Abstract
Schizophrenia is a severe psychiatric disorder characterized by a complex mode of inheritance. Forebrain-specific CNB knockout mice display a spectrum of behavioral abnormalities related to altered behaviors observed in schizophrenia patients. To examine whether calcineurin dysfunction is involved in schizophrenia etiology, we undertook studies of an initial subset of calcineurin-related genes, prioritizing ones that map to loci previously implicated in schizophrenia by linkage studies. Transmission disequilibrium studies in a large sample of affected families detected association of the PPP3CC gene, which encodes the calcineurin γ catalytic subunit, with disease. Our results identify PPP3CC, located at 8p21.3, as a potential schizophrenia susceptibility gene and support the proposal that alterations in calcineurin signaling contribute to schizophrenia pathogenesis.
Schizophrenia is a severe psychiatric condition that affects ≈1% of the population worldwide (1). Studies of the inheritance of schizophrenia have revealed that it is a multifactorial disease characterized by multiple genetic susceptibility elements, each contributing a modest increase in risk (2). Family linkage studies and studies of chromosomal abnormalities associated with schizophrenia have identified a number of schizophrenia susceptibility loci (2, 3). Such loci provide a basis for higher resolution genetic studies and a criterion for assessment of potential candidate genes. In addition to direct genetic analysis, a longstanding body of pharmacological studies has led to the prevailing hypotheses that dysfunction of dopaminergic or N-methyl-D-aspartate (NMDA) receptor-mediated signaling are major contributing factors in schizophrenia pathogenesis (4–6).
Calcineurin is a calcium-dependent serine/threonine protein phosphatase that is highly expressed in the CNS (7, 8). Calcineurin consists of a heterodimer composed of a regulatory subunit, CNB, and a catalytic subunit, CNA (7, 8). There are three different CNA isoforms encoded by distinct genes. Calcineurin activity plays a key role in the downstream regulation of dopaminergic signal transduction (9) and in the induction of certain forms of N-methyl-D-aspartate receptor-dependent synaptic plasticity (10, 11). Thus, calcineurin function could comprise a critical link between dopaminergic and glutamatergic signaling.
In an accompanying study, we report that forebrain-specific CNB knockout mice display a spectrum of behavioral abnormalities that is strikingly reminiscent of altered behaviors observed in schizophrenia patients (12). Based on these findings, we decided to further investigate the potential involvement of calcineurin dysfunction in schizophrenia etiology by directly testing for genetic association of calcineurin-related candidate genes with schizophrenia. We prioritized examination of genes encoding calcineurin subunits or calcineurin-related molecules that map to schizophrenia susceptibility loci. We present direct genetic evidence for association of the PPP3CC gene with schizophrenia. PPP3CC encodes the calcineurin γ catalytic subunit (CNAγ), and is located at chromosome 8p21.3, within a confirmed schizophrenia susceptibility locus (13–17). Our results identify the PPP3CC gene as a potential schizophrenia susceptibility gene, and support the idea that alterations in calcineurin signaling contribute to schizophrenia pathogenesis.
Methods
Patient Samples. The patient samples used in this study have been described (18, 19).
PCR/Sequencing. Sequence determination was accomplished by PCR amplification of relevant fragments from genomic DNA followed by fragment purification and sequencing. For procedural details, see supporting information, which is published on the PNAS web site, www.pnas.org.
Sequence Analysis. Sequence analysis was performed by using dnastar software. Patient sequences were compared with the human genome draft sequence, available at the University of California, Santa Cruz, web site, http://genome.ucsc.edu. Contigs including the patient sequences and the human draft sequence were constructed for each fragment, and polymorphisms were identified by comparison.
Genotyping. Polymorphisms used for genotyping were identified by direct sequencing or found in single-nucleotide polymorphism (SNP) databases including the NCBI SNP database (www.ncbi.nlm.nih.gov/SNP) and the Celera database (www.celera.com). Insertion/deletion polymorphisms were typed by PCR of genomic DNA from individual subjects and identified by altered fragment size as assessed by agarose gel electrophoresis. SNPs were typed either by PCR-restriction fragment length polymorphism genotyping or by fluorescence polarization template-directed dye-terminator incorporation genotyping as described (18, 19). See supporting information for genotyping primer sequences.
Association Analysis. For single and multiple marker haplotype transmission, the program transmit v2.5.2 (20) was used. This program uses the score test based on the Conditional on Parental Genotype (CPG) likelihood for the estimate of unknown haplotype phase and missing data. We obtained both global P values, which estimate the significance of transmission distribution for all of the haplotypes tested, and P values for the significance of transmission distortion of a specific haplotype.
