Abstract
Potato leafroll polerovirus (PLRV) genomic RNA acts as a polycistronic mRNA for the production of proteins P0, P1, and P2 translated from the 5′-proximal half of the genome. Within the P1 coding region we identified a 5-kDa replication-associated protein 1 (Rap1) essential for viral multiplication. An internal ribosome entry site (IRES) with unusual structure and location was identified that regulates Rap1 translation. Core structural elements for internal ribosome entry include a conserved AUG codon and a downstream GGAGAGAGAGG motif with inverted symmetry. Reporter gene expression in potato protoplasts confirmed the internal ribosome entry function. Unlike known IRES motifs, the PLRV IRES is located completely within the coding region of Rap1 at the center of the PLRV genome.
Initiation of protein biosynthesis in eukaryotes is generally governed by a scanning mechanism. The scanning complex, comprising the 40S ribosomal subunit and initiation factors (1, 2), binds to the 5′ end of mRNA and migrates downstream until the complex reaches an AUG codon embedded in an optimum context for the initiation of protein synthesis (3). For the majority of eukaryotic mRNAs, the first AUG encountered by the scanning complex serves as the translation initiation codon (first-AUG rule). However, in some instances, the scanning complex bypasses the first AUG (preferably when the AUG is in an unfavorable sequence context) and initiates protein synthesis at downstream AUGs (leaky scanning) (2). A second mechanism operating for internal initiation on polycistronic mRNAs is the reinitiation of released ribosomes after protein-synthesis termination at upstream stop codons. Hohn and coworkers (4) identified a “shunt” mechanism for discontinuous ribosome migration when they studied the translation of cauliflower mosaic virus 35S RNA. In this system, scanning is initiated at the 5′ end of the mRNA, but the scanning complex is forced by cis-acting viral sequences to bypass >300 nt before resuming scanning (for a review on translation in plants see ref. 5).
In contrast to these strategies, which are characterized by ribosome scanning, direct ribosome entry at the site of translation initiation mediated by a “ribosome landing pad” or an internal ribosomal entry site (IRES) is an alternative translation initiation mechanism. IRES-mediated translation initiation was first demonstrated for picornaviral protein synthesis (6–8). IRES sequences form complex secondary and tertiary structures, are interacting with the translational machinery, and are followed by an AUG start codon (reviewed in ref. 9; see refs. 10–14). IRES-mediated protein initiation has been reported, for example, with hepatitis C virus (15–18), murine leukemia virus (19), Moloney murine leukemia virus (20), potyviral RNAs (21, 22), crucifer-infecting tobamovirus (23), classical swine fever virus (24), cricket paralysis virus (25), and some nonviral mRNAs (for reviews see refs. 14 and 26). All the identified IRES signals are located at the 5′ end of their respective mRNAs with the exception of the cricket paralysis virus IRES, which is located 6,024 nt from the 5′ end of the viral RNA.
In view of the small size of viral genomes, viruses exploit noncanonical translation mechanisms in an effort to “decompress” their limited genetic information for the synthesis of a maximal number of gene products. Such expression strategies (for a review see ref. 27) have been studied in detail with luteoviruses and include –1 ribosomal frameshifting (28–31), amber stop-codon suppression (32–34), and cap-independent translation initiation (35, 36).
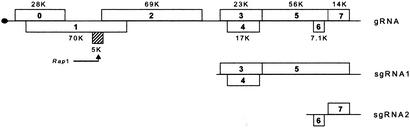
Potato leafroll virus (PLRV), the type species of the genus Polerovirus (previously subgroup 2) in the family Luteoviridae, has attracted special interest, because its host plant potato is easily transformed and thus offers the opportunity to study viral translational mechanisms in vivo. PLRV is a monopartite, single-stranded RNA virus that is aphid-transmitted in a persistent nonpropagative manner and restricted to the phloem tissue during replication (37). The PLRV genome consists of a 5.9-kb (+)-sense RNA that is covalently coupled to the small VPg protein (38, 39). Its eight large ORFs are organized into two gene clusters separated by a small intergenic region (Fig. 1). The 3′-located genes are translated from subgenomic RNAs [sgRNA1 (33) and sgRNA2 (40)], whereas genomic RNA serves as a template for the translation of genes in the 5′-proximal region (D.P., unpublished data).
Fig. 1.
Structure of the PLRV RNA genome. Major ORFs are shown with the apparent molecular weights of the corresponding gene products. The position of Rap1 is indicated in black. gRNA, genomic RNA; sgRNA, subgenomic RNA.
Here we describe the expression of the ORF replication-associated protein 1 (Rap1) located ≈1,500 nt downstream of the viral 5′ end within the P1 coding region but in a different reading frame. Evidence is presented to demonstrate that translation of Rap1 is a prerequisite for viral replication and that Rap1 translation occurs both in vitro and in vivo through internal ribosome entry involving an unusually structured IRES regulatory element.
Materials and Methods
Synthesis of Chimeric PLRV-β-Glucuronidase (GUS) Constructs. To study the expression of Rap1 in vitro, a 293-bp PLRV cDNA fragment [coordinates 1,481–1,773 of the PLRV sequence according to Keese et al. (41)] was isolated from pCPL3 (42) by limited AluI digestion and cloned into an EcoRV-digested pSP65-GUS vector (28) to yield construct pInt-mut2. The Rap1 ochre stop codon (coordinates 1,666–1,668) in pInt-mut2 was mutated by site-directed mutagenesis (altered site-directed mutagenesis system, Promega) to UAC to create a continuous Rap1-GUSC reading frame (pInt-mut1). Construct pInt-mut3 contained an additional ochre stop codon in the ORF1 coding sequences at position 1,483 of the PLRV sequence and was obtained from pInt-mut1 by site-directed mutagenesis.
