Abstract
Infected cell protein 0 (ICP0) of herpes simplex virus 1 expresses two E3 ubiquitin (Ub) ligase activities mapping in the domains encoded by exons 2 and 3, respectively. Site 1 (exon 3) is responsible for the degradation of the E2 Ub-conjugating enzyme cdc34 whereas site 2 (exon 2) is associated with a ring finger and has been shown to mediate the degradation of promyelocytic leukemia (PML) and Sp100 proteins and the dispersal of nuclear domain 10 (ND10). In in vitro assays site 2 polyubiquitylates the E2 enzymes UbcH5a and UbcH6 but not other (e.g., UbcH7) enzymes. In this article, we show that ectopic expression of dominant negative UbcH5a carrying the substitution C85A delayed or blocked the degradation of PML and Sp100 and dispersal of ND10 whereas ectopic expression of wild-type UbcH5a or dominant negative UbcH6 and UbcH7 carrying the substitutions C131A and C86A, respectively, had no effect. These results link the degradation of PML and Sp100 and the dispersal of ND10 to the E3 activities of ICP0 associated with the UbcH5a E2 enzyme.
Keywords: ubiquitin ligase, infected cell protein 0, ring finger
Infected cell protein 0 (ICP0) of herpes simplex virus 1 (HSV-1) is a multifunctional protein whose synthesis is directed by a spliced mRNA (1). The 775 codons encoding the protein are distributed in three exons. ICP0 is associated with two sites of functions. The first centers on the observation that ICP0 by itself acts as a transactivator of genes introduced into cells by infection or transfection (2). In most cell lines mutants lacking the gene encoding ICP0 require high multiplicity of infection for efficient viral replication (3). The molecular basis for the enhancing activity of ICP0 are not known. ICP0 is also associated with stabilization of cyclin D3 (4, 5) and the disappearance of nuclear structures known as Kr bodies, promyelocytic oncogenic domains, nuclear domain 10 (ND10), etc. (6, 7), and the degradation of sumoylated forms of promyelocytic leukemia (PML) protein (8), Sp100 (8), centromeric protein A and C (9, 10), and the DNA-dependent protein kinase (11). The molecular basis of the latter events have been thoroughly investigated in recent years and appear to be associated with newly discovered E3 ubiquitin (Ub) ligase activities of the protein (12–15). In in vitro assays, ICP0 exhibited two Ub ligase sites. The first, designated HSV Ub ligase 1 (HUL-1), maps in exon 3 between amino acids 543 and 680 (12). The second, HUL-2, maps to the ring finger domain encoded in exon 2. This article concerns the Ub ligase activities of ICP0 (13, 15).
HUL-1 targets for destruction the Ub-conjugating enzyme UbcH3, also known as cdc34. The evidence in support of this conclusion is as follows: (i) In wild-type (wt) virus-infected cells, cdc34 is degraded by ICP0 in a proteasome-dependent manner (14). (ii) Cdc34 binds to ICP0 at two sites. Thus the substitution D199A in exon 2 decreases the affinity of ICP0 for cdc34. Deletion of amino acids 621–625 abrogates the binding of cdc34 to the polypeptide encoded by exon 3 (12, 14). (iii) The domain of exon 3 encompassing amino acids 543–680 is sufficient to mediate ATP-dependent polyubiquitylation of cdc34 in vitro. Deletion of amino acids 621–625 abolishes this activity. (iv) Cdc34 is not degraded in cells infected with viral mutants in which ICP0 amino acids 621–625 had been deleted. Finally, the targeting of cdc34 for destruction can be rationalized from the observation that HSV stabilizes both cyclin D3 and D1 although its primary interaction is with cyclin D3 (4). In cells overexpressing a dominant negative (dn) cyclin D3 ICP0 is retained in the nucleus and is not transported into the cytoplasm late in infection (4). Cdc34 has been reported to be the Ub-conjugating enzyme involved in the turnover of cyclin D1 and conceivably also cyclin D3 (16, 17).
The disappearance of ND10 and degradation of PML, Sp100, and other proteins listed above appear to be mediated by HUL-2. In in vitro assays, HUL-2 has been shown to polyubiquitylate UbcH5a and UbcH6 but not a number of other Ub-conjugating enzymes exemplified by UbcH7 (13, 15). A direct connection between the E3 ligase activity of HUL-2 and degradation of PML protein has not been established. The conserved cysteine residue in the active center of E2-conjugating enzymes is critical for binding Ub and transferring the thioester bond (18). A substitution of this cysteine to alanine is sufficient to abolish the binding of Ub in vitro (19). We expressed UbcH5a, UbcH6, and UbcH7 carrying C85A, C131A, and C86A, respectively, in human neuroblastoma SK-N-SH cells and show that in infected cells the degradation of PML and Sp100 proteins are directly linked to the UbcH5a Ub-conjugating enzyme.