Brain Expression. Adult total human brain, fetal total human brain, and human testis cDNA consisted of marathon-ready cDNAs purchased from CLONTECH. Primer pair one consists of a forward primer from PPP3CC exon 1, 5′-GCGCTTCCACCTCTCCACC-3′ and a reverse primer from PPP3CC exon 2, 5′-CTATCATAGTCTTCTCTTGCCTC-3′. Primer pair 2 consists of a forward primer, 5′-CCCATTCATGACTTAGAGTCC-3′ and a reverse primer, 5′-CCCCTTTATAGCACAAGACTTC-3′ from PPP3CC exon 14 (3′ UTR). These primers were designed to differ from PPP3CA and PPP3CB sequence, particularly at the 3′ end, to be PPP3CC specific. The adult human brain region panel was purchased from Origene (Rockville, MD). Fragments were amplified in a 25-μl reaction mixture containing ≈0.25 ng cDNA (CLONTECH) or 1.0 ng (Origene), each primer at 400 nM concentration, each dNTP at 200 μM concentration and 1.5 units of Taq polymerase (Sigma) in OptiPrime (Stratagene) buffer 6 conditions. Reactions were performed by touchdown PCR amplification as follows: an initial denaturation step at 94°C for 2 min, followed by 20 amplification cycles: 30 sec at 94°C; 45 sec at 68°C initially, 45 sec at 72°C (minus 1°C at each cycle) followed by 15 amplification cycles: 30 sec at 94°C; 45 sec at 53°C, 45 sec at 72°C, followed by a final extension step at 72°C for 7 min. Products were subjected to 2% agarose gel electrophoresis, visualized by ethidium bromide staining and photographed by using an eagle eye apparatus (Stratagene).
Results
Collectively, the behavioral abnormalities observed in forebrain-specific CNB knockout mice (see accompanying article) suggest that calcineurin dysfunction could be involved in schizophrenia pathogenesis. To determine whether alterations in calcineurin-related candidate genes could contribute to schizophrenia susceptibility, we are undertaking a comprehensive approach including genomic sequencing analysis of these genes in DNA from schizophrenia patients to identify polymorphisms, and transmission disequilibrium studies to examine the association of these genes with disease in a large sample of affected families. We prioritized examination of calcineurin subunit genes and genes encoding proteins that interact with calcineurin that map to putative schizophrenia susceptibility loci identified by linkage studies. The coincidence of candidate genes from the calcineurin pathway and susceptibility loci is outlined in Table 1 and includes loci with variable statistical support and among them two loci that found strong support in a multicenter study (6p) (31) and in a recent metaanalytical survey (8p) (37). We report here results from our analysis of four such genes: PPP3R1, PPP3CA, PPP3CC, and FKBP-5 located at 2p14, 4q24, 8p21.3, and 6p21.31, respectively.
Table 1. Coincidence of genes encoding calcineurin subunits and calcineurin-interacting proteins with locations of previously identified putative schizophrenia susceptibility loci.
| Gene name | Protein description | Gene location | Susceptibility loci | Refs. |
|---|---|---|---|---|
| Calcineurin subunits | ||||
| PPP3R1 | Calcineurin B subunit | 2p14 | 2p13-14 | 17,21,22 |
| PPP3CA | Calcineurin A α subunit | 4q24 | 4q22-26 | 16,17,23,24 |
| PPP3CC | Calcineurin A γ subunit | 8p21.3 | 8p21-22 | 13-17 |
| PPP3CB | Calcineurin A β subunit | 10q22.3 | 10q22-3 | 25 |
| Calcineurin binding proteins | ||||
| CABIN (=CAIN) | Calcineurin binding protein 1 | 22q11.23 | 22q11 | 2,3 |
| CHP | Calcium binding protein P22 | 15q15.1 | 15q15 | 26 |
| CS-1 (=MYOZZ) | Calcineurin binding protein calsarcin 1 | 4q26 | 4q25-26 | 16,17,23,24 |
| CS-2 (=MYOZ1) | Calcineurin binding protein calsarcin 2 | 10q22.2 | 10q22-3 | 25 |
| CS-3 (=MYOZ3) | Calcineurin binding protein calsarcin 3 | 5q33.1 | 5q33.2 | 16 |
| AKAP5 (=AKAP79) | A kinase (PRKA) anchor protein 5 | 14q23.3 | 14q22-24 | 25,27,28 |
| FKBP5 (=FKBP51) | FK506 binding protein 5 | 6p21.31 | 6p21.3; 6p22-24 | 29-31 |
| Proteins functionally coupled to calcineurin | ||||
| ITPR1 | Inositol 1,4,5-triphosphate receptor, Type 1 (IP3 receptor 1) | 3p26.1 | 3p24-26 | 13 |
| RYR3 | Ryanodine receptor type 3 | 15q13.3-15q14 | 15q14 | 3,32 |
| ILF2 (=NF45) | Subunit of nuclear factor of activated T cells (NFAT) | 1q21.3 | 1q21.3 | 33 |
| CAMLG | Calcium modulating ligand | 5q31.1 | 5q23.3-31.1 | 17,34-36 |
| NFATC2 | Nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 2 | 20q13.2 | 20q13 | 32 |
Gene locations are according to the November 2002 human draft sequence
To identify potential functional polymorphisms in these four genes that could contribute to schizophrenia susceptibility, as well as polymorphisms that could be used for association studies, we determined the sequence of coding and noncoding exons, splice donor and acceptor sites, and some intronic and promoter regions of these genes in genomic DNA isolated from 12 independent schizophrenia patients (U.S. schizophrenia sample) (18, 19). The obtained sequences were compared with the human draft sequence to identify polymorphisms.