Insertion of a stable stem–loop structure at the 5′ end of the GUS gene was performed by cloning of five BamHI linkers into the NcoI site of pInt-mut3 following the strategy described by Pelletier and Sonenberg (43). All further mutations were performed on pInt-mut3 by using the following oligonucleotides.
Initiation codon analysis: pInt-mut5 1513 5′-ACTGGGCTGATGAATTATGACTCCG-3′ 1536; pInt-mut6 1519 5′-CTGATGATTATGAACTCCGATGAGG-3′ 1542; pInt-mut7 1528 5′-ATGACTCCGATGAAGGATTACGGTC-3′ 1551; and pInt-mut8 1564 5′-CTGCAACAAATCCGCCCGCAGAG-3′ 1586.
Signal mutagenesis: pInt-mut9 1538 5′-TGAGGATTACGGTCTCCTCTGAGAGGCTGCAA-3′ 1569; pInt-mut10 1547 5′-CGGTCTGGAGAGTCTCCCTGCAACAAATGCGC-3′ 1578; and pInt-mut11 1538 5′-TGAGGATTACGGTCTCCTCTCTCTCCCTGCAACAAATGCGC-3′ 1578.
Analysis of the spacing region: pInt-mut12 1516, 5′-GGGCTGATGATTATGAGGAGAGAGAGGCTGCAA-3′ 1569; pInt-mut13–20 1537 5′-ATGAGGAT TACGGTC(AAA)nTGGAGAGAGAGGCTG-3′ 1566; and pInt-mut21 1537 5′-ATGAGGATTACGGTCGACTGGAGAGAGAGGTGGAGAGAGAGGCTGCAA-3′ 1569.
The integrity of all constructs was verified by restriction and sequence analysis.
Constructs for studying the expression of Rap1 in vivo were prepared as follows. For the construction of PLRV-Rap1, an XbaI fragment (coordinates 971–3,020) was isolated from pBINcDNACanadian (44) and cloned into the XbaI site of the pBluescript SK(+) vector (Stratagene). The amber stop codon was introduced by site-directed mutagenesis altering a single G to an A residue at position 1,553 of the PLRV sequence by using the oligonucleotide 1540 5′-AGGATTACGGTCTAGAGAGAGAGGCTGCAACAA-3′ 1572.
After verification of the entire XbaI fragment by sequence analysis, the fragment was cloned back into pBINcDNACanadian.
For plant transformation experiments, a HindIII fragment of PLRV cDNA (PLRV coordinates 1,480–2,394) was subcloned into the appropriate restriction site of the mutagenesis vector pSelect (Promega). To create a continuous reading frame between Rap1 and ORF1, an additional cytosine residue was introduced at position 1,667 by site-directed mutagenesis with the oligonucleotide 5′-AACAAACAAGCCTTTCAAATGGGCAAGCGGC-3′. From the resulting construct, an SwaI–EcoRV fragment (PLRV coordinates 1,494–2,293) was isolated and subcloned into an EcoRI-digested, blunt-end pRT104GUS vector (45) to give construct pGUS/Rap1-P1C. Subsequently, a fragment containing the bicistronic construct under the control of the 35S promoter was isolated from pGUS/Rap1-P1C and subcloned into the HindIII site of pBIN19 (46). The final construct was designated pBGUS/Rap1-P1C.
For protoplast transfection a Rap1-P1C fusion protein was constructed. Rap1 was fused by a newly introduced EcoRV site to the C terminus of ORF1 by using the following oligonucleotides: Rap1FW 1513 5′-ATTGGTACCACTGGGCTGATGATTATGACT-3′ 1533 (KpnI site in bold); Rap1BW 1665 5′-AGAGATATCAAGGCTTGTTTGTTGGAGCAG-3′ 1645 (EcoRV site in bold); P1CFW 1668 5′-CGCGATATCAATGGGCAAGCGGCACCGTCC-3′ 1688 (EcoRV site in bold); and P1CBW 1850 5′-CGCGATATCCTTCTGCAGGGCTTTCTGAGA-3′ 1830 (EcoRV site in bold).
Rap1 and P1C were ligated after digestion with EcoRV. Rap1 was expressed in Rap1-P1C without stop codon in the same reading frame with P1C. To obtain mut22, mut23, and mut24 a DNA copy of Rap1-P1C was obtained by PCR with the oligonucleotides Rap1FW and: mut22BW 1728 5′-ACTGTCGACTAGCGGCGTCGGGGATGT-3′ 1711 (SalI site in bold); mut23BW 1704 5′-ACTGTCGACTGCCGTTTGTTTTGGCGGA-3′ 1686 (SalI site in bold); and mut24BW 1689 5′-ACTGTCGACCGGACGGTGCCGCTTGCC-3′ 1672 (SalI site in bold).
The amplified fragments were digested with KpnI/SalI and subsequently subcloned together with a SalI/XbaI GUS (without AUG) fragment from pO7-GUS (40) in a two-fragment ligation into a KpnI/XbaI-linearized 35S vector. Peter Ivanov (Moscow State University, Moscow) kindly provided the vectors 35SHP, 35SHP-GUS, and 35SHP-encephalomyocarditis (EMCV)/GUS. The hairpin used in this construct is an ApaI, XhoI, SalI, ClaI, SalI, XhoI, and ApaI followed by one single KpnI. For the construction of 35S HP-mut22/GUS, HP-mut23/GUS, and HP-mut24/GUS, the 35SHP was digested with KpnI/PstI and ligated with the mut22–24/GUS KpnI/PstI insert.
In Vitro Transcription/Translation, Transfection of Potato Protoplasts, and Western Blot Analysis. Single-stranded RNAs were obtained by in vitro transcription of HindIII-linearized pInt-mut plasmid DNAs by bacteriophage SP6 polymerase in the presence of the cap analogue m7GpppG as described (47). Synthetic RNAs were translated in rabbit reticulocyte lysate or wheat germ extract (Promega) in the presence of [35S]methionine under conditions recommended by the supplier. In vitro products were analyzed on 12.5% SDS/polyacrylamide gels and detected by fluorography (48).