Materials and Methods
Cells and Virus. SK-N-SH, HEp-2, HeLa, and Vero cells (American Type Culture Collection) or Sf9 cells (PharMingen) were maintained as described (20, 21). HSV-1 strain F [HSV-1(F)] is the prototype HSV-1 strain used in this laboratory. Baculoviruses were produced and titered in Sf9 cells. Except during the 1-h exposure of mammalian cells to either HSV or baculoviruses, the infected cells were maintained in DMEM supplemented with 10% FBS.
Construction of Recombinant Baculoviruses. The construction of the MTS1 baculovirus transfer vector, which contains the human cytomegalovirus immediate early promoter/enhancer sequences, inserted into the XhoI/EcoRI sites of pAcSG2 (PharMingen) has been described (22). A myc tag was inserted between EcoRI and StuI sites of MTS1 by annealing primers (5′-GCAGAATTCATGGCATCAATGCAGAAGCTGATCTCAGAG-3′ and 5′-GCAAGGCCTCAGGTCCTCCTCTGAGATCAGCTTCTGCAT-3′). The annealed primers were end-filled with Klenow, digested with EcoRI and StuI, and then ligated with EcoRI + StuI double-digested MTS1. This generated a new vector plasmid, MTS1-Myc. Plasmids pET-3a-UbcH5 and PC86-UbcH5(C-A) (18) were generous gifts from Peter Howley (Harvard Medical School, Boston). Primers 5′-GCAAGGCCTATGGCGCTGAAGAGGATTCAG-3′ and 5′-GCACTGCAG TTACATTGCATATTTCTGAG-3′ were used to generate by PCR DNA fragments containing wt and dnUbcH5a, using pET-3a-UbcH5 and PC86-UbcH5(C-A) as PCR templates. Plasmids pET-22b-UbcH6 and pET-3a-UbcH7 were kindly provided by Martin Scheffner (University of Cologne, Cologne, Germany) (23). Primers 5′-GCAAGGCCTATGTCGGATGACGATTCGAGG-3′ and 5′-GCACTGCAGTTATGTAGCGTATCTCTTGG-3′ were used to generate wtUbcH6 DNA fragment from pET-22b-UbcH6 by PCR. Primers 5′-GCAAGGCCTATGGCGGCCAGCAGGAGGC TG-3′ and 5′-GCACTGCAGTTAGTCCACAGGTCGCTTTTC-3′ were used to generate wtUbcH7 DNA fragment from pET-3a-UbcH7 by PCR. The PCR products of wtUbcH5a, dnUbcH5a, wtUbcH6, and wtUbcH7 fragments were double-digested with StuI + PstI and gel-purified before being ligated with StuI + PstI double-digested MTS1-Myc. The new plasmids were designated MTS1-Myc-wtUbcH5a, MTS1-Myc-dnUbcH5a, MTS1-Myc-wtUbcH6, and MTS1-Myc-wtUbcH7. The dn UbcH6 and UbcH7 were generated with a QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's protocols, Primers 5′-CAGTCAAGGTGTTATTGCCTTGGACATATTGAAAG-3′ and 5′-CTTTCAATATGTCCAAGGCAATAACACCTTGACTG-3′ were used for UbcH6, and primers 5′-CGAAAAGGGGCAGGTCGCTCTGCCAGTAATTAGTG-3′ and 5′-CACTAATTACTGGCAGAGCGACCTGCCCCTTTTCG-3′ were used for UbcH7 to switch cysteine to alanine in the active centers. The mutants generated this way were named MTS1-Myc-dnUbcH6 and MTS1-Myc-dnUbcH7.
Baculoviruses encoding wt or dn forms of UbcH5a, UbcH6, or UbcH7 were generated by cotransfecting Sf9 cells with linearized baculovirus DNA (BaculoGold, PharMingen) and the MTS1 constructs described above. In mammalian cells infected with the recombinant baculoviruses, the expression of myc-tagged wt or dn UbcH5a, UbcH6, or UbcH7 was directed by the cytomegalovirus promoter.
Infection of Cells. SK-N-SH cells grown to 80–90% confluency in 25-cm2 flasks were exposed to 10 plaque-forming units (PFU) of recombinant baculoviruses per cell. After incubation at 37°C for time intervals indicated in Results in medium supplemented with 10% FBS, 5 mM sodium butyrate, and where indicated in Results, 1,000 units of IFN-γ (Research Diagnostics, Flanders, NJ) per ml, the cells were exposed to HSV-1(F) (5 PFU per cell).