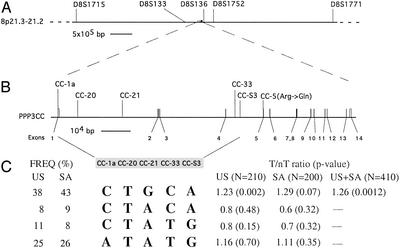
Nineteen polymorphisms were found in the PPP3R1 gene, 12 were found in the PPP3CA gene, 16 were found in the PPP3CC gene, and 4 were found in the FKBP-5 gene (see supporting information for sequences and locations of these polymorphisms). Among these identified polymorphisms, only one caused a coding sequence alteration. This polymorphism (CC-5, Fig. 1) is situated in exon 5 of the PPP3CC gene and results in a nonconservative change in the amino acid sequence of the encoded protein from a charged arginine residue at position 163 to a neutral glutamine residue. Further analysis found the CC-5 polymorphism in 3 of 210 tested patients and in 0 of 75 unaffected Caucasian controls from the Coriell Cell Repository. Assessment of the significance of this mutation will require examination of expanded samples.
Fig. 1.
PPP3CC gene locus. (A) The location of the PPP3CC gene in the 8p21.3 region is depicted in relation to relevant markers from linkage studies. D8S136 (13); D8S1771 (15, 16); D8S1752 (15); and D8S1715 and D8S133 (14). (B) An expanded view of the PPP3CC gene is presented, including the exon/intron structure and the locations of the SNPs used for our association studies and of the coding sequence mutation identified in exon 5. This mutation changes a G to an A at position 824 of the mRNA (GenBank accession no. NM_005605). Distances and positions in this figure are according to the November 2002 human draft sequence. (C) Haplotype distribution and transmission at the PPP3CC locus. Only four haplotypes with frequencies ≥5% were observed in both U.S. and SA samples and are shown here. The most common PPP3CC haplotype is consistently overtransmitted in both samples. T/nT, transmitted/nontransmitted.
To further investigate the involvement of these four genes in schizophrenia pathogenesis, we have undertaken linkage disequilibrium (LD) studies in large family samples (triads) that test for preferential transmission of common (>10% frequency) variants and multivariant haplotypes from parents to affected individuals. For this study we used a subset of the polymorphisms that we identified by direct sequencing, supplemented with additional SNPs obtained from the NCBI or Celera databases. See supporting information for further details of the examined polymorphisms.
We initially examined each allele of each marker, or a combination of two adjacent markers, for evidence of transmission disequilibrium by using the TDT test as implemented in the program TRANSMIT (20), in a sample of 210 triads collected from the United States (U.S. sample). The results of the association screen are indicated in Table 2 along with the pairwise disequilibrium coefficients (D′) (38) for all marker pairs within each gene calculated for the nontransmitted chromosomes (Table 3). Analysis of the transmission revealed nominally significant association between schizophrenia and a subset of PPP3CC SNPs and two-SNP haplotypes. No other significant associations were observed (Table 2).