Protoplasts were isolated from Solanum tuberosum (cv. Désirée), and Ca(NO3)2/polyethylene glycol-mediated DNA transfer was performed as described by Negrutiu et al. (49) by using 3.3 × 105 protoplasts and 10 μg of plasmid DNA per transfection. GUS activity in protein extracts from transfected protoplasts was determined by a fluorometric assay (50). Western blot analysis of crude extracts from potato protoplasts was carried out as described by Prüfer et al. (51) by using either a polyclonal antiserum against P1 (52) or a monoclonal antiserum against the coat protein (ref. 53 and L.K., unpublished results).
Results
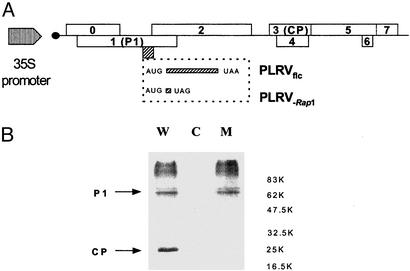
Analysis of Rap1 Function in Vivo. The biological role of Rap1 was studied by a reverse genetic approach with an infectious, full-length cDNA copy (PLRVflc) synthesized for a Canadian PLRV isolate (54). PLRVflc was mutated by site-directed mutagenesis resulting in the construct PLRV-Rap1 (see Materials and Methods and Fig. 2A). The introduced amber stop codon was located within the Rap1 reading frame without changing the amino acid composition of P1. Therefore, neither the function of P1 nor of P1/P2 (the transframe protein) (28) was predicted to be affected by this mutation.
Fig. 2.
Multiplication of wild-type and Rap1-deficient PLRV full-length constructs in potato protoplasts. (A) Schematic representation of the wild-type and mutant PLRV genome. A Rap1-deficient full-length clone was obtained by insertion of an amber stop codon immediately downstream of the Rap1 initiation codon without altering the ORFs for P1 and P1/P2. (B) Immunodetection of P1 and the coat protein (CP) in crude extracts of protoplasts 48 h after transfection (C = control) with PLRVflc (W) or PLRV–Rap1 (M). The positions of P1 and the coat protein are indicated.
Wild-type PLRVflc and PLRV-Rap1 DNAs were transfected into potato protoplasts in an effort to study the role of Rap1 during viral multiplication. Fig. 2B shows a Western blot of potato protoplasts transfected with either a wild-type (lane 1, W) or the Rap1-deficient full-length PLRV cDNA clone (lane 3, M). As expected, P1 expression was detected for both constructs because of the high, constitutive expression of (+)-sense viral RNA (the template for P1 translation) under the control of the 35S promoter with no expression in the mock-inoculated control (Fig. 2B, lane 2). Expression of the coat protein (P3) was predicted to occur only after transcription of viral (-) strand by the PLRV replicase complex followed by the recognition of the subgenomic RNA1 promoter on this RNA template (see Fig. 1). As shown, PLRV coat protein could be detected after transfection by the wild-type (Fig. 2B, lane 1) but not by the Rap1-deficient clone (Fig. 2B, lane 3). Furthermore, genomic and subgenomic viral RNAs were detected by Northern analysis only in PLRV wild-type-transfected potato protoplasts (data not shown). These transfection experiments with Rap1-deficient viral RNA indicated that Rap1 is involved in PLRV RNA multiplication.
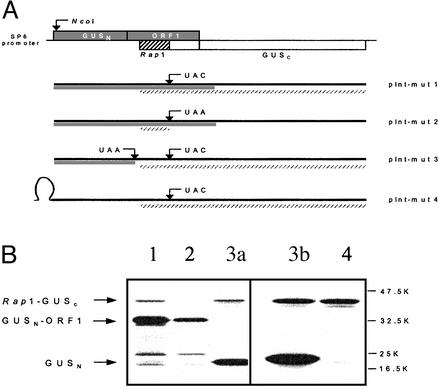
Localization of the IRES Signal. As deduced from the position of the Rap1 ORF, Rap1 translation initiation was expected to occur via a noncanonical translation mechanism in that the internal entry of ribosomes was supposedly guided by specific sequence structures. To identify this putative IRES element, the appropriate region of the PLRV genome was inserted into an internal position within the Escherichia coli GUS gene (Fig. 3A). To facilitate detection by PAGE, the small Rap1 coding region was enlarged by generating a continuous ORF between Rap1, 34 downstream amino acids, and a C-terminal section of the GUS gene. For this, the Rap1 stop codon was mutated to UAC by site-directed mutagenesis (see Materials and Methods). In this construct (pInt-mut1, Fig. 3A) the coding sequence resulting from the fusion of the N-proximal half of the GUS gene (GUSN) to the residual ORF1 fragment was expected to yield a chimeric GUSN-ORF1 protein of ≈33 kDa. In contrast, internal initiation at a Rap1 AUG would allow translation to proceed to the GUS C terminus (GUSC) and produce a Rap1-GUSC chimeric protein with a size of ≈43 kDa. Additionally, in the pInt-mut2 construct, where the Rap1 stop codon is still present, only the GUSN-ORF1 should be detected. In vitro transcription/translation experiments (Fig. 3B) with pInt-mut1 showed that in addition to the synthesis of the GUSN-ORF1 product, the ≈43-kDa Rap1-GUSC fusion protein was produced by internal initiation at an appropriate Rap1 AUG (Fig. 3B, lane 1). In contrast, a Rap1-GUSC protein was not detected for pInt-mut2.
Fig. 3.