Immunoblots. SK-N-SH cells grown in 25-cm2 flasks were infected as described above. At the times indicated in Results, the cells were collected by scraping, rinsed twice with ice-cold PBS, resuspended in 200 μl of PBS containing 0.5 mM PMSF, and sonicated briefly. The total protein concentrations were measured by Bradford reagent (Bio-Rad). Total protein lysates (40 μg per lane) were then mixed with 1/3 volume of 4× SDS-loading buffer, boiled for 5 min, electrophoretically separated in denaturing polyacrylamide gels, transferred to preequilibrated poly(vinylidene fluoride) membrane (Millipore), blocked in TBST (20 mM Tris, pH 7.5/150 mM NaCl/0.5% Tween 20) containing 5% nonfat dry milk at room temperature for 1 h, and reacted at 4°C overnight with either rabbit polyclonal antibody to PML (1:400, Santa Cruz Biotechnology), Sp100 (1:2,000, Chemicon), ICP0 (1:1,000; ref. 5), or mAb to c-Myc (1:1,000, Santa Cruz Biotechnology) and actin (1:400, Sigma). After extensive rinsing, the membranes were reacted with appropriate goat secondary antibody. The antibody–antigen complexes were visualized by enhanced chemiluminescence (Amersham Pharmacia Biosciences) according to the manufacturer's instructions.
Confocal Microscopy. SK-N-SH cells were seeded onto 4-well glass slides (Cell-Line, Newfield, NJ) at 4 × 104 cells per well. One day after the seeding, the cells were exposed to recombinant baculovirus (40 PFU per cell) and replenished with 150 μl of medium supplemented with 5 mM sodium butyrate and IFN-γ as indicated in Results. After 24 h of incubation at 37°C, the cells were exposed to HSV-1(F) (10 PFU per cell). After 4 h of incubation at 37°C, the cells were fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, blocked in PBS containing 20% human serum for 1 h, and reacted at 4°C overnight in PBS containing 10% human serum and polyclonal rabbit sera to PML (1:400), Sp100 (1:2,000), or ICP0 (1:1,000), or mouse mAb to c-Myc (1:1,000). After extensive rinsing, the cells were reacted with FITC-conjugated goat anti-rabbit (Sigma) and Texas red-conjugated goat anti-mouse (Molecular Probes) as recommended by the manufacturers. Digital images were acquired with a Zeiss confocal microscope.
Results
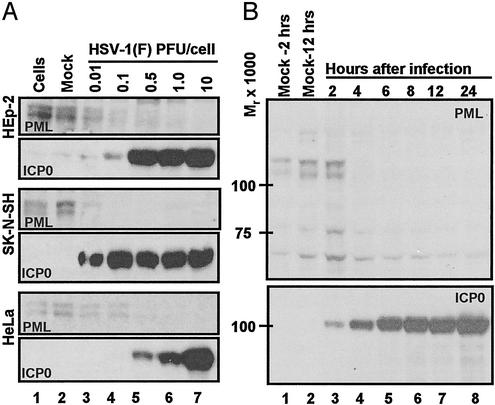
Degradation of PML Is Rapid and Efficient in SK-N-SH Cells Infected with HSV-1(F). The objective of this series of experiments was to identify a cell line in which PML was efficiently degraded after infection with wt virus. As shown in Fig. 1A, replicate cultures of HEp-2, SK-N-SH, or HeLa cells were untreated, mock-infected, or exposed to various ratios of PFU per cell. The cells were harvested at 24 h after infection and processed as described in Materials and Methods. The results (Fig. 1 A) were as follows:
Fig. 1.
Expression of ICP0 and accumulation of PML in infected cells. (A) Accumulation of PML and ICP0 in three cells lines exposed to ratios of PFU per cell shown, incubated for 24 h after virus exposure, and processed as described in Materials and Methods. The 6% denaturing polyacrylamide gels were loaded with 40 μg of total protein per lane. The HSV-1(F) stock was titered in Vero cells. Mock, mock-infected cells; cells, cells maintained in growth medium and neither mock-infected nor infected. The electrophoretically separated proteins were reacted with antibodies as described in Materials and Methods. (B) PML degradation in SK-N-SH cells. Zero time is 1 h after initial exposure of cells to virus. Cells were harvested at 2 and 12 h after mock infection and at times shown after HSV-1(F) infection (0.5 PFU per cell). The harvested cells were processed as described in Materials and Methods and in the legend to A.
The accumulation of ICP0 as a function of multiplicity of infection is an indicator of the susceptibility of the cells to viral infection. Of the cell lines tested, SK-N-SH cells accumulated significant amounts of ICP0 after exposure to 0.01 PFU per cell. This amount was similar to that recovered in HeLa cells exposed to 100-fold more virus (10 PFU per cell). Significantly higher amounts of virus were required to yield the highest accumulations of ICP0 in HEp-2 cells (0.5 PFU per cell) or HeLa (10 PFU per cell).