Table 2. TDT results in the U.S. sample.
|
P values
|
||||
|---|---|---|---|---|
| Gene | Distance, kb | SNP | One SNP | Two SNP |
| PPP3RI | RIPI | 0.381 | ||
| 0.14 | 0.810 | |||
| RIS1 | 0.649 | |||
| 21 | 0.604 | |||
| RI24 | 0.206 | |||
| 25 | 0.603 | |||
| RI28 | 0.297 | |||
| 27 | 0.471 | |||
| RIS3 | 0.976 | |||
| PPP3CA | CAS6 | 0.556 | ||
| 159 | 0.710 | |||
| casFP1 | 0.470 | |||
| 76 | 0.296 | |||
| casFP2 | 0.221 | |||
| 0.14 | 0.216 | |||
| CA31 | 0.745 | |||
| FKBP5 | FK-S1 | 0.118 | ||
| 46 | 0.285 | |||
| FK-33 | 0.454 | |||
| 32 | 0.822 | |||
| FK-35 | 0.544 | |||
| 24 | 0.485 | |||
| FK-36 | 0.793 | |||
| PPP3CC | CC1a | 0.777 | ||
| 6.9 | 0.752 | |||
| CC20 | 0.380 | |||
| 15 | 0.169 | |||
| CC21 | 0.038 | |||
| 39 | 0.013 | |||
| CC33 | 0.099 | |||
| 1.7 | 0.003 | |||
| CCS3 | 0.041 | |||
Table 3. Pairwise disequilibrium values for the U.S. sample.
| Genes | Pairwise D' values | |||
|---|---|---|---|---|
| PPP3RI | RIPI | RIS1 | RI24 | RI28 |
| RIS1 | 0.86 | |||
| RI24 | 0.96 | 0.95 | ||
| RI28 | 0.98 | 0.91 | 1 | |
| RIS3 | 1 | 0.95 | 0.96 | 0.99 |
| PPP3CA | CAS6 | casFP1 | casFP2 | |
| casFP1 | 0.56 | |||
| casFP2 | 0.25 | 0.07 | ||
| CA31 | 0.25 | 0.40 | 0.97 | |
| FKBP5 | FK-S1 | FK-33 | FK-35 | |
| FK-33 | 0.90 | |||
| FK-35 | 0.93 | 0.89 | ||
| FK-36 | 0.91 | 0.81 | 1 | |
| PPP3CC | CC1a | CC20 | CC21 | CC33 |
| CC20 | 0.72 | |||
| CC21 | 0.98 | 0.78 | ||
| CC33 | 0.89 | 0.39 | 0.67 | |
| CCS3 | 0.97 | 0.51 | 0.90 | 0.91 |
Pairwise D' values are calculated on nontransmitted chromosomes
To better characterize the nature of the PPP3CC locus variant contributing to schizophrenia susceptibility, we analyzed the U.S. sample for preferential transmission of specific haplotypes by using genotypes from all five PPP3CC SNPs. The PPP3CC locus shows limited haplotypic diversity over the tested region (62.8 kb), which is likely adequately captured by the SNPs genotyped here. Only four haplotypes with frequencies ≥5% were observed, in agreement with the observed strong linkage disequilibrium among the tested SNPs. A nominally significant global transmission distortion (P = 0.0038) was observed that could be primarily accounted for by a significant overtransmission to affected probands of the most common PPP3CC haplotype. This risk haplotype is present in ≈38% of the parental chromosomes and was overtransmitted at a ratio of T/nT: 1.23/1 (P = 0.0022). The allelic composition of this haplotype reflects the single-locus results.
Overall, our initial analysis provided evidence for a nominally significant association between the PPP3CC gene and schizophrenia in a U.S. sample and identified a common risk haplotype. We sought additional support for our findings by examining the pattern of the five-SNP haplotype transmissions in another family sample derived from an independently collected population consisting of 200 triads from the Tshwane (formerly known as Pretoria) and Cape Town area in South Africa (SA sample). It should be noted that previous linkage analysis in this founder population did not provide significant evidence for linkage at the 8p21 locus (D.H., J.A.G., G. Abecasis, and M.K., unpublished data), suggesting that the effect of a disease gene from this region would be, at best, small in this population. The frequency distribution of the multilocus haplotypes was almost identical in the two samples (heterogeneity χ2 test, P > 0.05). Although no significant global transmission distortion was observed in the SA sample, a trend for overtransmission of the same common risk haplotype (P = 0.07) with an almost identical transmission distortion ratio (1.29/1) as well as an overall similar pattern of multiallelic transmissions, was clearly evident. As expected in the combined sample of 410 families, transmission of the CTGCA risk haplotype significantly deviated from random (P = 0.00126) with the haplotype overtransmitted 1.26 times to 1. Moreover, several two-, three-, and four-SNP haplotypes showed significant association with schizophrenia in the combined sample (Table 4). The most significant of these associations remains significant at the 0.01 level after adjusting for multiple testing, even with a Bonferroni correction (for the >40 nonindependent tests we performed).