Localization of the IRES signal. (A) Constructs for the analysis of IRES activity. Part of the ORF1 coding region (including the Rap1 coding region) was cloned into the GUS gene such that ORF1 was in frame with the N terminus of the GUS gene (GUSN) and the Rap1 coding region in frame with the C terminus of the GUS gene (GUSC) as schematically shown. This construct was termed pInt-mut2. Point mutations were introduced into this construct by site-directed mutagenesis (as described in Materials and Methods) to generate mutants pInt-mut1 and pInt-mut3. Introduction of a stable stem–loop structure into the 5′-end pInt-mut3 yielded pInt-mut4. The solid bars indicate translation products expected for the 5′ translation initiation. The slashed bars indicate translation products expected for internal initiation. (B) PAGE analysis of in vitro pInt-mut1 to mut4 mRNAs. Plasmid DNAs of pInt-mut1 to mut4 were linearized by HindIII digestion and transcribed into mRNAs by SP6 RNA polymerase. Translation was achieved in the rabbit reticulocyte system (lanes 1–3a) and in the wheat germ system (lanes 3b and 4) in the presence of [35S]methionine. The products were separated on a 12.5% SDS-containing polyacrylamide gel and detected by autoradiography. Lane 1, construct pInt-mut1; lane 2, pInt-mut2; lanes 3a and 3b, pInt-mut3; lane 4, pInt-mut4.
The corresponding mRNAs transcribed from these constructs facilitated ribosomal recognition of the PLRV IRES region during translation of the GUSN-ORF1 reading frame. To avoid the possibility that translation of ORF1 would be required for translation initiation, we introduced either an ochre stop codon immediately downstream of the last GUSN amino acid (pInt-mut3; Fig. 3A) or a stable stem–loop structure at the 5′ end of the GUS gene (pInt-mut4; Fig. 3A). For both pInt-mut3 and pInt-mut4 the Rap1-GUSC fusion protein was still synthesized efficiently in rabbit reticulocyte (Fig. 3B, lane 3a) and wheat germ extracts (Fig. 3B, lanes 3b and 4), although GUSN synthesis was strongly reduced with the pInt-mut4 in comparison to pInt-mut3. Thus, translation of the ORF1 coding region is not necessary for the internal initiation process (Fig. 3B).
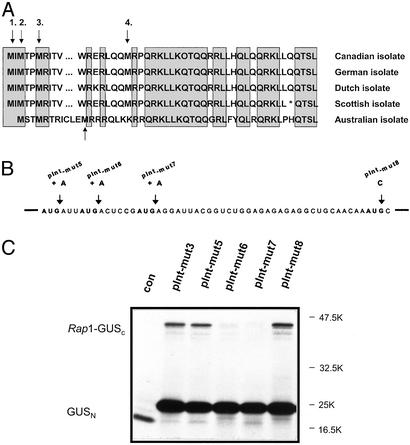
Identification of the Rap1 Start Codon. Sequence analysis of the Rap1 coding region identified four possible AUGs for translation initiation. The second and third AUG position is conserved among all PLRV isolates sequenced to date (Fig. 4A) (refs. 39, 41, and 55 and D.P., unpublished data). To identify the translation initiator AUG, insertion (out-of-frame mutations, pInt-mut5 to pInt-mut7) as well as substitution (in-frame mutation, pInt-mut8) mutagenesis was performed with pInt-mut3 as the starting material (Fig. 4B). In vitro transcription/translation experiments (Fig. 4C) demonstrated that translation initiated at the second AUG (AUG2) of Rap1, indicating the importance of this codon for all PLRV isolates. Furthermore, insertion mutations within AUG1 or the alteration of AUG4 to AUC did not have a significant effect on initiation efficiency. Mutations in AUG3 (pInt-mut7) did not result in translation of Rap1-GUSC, because with translation initiating at AUG2 the transcript was out of frame at the mutated AUG3 (Fig. 4).
Fig. 4.
Identification of the Rap1 translational start codon. (A) Rap1 sequence alignment for all known PLRV isolates. Black arrows indicate putative AUG initiation codons. The gray arrow indicates the nonconserved AUG codon in the Australian isolate. Shadowed boxes indicate conserved amino acid sequences. (B) Generation of Rap1 mutants for AUG initiation. Single adenosine residues were introduced into pInt-mut3, and the mutant constructs pInt-mut5 to pInt-mut7 were obtained. In mutant pInt-mut8, the fourth Rap1 AUG codon was changed to AUC. (C) PAGE analysis of products from in vitro translation of pInt-mut3 (3) and pInt-mut5 to pInt-mut8 (5–8) mRNAs. Analysis was performed as described in the Fig. 3 legend. C, mock translation.
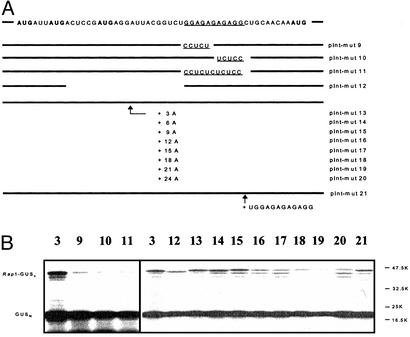
Analysis of the Inverted Repeat GGAGAGAGAGG. By using deletion analysis (data not shown), the inverted repeat motif GGAGAGAGAGG 22 nt downstream of AUG2 was identified as necessary for internal ribosome entry. To document the significance of this motif in the translation initiation process, several additional mutations were produced (pInt-mut9 to pInt-mut11; Fig. 5A) by using pInt-mut3 as the starting material. For this, the GGAGAGAGAGG sequence was either replaced partially by its complementary sequence CCTCTGAGAGG (pInt-mut9) and GGAGAGTCTCC (pInt-mut10) or replaced completely with CCTCTCTCTCC in pInt-mut11. As shown in Fig. 5B, all three of these mutations resulted in a dramatic reduction (>90%) of initiation efficiency.
Fig. 5.