The decrease in the accumulation of PML in infected relative to uninfected cells was concordant with the accumulation of ICP0. PML was virtually undetected in HEp-2 cells exposed to 1.0 PFU per cell, SK-N-SH exposed to 0.1 PFU per cell, or HeLa cells exposed to 1 PFU per cell.
PML Is Rapidly Degraded in SK-N-SH Cells Infected with wt Virus. Replicate cultures of SK-N-SH cells were mock-infected or exposed to 0.5 PFU of virus per cell. The cells were harvested at 2, 4, 6, 8, 12, or 24 h after infection and processed as described above. As shown in Fig. 1B, PML was present in cells harvested at 2 h after mock infection but was undetectable in cells harvested at 4 h or later times after infection with wt virus. These results indicate that in SK-N-SH cells the degradation of PML occurs rapidly after relatively low multiplicity of infection. In other experiments PML was shown to be degraded even faster in SK-N-SH cells infected at higher multiplicities of infection (data not shown). All subsequent experiments were done in SK-N-SH cells.
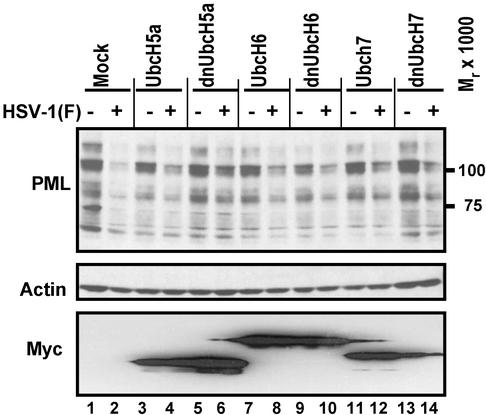
dnUbcH5a but Not UbcH6 or UbcH7 Ub-Conjugating Enzymes Blocks the Degradation of PML. As reported elsewhere, the degradation of PML is mediated by ICP0 and particularly by the ring finger domain encoded by exon 2 of the gene (24, 25). In in vitro assays a polypeptide encoded by exon 2 exhibited Ub ligase activities in that it mediated the polyubiquitylation of UbcH5a and UbcH6 but not that of UbcH7 or other Ub-conjugating enzymes (13, 15). To determine whether the interaction of ICP0 with UbcH5a or UbcH6 is responsible for the degradation of PML, we constructed dn E2-conjugating enzymes by substituting cysteines 85, 131, and 86 with alanines in UbcH5a, UbcH6, and UbcH7, respectively. Both wt and dn E2 Ub-conjugating enzymes were fused to a myc tag and cloned in baculoviruses under cytomegalovirus immediate early promoter. The recombinant baculoviruses were then used as vectors to express the E2 enzymes.
In the first series of experiments, SK-N-SH cells exposed to baculoviruses encoding wt or dn Ub-conjugating enzymes were incubated at 37°C for 24 h, then exposed to 5 PFU of HSV-1(F) per cell for 1 h. After 30 min of additional incubation, the cells were harvested and analyzed for the accumulation of PML and myc. Preliminary experiments have shown that in SK-N-SH cells exposed to 5 PFU of HSV-1(F) per cell PML disappears within a 30-min interval after the 1-h attachment/penetration interval. The accumulation of actin probed with antiactin antibody served as a loading control. As shown in Fig. 2, PML protein levels were reduced in HSV-1(F)-infected cells exposed to baculoviruses encoding wt or dn UbcH6, wt or dn UbcH7, or wtUbcH5a (Fig. 2, lanes 2–4 and 7–14) but not in cells exposed to baculoviruses encoding dnUbcH5a (Fig. 2, lanes 5 and 6).
Fig. 2.
Accumulation of PML in SK-N-SH cells mock-infected or infected with HSV-1(F) 24 h after they were mock-transduced with insect cell medium or transduced with baculoviruses expressing wt or dn UbcH5a, UbcH6, or UbcH7. The multiplicities of infection were 10 PFU of baculoviruses and 5 PFU of HSV-1(F) per cell. To enhance the expression of genes encoded in baculovirus vectors, the cultures were incubated in medium containing 5 mM sodium butyrate after transduction. The cells were harvested 30 min after the 1-h exposure to HSV-1(F), electrophoretically separated in a denaturing 4–20% gradient polyacrylamide gel, and reacted with antibodies as described in Materials and Methods. Actin served as a gel loading control.