Table 4. Haplotype transmission in the combined sample (n = 410 families).
|
P values
|
|||||
|---|---|---|---|---|---|
| Gene | SNP | Two SNP | Three SNP | Four SNP | Five SNP |
| PPP3CC | CC1a | ||||
| 0.587 | |||||
| CC20 | 0.169 | ||||
| 0.054 | 0.005 | ||||
| CC21 | 0.0008 | 0.00126 | |||
| 0.0013 | 0.0001 | ||||
| CC33 | 0.0003 | ||||
| 0.0004 | |||||
| CCS3 | |||||
P values represent global significance calculated from the global χ2 values from TRANSMIT program TDT analysis
One simple explanation for the observed pattern of association is that a putative causative variant(s) is present within the risk haplotype background. We have determined the sequence of all exons, all splice donor and acceptor sites, and 1,500 bp of promoter sequence from several patients that are homozygous for the risk haplotype and several that do not have the risk haplotype. This analysis did not identify any sequence alterations that segregate with the risk haplotype; therefore, it is likely that the putative causative variant(s) resides in an area of the PPP3CC locus that was not analyzed by sequencing. Because a potential causative variant linked to the risk haplotype is not present in the PPP3CC coding sequence, it is likely that the causative sequence variation affects PPP3CC transcript expression or processing. In this regard, studies to determine whether the risk haplotype correlates with altered PPP3CC transcript expression or processing are of considerable interest.
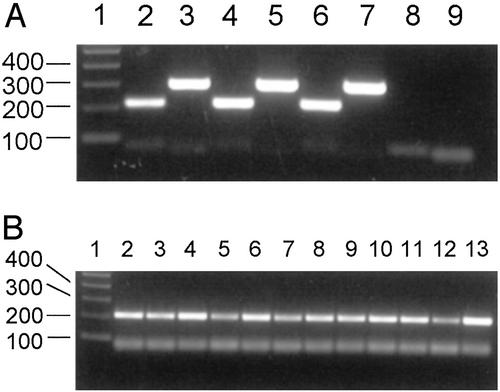
PPP3CC has been designated a testis-specific calcineurin catalytic subunit gene based primarily on its initial characterization in the mouse (39). To determine whether PPP3CC is expressed in the human brain, we first performed PCR amplification of cDNA from human total adult brain and from total fetal brain with PPP3CC-specific primers. As shown in Fig. 2A, PCRs with two different primer pairs indicate that PPP3CC is expressed in the human adult and fetal brain. To further analyze the expression of PPP3CC in the human brain, we performed PCR amplification of a panel of CNS region-specific cDNAs with one of the PPP3CC-specific primer pairs. As shown in Fig. 2B, PPP3CC expression is detected in multiple regions of adult human brain including frontal and temporal lobes, hippocampus, amygdala, thalamus, striatum, substantia nigra, hypothalamus, cerebellum, pons, and medulla. PPP3CC expression is also detected in spinal cord.
Fig. 2.
PPP3CC expression in human brain. (A) PCR amplification of cDNA from human adult total brain, fetal total brain, and testis. PCR was performed on ≈0.25 ng of cDNA with two primer pairs. Primer pair 1 amplifies a 218-bp fragment extending from exon 1 to exon 2. Primer pair 2 amplifies a 298-bp fragment from exon 14 consisting of 3′ UTR sequence. Lane 1, 100-bp marker; lane 2, adult brain, primer pair 1; lane 3, adult brain, primer pair 2; lane 4, fetal brain, primer pair 1; lane 5, fetal brain, primer pair 2; lane 6, testis, primer pair 1; lane 7, testis, primer pair 2; lane 8, no DNA control, primer pair 1; lane 9, no DNA control, primer pair 2. The products <100 bp in size are present in lanes 8 and 9 and are most likely primer-related amplification artifacts. (B) PCR amplification of cDNA from human adult brain regions. PCR was performed on ≈1.0 ng of cDNA with primer pair 1. Lane 1, 100-bp marker; lane 2, frontal lobe; lane 3, temporal lobe; lane 4, cerebellum; lane 5, hippocampus; lane 6, substantia nigra; lane 7, caudate nucleus; lane 8, amygdala; lane 9, thalamus; lane 10, hypothalamus; lane 11, pons; lane 12, medulla; lane 13, spinal cord.