Analysis of the GGAGAGAGAGG motif (underlined). (A) Mutants constructed with pInt-mut3 as the starting material. The N-terminal Rap1 sequence is shown at the top. Mutations were performed to study their effect on IRES activity with respect to the GGAGAGAGAGG sequence (pInt-mut9 to pInt-mut11 and pInt-mut21) and to its spacing from the Rap1 initiation codon (pInt-mut12 to pInt-mut20). (B) Translational products were analyzed as described in the Fig. 3 legend. Numbers indicate pInt-mut construct.
A further series of mutations (pInt-mut12 to pInt-mut20; Fig. 5A) was generated altering the spacing between AUG2 and the GGAGAGAGAGG motif. In mutation pInt-mut12, 21 nt of the entire spacer region were deleted, whereas in mutations pInt-mut13 to pInt-mut20, an increasing number of AAA triplets was inserted (see Fig. 5A). In vitro transcription/translation experiments (Fig. 5B) revealed a reduction (≈50%) in initiation efficiency for the deletion mutation pInt-mut12. A more dramatic reduction (>90%) was also observed for the AAA insertion mutations but only when more than nine A residues were introduced into the spacer region. In addition, duplication of the TGGAGAGAGAGG motif (pInt-mut21; Fig. 5) did not have a significant influence on the Rap1 translation initiation efficiency.
In summary, these results demonstrated that, in addition to the inverted repeat sequence GGAGAGAGAGG, the spacing between this motif and AUG2 plays an important role in the initiation event.
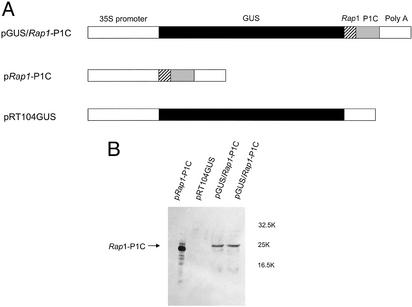
Functional Analysis of the PLRV IRES Region in Vivo. In addition to the in vitro experiments, the IRES function was studied in vivo with transgenic potato plants expressing the GUS gene alone (pRT104GUS), a bicistronic GUS/Rap1-P1C mRNA (pGUS/Rap1-P1C), or only the second gene Rap1-P1C (pRap1-P1C) of the bicistronic mRNA as shown in Fig. 6A. For a rapid identification of transgenic plants as well as a simple quantification of gene expression, the GUS gene was chosen as the first gene followed by a fragment representing the entire Rap1 coding region in frame to the C terminus of P1 (pGUS/Rap1-P1C; see Materials and Methods)as the second cistron. This strategy had the additional advantage of the P1 C terminus being easily detected by Western blot experiments with available monoclonal antibodies, because P1C is highly stable in PLRV-infected plants (52).
Fig. 6.
Stable expression of mono- and dicistronic mRNAs in transgenic potato plants. (A) Schematic representation of constructs used for plant transformation. In the dicistronic pGUS/Rap1-P1C construct, translation of Rap1-P1C is guided by the IRES sequence. Monocistronic constructs (pRap1-P1C and pRT104GUS) served as controls. (B) Western blot analysis of plants expressing the dicistronic and monocistronic mRNAs. The Rap1-P1C protein was detected with an monoclonal anti-prP1C-serum. The position of Rap1-P1C is indicated, and the apparent molecular weight of protein markers is given.
A representative Western blot analysis of plants expressing high levels of the GUS enzyme (data not shown) from the bicistronic mRNA is shown in Fig. 6B (lanes 3 and 4). The product of the 3′-located gene Rap1-P1C was readily detected in protein extracts from these plants (Fig. 6B, lanes 3 and 4). The observed translation product migrated a slightly smaller distance compared with a control plant transgenic only for the second gene (p Rap1-P1C; Fig. 6A, lane 1). This might be explained by the use of a different AUG for translation initiation in the mono- and dicistronic construct. A similar immunoreactive band could not be observed in transgenic plants expressing only the GUS gene (pRT104GUS; Fig. 6A, lane 2). The stability of the corresponding mRNAs for pRap1-P1C, pGUS/Rap1-P1C, and pRT104GUS was verified in a Northern Blot (data not shown).
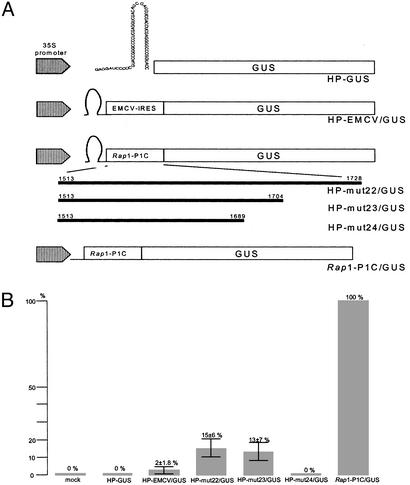
In addition the IRES function was studied by transfection of potato protoplasts with reporter gene constructs (Fig. 7A) containing a stable stem–loop structure to prevent leaky scanning and followed by the PLRV (Rap1-P1C), the EMCV, or no (negative control) IRES and a GUS gene. A construct lacking the stable stem–loop structure served as a positive control (Rap1-P1C/GUS). To identify the Rap1-IRES region, deletion analyses were performed at the 3′ end of Rap1-P1C with constructs HP-mut22/GUS, HP-mut23/GUS, and HP-mut24/GUS. Fig. 7B summarizes the results of seven independent transient-expression experiments in potato protoplasts: Internal ribosome entry in the expression of the GUS enzyme occurs at a frequency of ≈15 ± 6% for PLRV and 2 ± 1.8% for EMCV, as normalized with respect to the control constructs (mock; Rap1-P1C/GUS). In further experiments with a reporter construct in which the PLRV sequence upstream of nucleotide 1,704 had been deleted (HP-mut24/GUS), the expression of the GUS enzyme could not be detected. Thus, this evidence indicates that in vivo expression of Rap1 from IRES requires additional sequence information downstream of the coding region of Rap1.