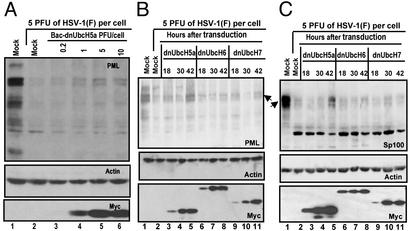
Next, SK-N-SH cells were exposed to 0.2, 1, 5, or 10 PFU of baculoviruses expressing dnUbcH5a per cell. After 24 h of incubation, the cells were exposed to 5 PFU of HSV-1(F) per cell as described above. As shown in Fig. 3A, the accumulation of PML was dnUbcH5a dose dependent. Thus the amount of PML detected in HSV-1(F)-infected cells was higher in cells exposed to 5 or 10 PFU of baculovirus per cells than in cells exposed to 0.2 or 1 PFU (Fig. 3A, lanes 5 and 6 vs. 2–4).
Fig. 3.
Accumulation of PML and Sp100 protein in cells transduced with dnUbcH5a. (A) Accumulation of PML in SK-N-SH cells transduced with various ratios of recombinant baculoviruses encoding dnUbcH5a per cell. The experiment was carried out as described in the legend to Fig. 2 except that the multiplicity of infection with baculovirus encoding dnUbcH5a was varied as shown. (B) Effect of the length of incubation of transduced cells on the accumulation of PML in SK-N-SH cells infected with HSV-1(F). The cells were transduced with 10 PFU of baculovirus encoding dnUbcH5a per cell. At the times shown the cells were infected with HSV-1(F). Lanes 1 and 2, the cells were mock-transduced by exposure to insect cell medium. (C) Effect of the time of gene expression of the transducing recombinant baculoviruses on the accumulation of Sp100 in SK-N-SH cells infected with HSV-1(F). The experiment was done as described in the legend to B except that the cells were treated with 1,000 units of IFN-γ per ml concurrent with 5 mM sodium butyrate to enable the visualization of Sp100 protein and reacted with antibodies as described in Materials and Methods.
The accumulation of proteins encoded by baculoviruses increases with time after transduction. SK-N-SH cells transduced with 10 PFU of recombinant baculovirus per cell were incubated for 18, 30, or 42 h and then exposed to 5 PFU of HSV-1(F) per cell. The cells were then harvested and processed as above. The results (Fig. 3B) show that the highest accumulation of PML was detected in cells transduced with dnUbcH5a 42 h before HSV-1(F) infection.
We conclude that the degradation of PML is blocked by dnUbcH5a but not by dn or wt UbcH6, dn or wt UbcH7, or wtUbcH5a. Moreover, the quantity of PML recovered in transduced cells appears to be dnUbcH5a dose dependent.
The Degradation of Sp100 Is Also Blocked by dnUbcH5a. Sp100 is also a component of ND10 degraded after HSV-1 infection (8, 26). Unlike PML, the Sp100 protein was barely detectable in uninduced SK-N-SH cells. To determine whether dnUbcH5a also blocks the degradation of Sp100, SK-N-SH cells were transduced with 10 PFU of recombinant baculovirus per cell and concurrently treated with 1,000 units of IFN-γ per ml of medium. At 18, 30, or 42 h after transduction the cells were exposed to 5 PFU of HSV-1(F) per cell. The cells were harvested and processed as above. The results shown in Fig. 3C were as follows. As expected, the mock-infected cells treated with IFN-γ (Fig. 3C, lane 1) accumulated readily detectable amounts of Sp100 protein. The latter protein accumulated in significantly lower amounts in nontransduced cells (Fig. 3C, lane 2) or cells transduced with dn UbcH6 or UbcH7 (Fig. 3C, lanes 6–11). Significant amounts of Sp100, however, were recovered in cells transduced with recombinant baculovirus expressing dnUbcH5a 42 h before HSV-1(F) infection (Fig. 3C, lane 5).