Discussion
The spectrum of behavioral abnormalities observed in forebrain-specific CNB knockout mice (12) prompted us to employ transmission studies to directly test for genetic association of genes encoding calcineurin-related molecules with schizophrenia. In our initial analysis of four such genes, we have found evidence for a nominally significant over-transmission of a common PPP3CC gene haplotype (found in ≈40% of human chromosomes) in a sample of 410 affected families. Our findings identify PPP3CC as a potential schizophrenia susceptibility gene and support the proposal that alterations in calcineurin signaling contribute to schizophrenia pathogenesis. The increase in disease risk associated with the risk haplotype is expected to be low (≈30%). However, because of its high frequency, the risk haplotype may correspond to a high population attributable risk affecting a large percentage of schizophrenics.
The PPP3CC gene is located within chromosome 8p21.3, a region that has been identified as a schizophrenia susceptibility locus by linkage studies in several independent samples derived from different populations (Fig. 1) (13–16), as well as by a recent metaanalysis of whole-genome linkage scans (37). Because the purpose of our analysis was to test the contribution of candidate genes from the calcineurin pathway, we did not employ finer mapping techniques by using a denser collection of markers at the 8p21.3 locus. Therefore, we cannot formally exclude the possibility that the observed association signal originates from genes in the vicinity of the PPP3CC locus. Such genes include SCAM-1 (src homology 3-containing adaptor protein 1), PDLIM2, and EGR3/PILOT, whose expression is modulated by neuronal activity (40), neuregulin signaling (41), and calcineurin activity (42). Nevertheless, it will be of interest to determine whether alterations in the PPP3CC gene account for the linkage results obtained for this region in these samples. Recently, the neuregulin (NRG1) gene, located at 8p12, has been identified as a potential schizophrenia susceptibility gene from the 8p locus, in Icelandic (43) and Scottish (44) populations. Markers representing the risk haplotype at the 5′ end of the NRG1 gene, that was found to be over-represented in Icelandic and Scottish patients, were genotyped for the identical samples used in the present study (D.H., J.A.G., and M.K., unpublished data). This analysis revealed a pattern of association directly opposite to the one observed for the PPP3CC gene with evidence for association in the SA sample and no evidence for association in the more diverse U.S. sample. This is consistent with the notion that more than one gene from the extended 8p region may be contributing to schizophrenia susceptibility, as already noted by others (37).
Our initial association analysis of four calcineurin-related genes has detected significant association for only the PPP3CC gene that needs to be replicated in additional samples. It remains possible that the other three genes confer a smaller disease risk in the U.S. population that is difficult to reveal by an association analysis with the number of SNPs per gene and the sample size used thus far. In this regard, examination of these genes in expanded samples and in different populations remains of interest. In particular, examination of potential association of PPP3R1 with schizophrenia in the Palauan population will be informative, because a 2p13–14 susceptibility locus was identified by linkage studies in this population (21, 22). In addition to the four genes examined here, other calcineurin-related genes map to putative schizophrenia susceptibility loci (Table 1). Further examination of members of this group of genes in relation to schizophrenia is required.
There are several possible mechanisms by which altered calcineurin function could contribute to schizophrenia pathogenesis. Calcium-dependent activation of calcineurin leads to dephosphorylation of DARPP-32, which is phosphorylated after dopamine D1 receptor activation, and of the related inhibitor of protein phosphatase-1, inhibitor-1 (10, 45). Normal calcineurin function may be required for calcium-dependent regulation of downstream events in the D1-mediated dopaminergic signaling cascade. In addition, calcineurin is required for certain types of N-methyl-D-aspartate receptor-dependent synaptic plasticity including long-term depression (LTD) (10, 11). Thus, altered calcineurin activity could affect the range of bidirectional synaptic modifiability (11). The involvement of calcineurin in dopaminergic and glutamatergic signaling events raises the possibility that calcineurin function is required as a critical link between these two neurotransmitter systems.
A complex of calcineurin and dynamin-1 has been shown to be involved in regulation of clathrin-mediated endocytosis (46). This process has been implicated in endocytosis of synaptic vesicles (46) and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors (47). Altered calcineurin activity might therefore result in abnormal calcium-dependent regulation of critical synaptic endocytotic events and consequent abnormal synaptic function.
An interaction of calcineurin with the ryanodine receptor type 3/inositol triphosphate receptor 1 (ITPR1) complex has been shown to regulate intracellular calcium release (48). Furthermore, calcineurin activity has been shown to be required for expression of the ITPR1 in neurons (49). Therefore, altered calcineurin activity could lead to abnormal neuronal calcium homeostasis. In addition to its role in regulation of intracellular calcium release, calcineurin has recently been shown to be involved in serotonin-dependent modulation of l-type calcium channel function (50), suggesting that altered calcineurin activity could also lead to abnormal serotonergic modulation of calcium entry.