Fig. 7.
Determination of internal ribosome entry efficiencies in potato protoplasts. (A) Schematic representation of constructs used for protoplast transfection. The sequence of the stable stem–loop is shown in the HP-GUS construct. In the constructs HP-EMCV/GUS, HP-mut22/GUS, HP-mut23/GUS, and HP-mut24/GUS, translation of the GUS gene is guided by an appropriate sequence. Rap1-1C/GUS (without hairpin) and a mock served as controls. (B) Efficiencies for IRES-mediated expression of the GUS enzyme. GUS activity of the Rap1-P1C/GUS construct was set at 100%. The percentages of internal initiation of the GUS gene were calculated for seven independent transfections each after subtraction of the background value measured for the mock control.
Discussion
In this study, we identify an IRES regulating the in vitro and in vivo synthesis of a small ORF (Rap1) of the PLRV genome and present an analysis of its putative biological function as a replication-associated protein. The biological relevance of Rap1 for PLRV multiplication was examined in vivo by using specific mutants of an infectious full-length PLRV cDNA clone. A Rap1-deficient full-length PLRV cDNA clone did not permit viral transcription in potato protoplasts, providing evidence for the role of Rap1 in viral multiplication. The PLRV IRES element for Rap1 translation is characterized by an unusual structure and location. In contrast to all other IRES elements described up to now, the IRES for PLRV Rap1 translation initiation is located internally within the PLRV RNA genome ≈1,500 nt downstream of its 5′ end. Furthermore, the core structural elements for internal ribosome entry include a conserved AUG codon and a downstream GGAGAGAGAGG motif with inverted symmetry. A distinct subgenomic RNA for Rap1 translation has not been detected in PLRV-infected plants (33, 40). Rap1 translation was examined by both in vitro and in vivo analyses. These in vitro transcription/translation experiments together with transient expression of PLRV-IRES-GUS reporter constructs in potato protoplasts provided evidence that Rap1 is translated by internal ribosome entry and that the second Rap1 AUG, the inverted repeat sequence GGAGAGAGAGG and further downstream PLRV sequences are required for optimal translation efficiency in vivo.
In contrast to most of the IRES elements described to date (13), the PLRV IRES sequence is located near the center of the viral genome and encompasses just sequence information down-stream of the start codon of the Rap1 reading frame. According to the picornaviral paradigm of internal initiation (13), a requirement for sequences from the coding region was unexpected. With the exception of cardioviruses (e.g., EMCV), where the ribosomal entry site is at the authentic initiation codon, internal initiation does not require downstream coding sequences. However, certain motifs, especially G-rich sequences, can inhibit internal initiation if located next to the AUG initiation codon (56). In polioviruses, extensive mutational analysis of the sequences downstream of a silent AUG triplet decreased in vitro internal initiation efficiency (57). To our knowledge, only the hepatitis C virus and classical swine fever virus IRESs extend into the coding sequence of the viral polyprotein (13), and only in the case of hepatitis C virus is the coding region important for efficient internal translation initiation (17).
In addition to its location, a further unexpected characteristic of the PLRV IRES sequence is its relatively short and simple structure compared with other picornaviral and viral IRES signals. Some similarities are found in the short IRES sequences of Moloney murine leukemia virus (20) and crucifer-infecting tobamovirus (crTMV) (23). Interestingly, the IRES sequences of PLRV and crTMV share a significant homology in that both sequences contain a purine-rich tract, although its function is still unclear (23). However, in contrast to the PLRV IRES identified here, the crTMV IRES tract is directly repeated and located upstream of the AUG initiation codon for the crTMV capsid protein.
For the picornaviral IRES, the AUG entry site codon is an important determinant of internal initiation, as is its distance from upstream elements such as the conserved oligopyrimidine tract (58, 59). In contrast, sequences downstream of the AUG are relatively unimportant. Detailed mutagenesis of the PLRV IRES identified a downstream purine tract GGAGAGAGAGG and its distance from AUG2 to be important for efficient internal initiation. The function of this motif for efficient internal initiation is still unclear. However, one can speculate that it is directly interacting with ribosomal RNAs or proteins and forcing ribosomes to translate this internal reading frame. In contrast, the downstream box of some prokaryotic mRNAs lacking an upstream Shine–Dalgarno sequence was postulated for many years to base-pair to the 16S RNA for enhanced translation. Recently, O'Connor et al. (60) could demonstrate that radical alterations in the 16S counterpart of the downstream box do not affect expression of downstream box-containing mRNAs, indicating that mRNA–rRNA base pairing is not essential for enhanced translation. Therefore, further studies are needed to clearly define the mechanism by which the GGAGAGAGAGG motif functions in internal translation initiation.
The PLRV IRES is located within a region of the PLRV RNA genome that is characterized by noncanonical translation mechanisms such as –1 ribosomal frameshifting (28, 31), which require a stable RNA structure for efficient frameshifting. Such structures are thought to slow down ribosomes and provide the time necessary for the frameshift event. Currently, we cannot exclude the possibility that these structures, which are located some 100 nt down-stream of the inverted repeat sequence GGAGAGAGAGG, are involved in efficient internal ribosome entry in vivo. Alternatively, the synthesis of Rap1 may be required for efficient –1 ribosomal frameshifting during the production of the P1/P2 transframe protein (the putative replicase) in the in vivo situation (28).