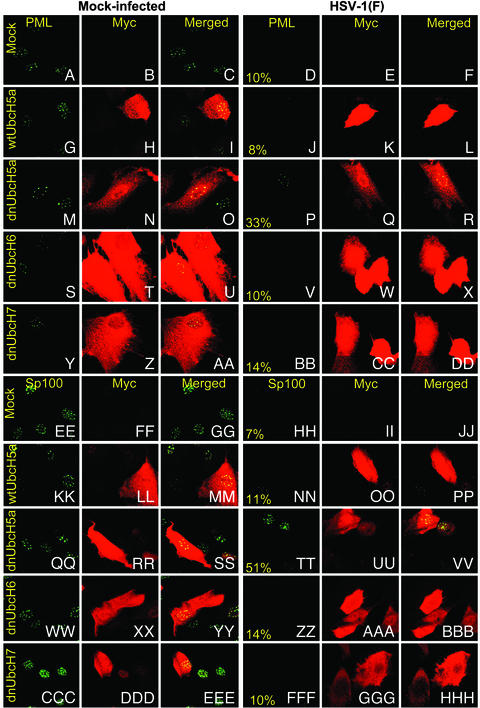
dnUbcH5a Increases the Retention of PML and Sp100 in ND10 Structures. In this series of experiments SK-N-SH cells grown on 4-well slides were either mock-transduced or transduced with baculoviruses (10 PFU per cell) expressing wtUbcH5a, dnUbcH5a, dnUbcH6, or dnUbchH7 before exposure to 5 PFU of HSV-1(F) per cell. The cells were fixed 4 h after infection with HSV-1(F) and reacted first with rabbit polyclonal anti-PML and mouse monoclonal anti-myc antibodies followed by exposure of the cells to FITC-conjugated goat antibody against rabbit IgG and Texas red-conjugated goat antibody against mouse IgG. Representative images collected with a Zeiss confocal microscope are shown in Fig. 4 A–DD. As expected, the ND10 structures characterized by the presence of PML rapidly disappeared from nuclei of HSV-1(F)-infected cells. At 4 h after infection only 8–14% of the cells transduced with either wtUbcH5a, dnUbcH6, or dnUbcH7 contained ND10 structures containing PML. This number increased to 33% of the cells in cultures transduced with dnUbcH5a and infected with HSV-1(F). These results are consistent with the conclusion of experiments described above showing that dnUbcH5a blocked or at least delayed the degradation of PML in HSV-1-infected cells.
Fig. 4.
Accumulation of PML (A–DD) or Sp100 proteins (EE–HHH) in SK-N-SH cells mock-transduced with insect cell medium or baculoviruses encoding wtUbcH5a or dnUbcH5a, dnUbcH6, or dnUbcH7. The cells were transduced with baculoviruses and incubated in medium containing sodium butyrate (A–DD) or both sodium butyrate and 1,000 units of IFN-γ per ml (EE–HHH). The cells were fixed 4 h after infection with HSV-1(F) and reacted with anti-PML or anti-Sp100 protein antibody (FITC) or anti-myc tag antibody (Texas red) as described in Materials and Methods. The left three columns are representative of mock-infected cells. The right three columns are representative of the HSV-1(F)-infected cells. The numbers in the fourth column indicate the percent of cells that contained PML or Sp100 proteins in discernible ND10 structures and that were either mock-transduced (A–F and EE–JJ) or transduced with baculoviruses encoding wt or dn E2 enzymes (G–DD or KK–HHH). The percentages are based on counts of ≈200 cells with the aid of high magnification (×63 objective) in a Zeiss confocal microscope.
The experiment described above was repeated with SK-N-SH cells transduced with the various wt and dn E2 enzymes and concurrently exposed to 1,000 units of IFN-γ per ml of medium. In this instance the cells were reacted with antibodies against Sp100 and myc. Representative images are shown in Fig. 4 EE–HHH. As in the case of PML protein, Sp100 protein was detected in ND10 structures of only 7–14% of cells transduced with wtUbcH5a, dnUbcH6, or dnUbcH7. The number of cells exhibiting ND10 structures containing Sp100 increased to 51% in cells transduced with dnUbcH5a.
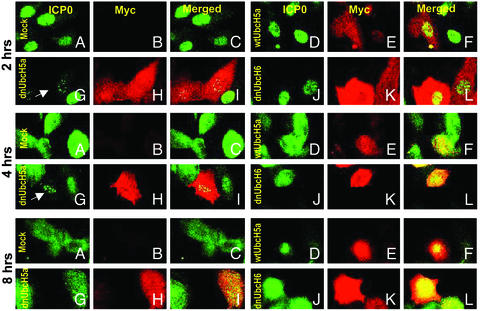
dnUbcH5a Causes a Retention of ICP0 in Nuclear Speckles Characteristic of ND10 Structures. To investigate the affects of ectopic overexpression of E2s on ICP0, SK-N-SH cells were mock-treated or transduced with wt or dn UbcH5a or dnUbcH6. At 2, 4, or 8 h after infection, the cells were reacted with anti-ICP0 and anti-myc antibodies as described in Materials and Methods. Representative images are shown in Fig. 5. The results were as follows: (i) In mock-transduced cells 2 h after infection, ICP0 was either still in the immediate vicinity of ND10 structures or began to fill the nucleus (Fig. 5A, 2 h). The dispersal of ICP0 throughout the nucleus was nearly complete by 4 h after HSV-1 infection (Fig. 5A, 4 h). At 8 h after ICP0 was also detected in the cytoplasm of infected cells (Fig. 5A, 8 h). This process was similar to that described elsewhere (27) and was also observed in cells transduced with wtUbcH5a or dnUbcH6. (ii) In cells transduced with dnUbcH5a, the ICP0 was retained in nuclear speckle-like structures at 2 h after infection (Fig. 5 G–I, 2 h, arrow) or at 4 h after infection (Fig. 5 G–I, 4 h, arrow). The 4 h-infected cells were counted to calculate the percentage of speckle-associated ICP0. In cells transduced with dnUbcH5a, 83% of the cells contained ICP0 restrained into nuclear speckles whereas no speckle-like ICP0 was observed in wtUbcH5a-transduced cells (Fig. 5 D–F, 4 h) and only 16% of the cells contained speckle-retained ICP0 in dnUbcH6-transduced cells. The speckled structures containing ICP0 colocalized with PML nuclear bodies (data not shown). At 8 h after infection ICP0 was dispersed throughout the nucleus of all HSV-1(F)-infected cells and the speckles containing ICP0 were no longer visible (Fig. 5 G–I, 8 h).