Lastly, calcineurin is required for the nuclear factor of activated T cell (NFAT)-mediated transcriptional response (51). At least one isoform of NFAT is expressed in the mammalian brain (52). Calcineurin activity has been shown to be required for the expression of specific genes in neurons (49), consistent with the possibility that altered calcineurin activity could lead to changes in calcium-dependent neuronal transcription that could have profound effects on neuronal function.
Conclusions
We have obtained several converging lines of evidence suggesting that altered calcineurin signaling could be a contributing factor in schizophrenia pathogenesis. Further investigation of the neuronal functions of calcineurin and related proteins, and continued investigation of the association of calcineurin-related candidate genes with schizophrenia should help to elucidate the involvement of altered calcineurin signaling in this condition. Hopefully such research will reveal new possibilities for therapeutic intervention.
Supplementary Material
Acknowledgments
We thank Junne Kamihara and David Housman for helpful advice and suggestions, and Shu Huang and Celina Lafaille for excellent technical assistance. This work was supported by National Institutes of Health Grant P50-MH58880 and a gift from Otsuka Maryland Research Institute, Inc. (to S.T.) and a National Alliance for Research on Schizophrenia and Depression Young Investigator Award (to T.M.), as well as National Institutes of Health Grant R01-MH61399 (to M.K.). J.A.G. is supported by grants from the McKnight Endowment Fund for Neuroscience, the EJLB Foundation, and the New York City Council Speaker's Fund.
Abbreviation: SNP, single-nucleotide polymorphism.
References
- 1.Lewis, D. A. & Lieberman, J. A. (2000) Neuron 28, 325-334. [DOI] [PubMed] [Google Scholar]
- 2.Karayiorgou, M. & Gogos, J. A. (1997) Neuron 19, 967-979. [DOI] [PubMed] [Google Scholar]
- 3.Thaker, G. K. & Carpenter, W. T., Jr. (2001) Nat. Med. 7, 667-671. [DOI] [PubMed] [Google Scholar]
- 4.Seeman, P. (1987) Synapse 1, 133-152. [DOI] [PubMed] [Google Scholar]
- 5.Tsai, G. & Coyle, J. T. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 165-179. [DOI] [PubMed] [Google Scholar]
- 6.Carlsson, A., Waters, N., Holm-Waters, S., Tedroff, J., Nilsson, M. & Carlsson, M. L. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 237-260. [DOI] [PubMed] [Google Scholar]
- 7.Klee, C. B., Ren, H. & Wang, X. (1998) J. Biol. Chem. 273, 13367-13370. [DOI] [PubMed] [Google Scholar]
- 8.Shibasaki, F., Hallin, U. & Uchino, H. (2002) J. Biochem. (Tokyo) 131, 1-15. [DOI] [PubMed] [Google Scholar]
- 9.Greengard, P. (2001) Science 294, 1024-1030. [DOI] [PubMed] [Google Scholar]
- 10.Mulkey, R. M., Endo, S., Shenolikar, S. & Malenka, R. C. (1994) Nature 369, 486-488. [DOI] [PubMed] [Google Scholar]
- 11.Zeng, H., Chattarji, S., Barbarosie, M., Rondi-Reig, L., Philpot, B. D., Miyakawa, T., Bear, M. F. & Tonegawa, S. (2001) Cell 107, 617-629. [DOI] [PubMed] [Google Scholar]
- 12.Miyakawa, T., Leiter, L. M., Gerber, D. J., Gainetdinov, R. R., Sotnikova, T. D., Zeng, H., Caron, M. G. & Tonegawa, S. (2003) Proc. Natl. Acad. Sci. USA 100, 8987-8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pulver, A. E., Lasseter, V. K., Kasch, L., Wolyniec, P., Nestadt, G., Blouin, J. L., Kimberland, M., Babb, R., Vourlis, S., Chen, H., et al. (1995) Am. J. Med. Genet. 60, 252-260. [DOI] [PubMed] [Google Scholar]
- 14.Kendler, K. S., MacLean, C. J., O'Neill, F. A., Burke, J., Murphy, B., Duke, F., Shinkwin, R., Easter, S. M., Webb, B. T., Zhang, J., et al. (1996) Am. J. Psychiatry 153, 1534-1540. [DOI] [PubMed] [Google Scholar]