The specific function of the Rap1 protein in viral multiplication is still unclear. According to its size and localization within the replicase genes, Rap1 protein may be an integral part of the PLRV replicase complex and modulate the specificity of the replication machinery. For example, the specificity of E. coli RNA polymerase activity is controlled by a variety of ancillary factors, assisting or interfering with enzyme activity. Proteins such as the sigma factor are important in changing the function of the core enzyme, and substitution of a sigma factor may cause the transition from expression of one set of genes to expression of another set as seen during the lytic cycle of bacteriophage infection (for a review see ref. 61). In addition to its function in the multiplication of genomic RNA, the PLRV replicase also has to serve for the transcription of at least two subgenomic RNAs (39, 40, 62). The change from replicating genomic RNA to the activation of the subgenomic promoters might be regulated by factors encoded in the PLRV region necessary for RNA replication such as Rap1. Initial binding experiments with bacterially expressed Rap1 protein showed a strong interaction between the Rap1 and the protein encoded by ORF1 (63). Further studies with full-length infectious PLRV transcripts (42, 44, 54) and Rap1 mutants will help to better define the function of Rap1 in the viral life cycle.
Acknowledgments
We thank Dr. R. R. Martin for the Canadian PLRV full-length infectious clone and F. Kulcsar and K. Toohey for technical assistance. Part of this work was supported by Deutsche Forschungsgemeinschaft Grant Ro 330/9-1 (to W.R.).
Abbreviations: IRES, internal ribosomal entry site; PLRV, potato leafroll polerovirus; Rap1, replication-associated protein 1; GUS, β-glucuronidase; EMCV, encephalomyocarditis.
References
- 1.Kozak, M. (1986) Adv. Virus Res. 31, 229–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozak, M. (1989) J. Mol. Biol. 108, 229–241. [Google Scholar]
- 3.Lütcke, H. A., Chow, K. C., Mickel, F. S., Moss, K. A., Kern, H. F. & Scheele, G. A. (1987) EMBO J. 1, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fütterer, J., Kiss-Laszlo, Z. & Hohn, T. (1993) Cell 73, 789–802. [DOI] [PubMed] [Google Scholar]
- 5.Fütterer, J. & Hohn, T. (1996) Plant Mol. Biol. 32, 159–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jang, S. K., Krausslich, H. G., Nicklin, M. J. H., Duke, G. M., Palmenberg, A. C. & Wimmer, E. (1988) J. Virol. 62, 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelletier, J. & Sonenberg, N. (1988) Nature 334, 320–325. [DOI] [PubMed] [Google Scholar]
- 8.Jackson, R. J., Howell, M. T. & Kaminski, A. (1990) Trends Biochem. Sci. 15, 477–483. [DOI] [PubMed] [Google Scholar]
- 9.Agol, V. I. (1991) Adv. Virus Res. 40, 103–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wimmer, E., Hellen, C. U. T. & Cao, X. (1993) Annu. Rev. Genet. 27, 353–436. [DOI] [PubMed] [Google Scholar]
- 11.Sonenberg, N. & Pelletier, J. (1989) BioEssays 11, 128–132. [DOI] [PubMed] [Google Scholar]
- 12.Belsham, G. J. & Sonenberg, N. (1996) Microbiol. Rev. 60, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Salas, E., Ramos, R., Lafuente, E. & Lopez deq Uinto, S. (2001) J. Gen. Virol. 82, 973–984. [DOI] [PubMed] [Google Scholar]
- 14.Hellen, C. U. T. & Sarnow, P. (2001) Genes Dev. 15, 1593–1612. [DOI] [PubMed] [Google Scholar]
- 15.Tsukiyama-Kohara, K., Iizuka, N., Kohara, M. & Nomoto, A. (1992) J. Virol. 66, 1476–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, C., Sarnow, P. & Siddiqui, A. (1993) J. Virol. 67, 3338–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reynolds, J. E., Kaminski, A., Kettinen, H. J., Grace, K., Clarke, B. E., Carroll, A. R., Rowlands, D. J. & Jackson, R. J. (1995) EMBO J. 14, 6010–6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rijnbrand, R., Bredenbeek, P., van der Straaten, T., Whetter, L., Inchauspe, G., Lemon, S. & Spaan, W. (1995) FEBS Lett. 365, 115–119. [DOI] [PubMed] [Google Scholar]
- 19.Berlioz, C. & Darlix, J. L. (1995) J. Virol. 69, 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vagner, S., Waysbort, A., Marenda, M., Gensac, M.-C., Amalric, F. & Prats, A.-C. (1995) J. Biol. Chem. 270, 20376–20383. [DOI] [PubMed] [Google Scholar]
- 21.Levis, C. & Astier-Manifacier, S. (1993) Virus Genes 7, 367–379. [DOI] [PubMed] [Google Scholar]
- 22.Basso, J., Dallaire, P., Charest, P. J., Daventier, Y. & Laliberte, J. F. (1994) J. Gen. Virol. 75, 3157–3165. [DOI] [PubMed] [Google Scholar]
- 23.Ivanov, P. A., Karpova, O. V., Skulachev, M. V., Tomashevskaya, O. L., Rodinova, N. P., Dorokhov, Yu, L. & Atabekov, J. G. (1997) Virology 232, 32–43. [DOI] [PubMed] [Google Scholar]
- 24.Pestova, T. V., Shatsky, I. N., Fletcher, S. P., Jackson, R. T. & Hellen, C. U. T. (1998) Genes Dev. 12, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson, J. E., Powell, M. J., Hoover, S. E. & Sarnow, P. (2000) Mol. Cell. Biol. 20, 4990–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Commandeur, U., Rohde, W., Fischer, R. & Prüfer, D. (2002) in Plant Viruses as Molecular Pathogens, eds. Khan, J. A. & Dijkstra, J. (Haworth, New York), pp. 175–202.