Fig. 5.
Distribution of ICP0 in SK-N-SH cells exposed to insect cell medium or baculoviruses encoding wtUbcH5a, dnUbcH5a, or dnUbcH6 and infected with HSV-1(F). The procedure for the treatment of the slide cultures was the same as described in Fig. 4 except that the cells were fixed at 2, 4, or 8 h after HSV-1(F) infection and reacted with anti-ICP0 (FITC) and anti-myc (Texas red) antibodies. The arrows point to cells containing ICP0 in speckled nuclear structures. In other assays these speckled structures were juxtaposed to or overlapped ND10 structures (data not shown).
Discussion
In in vitro assays ICP0 has been shown to contain two ligase sites located in the domains encoded by exon 3 (HUL-1 E3) and exon 2 (HUL-2 E3). Extensive studies have identified cdc34 as the target of the HUL-1 site, and several lines of evidence have shown that cdc34 interacts with ICP0 and directly linked a 4-aa sequence essential for the HUL-1 E3 activity to the proteasome-dependent destruction of cdc34. The HUL-2 site interacts with UbcH5a and UbcH6 E2 enzymes (13, 15), mediates the degradation of PML and Sp100 proteins, and causes the dispersal of the components of ND10 structures (8, 26). A direct link between ICP0 HUL-2 activity deduced from the polyubiquitylation in vitro of UbcH5a and UbcH6 with the degradation of PML, Sp100, or other proteins has not been demonstrated. To establish whether such a link exists, we tested three human cell lines with respect to their susceptibility to infection as defined in terms of the amount of virus required to initiate viral gene expression. These studies led to the identification of SK-N-SH as the most susceptible of the cell lines tested. In this cell line PML protein was rapidly lost from wt virus-infected cells in a dose-dependent manner. Transduction of these cells with wt or dn UbcH5a, UbcH6, or UbcH7 showed that the dnUbcH5a reduced or delayed the degradation of PML in a dose-dependent manner. Other experiments established a similar reduction or delay in the degradation of Sp100 protein. In immunofluorescence studies ectopic expression of dnUbcH5a increased the number of infected cells with discernible ND10 structures containing PML and Sp100 protein. Our results link therefore the destruction of PML and Sp100 and the dispersal of ND10 with the functional interaction of the domain encoded by exon 2 of ICP0 with UbcH5a.
The experiments described in this article leave several questions unanswered. For example, the destruction by HUL-1 of cdc34, an E2 enzyme involved in normal degradation of cyclin D, has been linked to the need of the infected cell to block the turnover of cyclin D3. In the process, both cyclin D3 and D1 are stabilized even though there is no evidence pointing to the need for cyclin D1 (4). The HUL-2 site has been reported to mediate the degradation of several proteins, including PML and Sp100. Because both of these proteins have been linked to the interaction of HUL-2 site with UbcH5a enzyme, the question arises as to whether the virus targets for the destruction of both proteins or whether the true target is only one of these proteins. Available data indicate that destruction of PML is required to block the effect of exogenous IFN (28). These data also help explain why ectopic expression of PML in the absence of IFN failed to have an effect on viral replication (21) and may also explain why expression of dnUbcH5a had no effect on viral replication (unpublished results). No such data are available for Sp100.
A second unresolved question relates to the significance of the interaction of HUL-2 site with the UbcH6 enzyme. Does the interaction of ICP0 with UbcH6 lead to degradation of specific proteins?
Lastly, a characteristic of ICP0 is its peregrinations from a site near PML in ND10 to fill the entire nucleus and from there to the cytoplasm. dnUbcH5a delayed the translocation of ICP0 from the ND10 structures. In other studies we have shown that ectopic overexpression of PML results in stabilization of ND10 structures (21). In these cells the peregrination of ICP0 could not be differentiated from that taking place of control-infected cells. These results suggest that the delay observed in the current studies may be caused by the direct interaction of ICP0 with dnUbcH5a rather than the presence of PML in ND10 structures.
