuvink, I., Motoyama, A. B., Daly, J. M., Neve, R. M. & Hynes, N. E. (2000) Mol. Cell. Biol. 20, 3210–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yakes, F. M., Chinratanalab, W., Ritter, C. A., King, W., Seelig, S. & Arteaga, C. L. (2002) Cancer Res. 62, 4132–4141. [PubMed] [Google Scholar]
- 20.Munster, P. N., Marchion, D. C., Basso, A. D. & Rosen, N. (2002) Cancer Res. 62, 3132–3137. [PubMed] [Google Scholar]
- 21.Naidu, R., Yadav, M., Nair, S. & Kutty, M. K. (1998) Br. J. Cancer 78, 1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chow, N. H., Chan, S. H., Tzai, T. S., Ho, C. L. & Liu, H. S. (2001) Clin. Cancer Res. 7, 1957–1962. [PubMed] [Google Scholar]
- 23.Guy, P. M., Platko, J. V., Cantley, L. C., Cerione, R. A. & Carraway, K. L., III (1994) Proc. Natl. Acad. Sci. USA 91, 8132–8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, H. H., Vijapurkar, U., Hellyer, N. J., Bravo, D. & Koland, J. G. (1998) Biochem. J. 334, 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motoyama, A. B., Hynes, N. E. & Lane, H. A. (2002) Cancer Res. 62, 3151–3158. [PubMed] [Google Scholar]
- 26.Basso, A. D., Solit, D. B., Munster, P. N. & Rosen, N. (2002) Oncogene 21, 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fedi, P., Pierce, J. H., Di Fiore, P. P. & Kraus, M. H. (1994) Mol. Cell. Biol. 14, 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prigent, S. A. & Gullick, W. J. (1994) EMBO J. 13, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graus-Porta, D., Beerli, R. R. & Hynes, N. E. (1995) Mol. Cell. Biol. 15, 1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beerli, R. R., Wels, W. & Hynes, N. E. (1994) J. Biol. Chem. 269, 23931–23936. [PubMed] [Google Scholar]
- 31.Beerli, R. R., Dreier, B. & Barbas, C. F., III (2000) Proc. Natl. Acad. Sci. USA 97, 1495–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beerli, R. R. & Barbas, C. F., III (2002) Nat. Biotechnol. 20, 135–141. [DOI] [PubMed] [Google Scholar]
- 33.Kafri, T., van Praag, H., Ouyang, L., Gage, F. H. & Verma, I. M. (1999) J. Virol. 73, 576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naviaux, R. K., Costanzi, E., Haas, M. & Verma, I. M. (1996) J. Virol. 70, 5701–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andjelkovic, M., Alessi, D. R., Meier, R., Fernandez, A., Lamb, N. J., Frech, M., Cron, P., Cohen, P., Lucocq, J. M. & Hemmings, B. A. (1997) J. Biol. Chem. 272, 31515–31524. [DOI] [PubMed] [Google Scholar]
- 36.Traxler, P., Bold, G., Buchdunger, E., Caravatti, G., Furet, P., Manley, P., O'Reilly, T., Wood, J. & Zimmermann, J. (2001) Med. Res. Rev. 21, 499–512. [DOI] [PubMed] [Google Scholar]
- 37.Siegel, P. M., Ryan, E. D., Cardiff, R. D. & Muller, W. J. (1999) EMBO J. 18, 2149–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ram, T. G., Schelling, M. E. & Hosick, H. L. (2000) Cell Growth Differ. 11, 173–183. [PubMed] [Google Scholar]
- 39.Schrager, C. A., Schneider, D., Gruener, A. C., Tsou, H. C. & Peacocke, M. (1998) Hum. Pathol. 29, 47–53. [DOI] [PubMed] [Google Scholar]
- 40.Rhei, E., Kang, L., Bogomolniy, F., Federici, M. G., Borgen, P. I. & Boyd, J. (1997) Cancer Res. 57, 3657–3659. [PubMed] [Google Scholar]
- 41.Mills, G. B., Lu, Y., Fang, X., Wang, H., Eder, A., Mao, M., Swaby, R., Cheng, K. W., Stokoe, D., Siminovitch, K., et al. (2001) Semin. Oncol. 28, 125–141. [DOI] [PubMed] [Google Scholar]
- 42.Hill, M. M. & Hemmings, B. A. (2002) Pharmacol. Ther. 93, 243–251. [DOI] [PubMed] [Google Scholar]
- 43.Bellacosa, A., de Feo, D., Godwin, A. K., Bell, D. W., Cheng, J. Q., Altomare, D. A., Wan, M., Dubeau, L., Scambia, G., Masciullo, V., et al. (1995) Int. J. Cancer 64, 280–285. [DOI] [PubMed] [Google Scholar]
- 44.Shayesteh, L., Lu, Y., Kuo, W. L., Baldocchi, R., Godfrey, T., Collins, C., Pinkel, D., Powell, B., Mills, G. B. & Gray, J. W. (1999) Nat. Genet. 21, 99–102. [DOI] [PubMed] [Google Scholar]
- 45.Xu, F., Yu, Y., Le, X. F., Boyer, C., Mills, G. B. & Bast, R. C., Jr. (1999) Clin. Cancer Res. 5, 3653–3660. [PubMed] [Google Scholar]
- 46.Agus, D. B., Akita, R. W., Fox, W. D., Lewis, G. D., Higgins, B., Pisacane, P. I., Lofgren, J. A., Tindell, C., Evans, D. P., Maiese, K., et al. (2002) Cancer Cell 2, 127–137. [DOI] [PubMed] [Google Scholar]