- 27.Rohde, W., Gramstat, A., Schmitz, J., Tacke, E. & Prüfer, D. (1994) J. Gen. Virol. 75, 2141–2149. [DOI] [PubMed] [Google Scholar]
- 28.Prüfer, D., Tacke, E., Schmitz, J., Kull, B., Kaufmann, A. & Rohde, W. (1992) EMBO J. 11, 1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brault, V. & Miller, W. A. (1992) Proc. Natl. Acad. Sci. USA 89, 2262–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia, A., van Duin, J. & Pleij, C. W. A. (1993) Nucleic Acids Res. 21, 401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kujawa, A. B., Drugeon, G., Hulanicka, D. & Haenni, A.-L. (1993) Nucleic Acids Res. 21, 2165–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bahner, I., Lamb, J., Mayo, M. A. & Hay, R. T. (1990) J. Gen. Virol. 71, 2251–2256. [DOI] [PubMed] [Google Scholar]
- 33.Tacke, E., Prüfer, D., Salamini, F. & Rohde, W. (1990) J. Gen. Virol. 71, 2265–2272. [DOI] [PubMed] [Google Scholar]
- 34.Brown, C. M., Dinesh-Kumar, S. P. & Miller, W. A. (1996) J. Virol. 9, 5884–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, S. & Miller, W. A. (1995) J. Biol. Chem. 270, 13446–13452. [DOI] [PubMed] [Google Scholar]
- 36.Wang, S., Browning, K. & Miller, W. A. (1997) EMBO J. 13, 4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Francki, R. I. B., Milne, R. G. & Hatta, T. (1985) Atlas of Plant Viruses (CRC, Boca Raton, FL).
- 38.Mayo, M. A., Barker, H., Robinson, D. J., Tamada, T. & Harrison, B. D. (1982) J. Gen. Virol. 59, 163–167. [Google Scholar]
- 39.Mayo, M. A., Robinson, D. J., Jolly, C. A. & Hyman, L. (1989) J. Gen. Virol. 70, 1037–1051. [DOI] [PubMed] [Google Scholar]
- 40.Ashoub, A., Rohde, W. & Prüfer, D. (1998) Nucleic Acids Res. 26, 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keese, P., Martin, R. R., Kawchuk, L. M., Waterhouse, P. & Gerlach, W. L. (1990) J. Gen. Virol. 71, 719–724. [DOI] [PubMed] [Google Scholar]
- 42.Prüfer, D., Schmitz, J., Tacke, E., Kull, B. & Rohde, W. (1997) Mol. Gen. Genet. 253, 609–614. [DOI] [PubMed] [Google Scholar]
- 43.Pelletier, J. & Sonenberg, N. (1985) Cell 40, 515–526. [DOI] [PubMed] [Google Scholar]
- 44.Töpfer, R., Pröls, M., Schell, J. & Steinbiss, H. H. (1988) Plant Cell Rep. 7, 225–228. [DOI] [PubMed] [Google Scholar]
- 45.Bevan, M. (1984) Nucleic Acids Res. 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franco-Lara, L. F., McGeachy, K. D., Commandeur, U., Martin, R. R., Mayo, M. A. & Barker, H. (1999) J. Gen. Virol. 80, 2813–2823. [DOI] [PubMed] [Google Scholar]
- 47.Melton, D. A., Krieg, P. A., Rebagliati, M. R., Maniatis, T., Zinn, K. & Green, M. R. (1984) Nucleic Acids Res. 12, 7035–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonner, W. M. & Laskey, R. A. (1974) Eur. J. Biochem. 46, 83–88. [DOI] [PubMed] [Google Scholar]
- 49.Negrutiu, I., Hinnisdaels, S., Cammaerts, D., Cherdshewasart, W., Gharti-Chhetri, G. & Jacobs, M. (1992) Int. J. Dev. Biol. 36, 73–84. [PubMed] [Google Scholar]
- 50.Jefferson, R. A., Kavanagh, T. A. & Bevan, M. W. (1987) EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prüfer, D., Wipf-Scheibel, C., Richards, K., Guilley, H., Lecoq, H. & Jonard, G. (1995) Virology 214, 150–158. [DOI] [PubMed] [Google Scholar]
- 52.Prüfer, D., Kawchuk, L., Monecke, M., Nowok, S., Fischer, R. & Rohde, W. (1999) Nucleic Acids Res. 27, 421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin, R. R. & Stace-Smith, R. (1984) Can. J. Plant Pathol. 6, 206–210. [Google Scholar]
- 54.Kawchuk, L., Jaag, H. M., Toohey, K., Rohde, W., Fischer, R. & Prüfer, D. (2002) Can. J. Plant Pathol. 24, 239–243. [Google Scholar]
- 55.van der Wilk, F., Huismann, M. J., Cornelissen, B. J. C., Huttinga, H. & Goldbach, R. (1989) FEBS Lett. 245, 51–56. [DOI] [PubMed] [Google Scholar]
- 56.Hunt, S. L., Kaminski, A. & Jackson, R. J. (1993) Virology 197, 801–807. [DOI] [PubMed] [Google Scholar]
- 57.Meerovitch, K., Nicholson, R. & Sonenberg, N. (1991) J. Virol. 65, 5895–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pilipenko, E. V., Gmyl, A. P., Maslova, S. V., Svitkin, Y. V., Sinyakov, A. N. & Agol, V. I. (1992) Cell 68, 119–131. [DOI] [PubMed] [Google Scholar]
- 59.Kaminski, A., Belsham, G. J. & Jackson, R. J. (1994) EMBO J. 13, 1673–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O'Connor, M., Asai, T., Squires, C. L. & Dahlberg, A. E. (1999) Proc. Natl. Acad. Sci. USA 96, 8973–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Helmann, J. D. & Chamberlin, M. J. (1988) Annu. Rev. Biochem. 57, 839–872. [DOI] [PubMed] [Google Scholar]
- 62.Tacke, E., Schmitz, J., Prüfer, D. & Rohde, W. (1993) Virology 197, 274–282. [DOI] [PubMed] [Google Scholar]
- 63.Kierdorf, M. (1999) Diploma thesis (University of Cologne, Cologne, Germany).