Acknowledgments
We thank Dr. Peter Howley and Dr. Martin Scheffner for the E2 plasmids and Ryan Hagglund for useful discussions. These studies were aided by grants from the National Cancer Institute (CA87661, CA83939, CA71933, CA78766, and CA88860) and the U.S. Public Health Service.
Abbreviations: ICP0, infected cell protein 0; HSV-1, herpes simplex virus 1; Ub, ubiquitin; HUL, HSV Ub ligase; HSV-1(F), HSV-1 strain F; ND10, nuclear domain 10; PML, promyelocytic leukemia; dn, dominant negative; wt, wild type; PFU, plaque-forming units.
References
- 1.Carter, K. L. & Roizman, B. (1996) Proc. Natl. Acad. Sci. USA 93, 12535-12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roizman, B. & Knipe, D. M. (2001) in Fields Virology, eds. Knipe, D. M., Howley, P., Griffin, D. E., Lamb, R. A. Martin, M. A., Roizman, B. & Straus, S. E. (Lippincott-Williams and Wilkins, New York), 4th Ed., pp. 2399-2459.
- 3.Sacks, W. R. & Schaffer, P. A. (1987) J. Virol. 61, 829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Sant, C., Lopez, P., Advani, S. J. & Roizman, B. (2001) J. Virol. 75, 1888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawaguchi, Y., Van Sant, C. & Roizman, B. (1997) J. Virol. 71, 7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maul, G. G., Negorev, D., Bell, P. & Ishov, A. M. (2000) J. Struct. Biol. 129, 278-287. [DOI] [PubMed] [Google Scholar]
- 7.Negorev, D. & Maul, G. G. (2001) Oncogene 20, 7234-7242. [DOI] [PubMed] [Google Scholar]
- 8.Chelbi-Alix, M. K. & de The, H. (1999) Oncogene 18, 935-941. [DOI] [PubMed] [Google Scholar]
- 9.Everett, R. D., Earnshaw, W. C., Findlay, J. & Lomonte, P. (1999) EMBO J. 18, 1526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lomonte, P., Sullivan, K. F. & Everett, R. D. (2001) J. Biol. Chem. 276, 5829-5835. [DOI] [PubMed] [Google Scholar]
- 11.Parkinson, J., Lees-Miller, S. P. & Everett, R. D. (1999) J. Virol. 73, 650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagglund, R. & Roizman, B. (2002) Proc. Natl. Acad. Sci. USA 99, 7889-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagglund, R., Van Sant, C., Lopez, P. & Roizman, B. (2002) Proc. Natl. Acad. Sci. USA 99, 631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Sant, C., Hagglund, R., Lopez, P. & Roizman, B. (2001) Proc. Natl. Acad. Sci. USA 98, 8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boutell, C., Sadis, S. & Everett, R. D. (2002) J. Virol. 76, 841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weissman, A. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 169-178. [DOI] [PubMed] [Google Scholar]
- 17.King, R. W., Deshaies, R. J., Peters, J. M. & Kirschner, M. W. (1996) Science 274, 1652-1659. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, S., Kao, W. H. & Howley, P. M. (1997) J. Biol. Chem. 272, 13548-13554. [DOI] [PubMed] [Google Scholar]
- 19.Ciechanover, A. (1994) Cell 79, 13-21. [DOI] [PubMed] [Google Scholar]
- 20.Zhou, G., Galvan, V., Campadelli-Fiume, G. & Roizman, B. (2000) J. Virol. 74, 11782-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez, P., Jacob, R. J. & Roizman, B. (2002) J. Virol. 76, 9355-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sciortino, M. T., Taddeo, B., Poon, A. P., Mastino, A. & Roizman, B. (2002) Proc. Natl. Acad. Sci. USA 99, 8318-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nuber, U., Schwarz, S., Kaiser, P., Schneider, R. & Scheffner, M. (1996) J. Biol. Chem. 271, 2795-2800. [DOI] [PubMed] [Google Scholar]
- 24.Everett, R. D. & Maul, G. G. (1994) EMBO J. 13, 5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maul, G. G. & Everett, R. D. (1994) J. Gen. Virol. 75, 1223-1233. [DOI] [PubMed] [Google Scholar]
- 26.Muller, S. & Dejean, A. (1999) J. Virol. 73, 5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez, P., Van Sant, C. & Roizman, B. (2001) J. Virol. 75, 3832-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chee, A. V., Lopez, P., Pandolfi, P. P. & Roizman, B. (2003) J. Virol. 77, 7101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]