- 15.Blouin, J. L., Dombroski, B. A., Nath, S. K., Lasseter, V. K., Wolyniec, P. S., Nestadt, G., Thornquist, M., Ullrich, G., McGrath, J., Kasch, L., et al. (1998) Nat. Genet. 20, 70-73. [DOI] [PubMed] [Google Scholar]
- 16.Gurling, H. M., Kalsi, G., Brynjolfson, J., Sigmundsson, T., Sherrington, R., Mankoo, B. S., Read, T., Murphy, P., Blaveri, E., McQuillin, A., et al. (2001) Am. J. Hum. Genet. 68, 661-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Straub, R. E., MacLean, C. J., Ma, Y., Webb, B. T., Myakishev, M. V., Harris-Kerr, C., Wormley, B., Sadek, H., Kadambi, B., O'Neill, F. A., et al. (2002) Mol. Psychiatry 7, 542-559. [DOI] [PubMed] [Google Scholar]
- 18.Liu, H., Heath, S. C., Sobin, C., Roos, J. L., Galke, B. L., Blundell, M. L., Lenane, M., Robertson, B., Wijsman, E. M., Rapoport, J. L., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 3717-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, H., Abecasis, G. R., Heath, S. C., Knowles, A., Demars, S., Chen, Y. J., Roos, J. L., Rapoport, J. L., Gogos, J. A. & Karayiorgou, M. (2002) Proc. Natl. Acad. Sci. USA 99, 16859-16864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clayton, D. (1999) Am. J. Hum. Genet. 65, 1170-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coon, H., Myles-Worsley, M., Tiobech, J., Hoff, M., Rosenthal, J., Bennett, P., Reimherr, F., Wender, P., Dale, P., Polloi, A. & Byerley, W. (1998) Mol. Psychiatry 3, 521-527. [DOI] [PubMed] [Google Scholar]
- 22.Camp, N. J., Neuhausen, S. L., Tiobech, J., Polloi, A., Coon, H. & Myles-Worsley, M. (2001) Am. J. Hum. Genet. 69, 1278-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levinson, D. F., Mahtani, M. M., Nancarrow, D. J., Brown, D. M., Kruglyak, L., Kirby, A., Hayward, N. K., Crowe, R. R., Andreasen, N. C., Black, D. W., et al. (1998) Am. J. Psychiatry 155, 741-750. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy, J. L., Basile, V. S. & Macciardi, F. M. (1999) Am. J. Med. Genet. 88, 224-228. [DOI] [PubMed] [Google Scholar]
- 25.Bailer, U., Leisch, F., Meszaros, K., Lenzinger, E., Willinger, U., Strobl, R., Gebhardt, C., Gerhard, E., Fuchs, K., Sieghart, W., et al. (2000) Neuropsychobiology 42, 175-182. [DOI] [PubMed] [Google Scholar]
- 26.Stober, G., Saar, K., Ruschendorf, F., Meyer, J., Nurnberg, G., Jatzke, S., Franzek, E., Reis, A., Lesch, K. P., Wienker, T. F. & Beckmann, H. (2000) Am. J. Hum. Genet. 67, 1201-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faraone, S. V., Matise, T., Svrakic, D., Pepple, J., Malaspina, D., Suarez, B., Hampe, C., Zambuto, C. T., Schmitt, K., Meyer, J., et al. (1998) Am. J. Med. Genet. 81, 290-295. [PubMed] [Google Scholar]
- 28.Craddock, N. & Lendon, C. (1999) Am. J. Med. Genet. 88, 244-254. [PubMed] [Google Scholar]
- 29.Chowdari, K. V., Xu, K., Zhang, F., Ma, C., Li, T., Xie, B. Y., Wood, J., Trucco, M., Tsoi, W. F., Saha, N., et al. (2001) Hum. Immunol. 62, 714-724. [DOI] [PubMed] [Google Scholar]
- 30.Wright, P., Donaldson, P. T., Underhill, J. A., Choudhuri, K., Doherty, D. G. & Murray, R. M. (1996) Am. J. Psychiatry 153, 1530-1533. [DOI] [PubMed] [Google Scholar]
- 31.Schizophrenia Linkage Collaborative Group for Chromosomes 3, 6 and 8 (1996) Am. J. Med. Genet. 67, 580-594. [DOI] [PubMed] [Google Scholar]
- 32.Freedman, R., Leonard, S., Olincy, A., Kaufmann, C. A., Malaspina, D., Cloninger, C. R., Svrakic, D., Faraone, S. V. & Tsuang, M. T. (2001) Am. J. Med. Genet. 105, 794-800. [DOI] [PubMed] [Google Scholar]
- 33.Brzustowicz, L. M., Hodgkinson, K. A., Chow, E. W., Honer, W. G. & Bassett, A. S. (2000) Science 288, 678-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwab, S. G., Eckstein, G. N., Hallmayer, J., Lerer, B., Albus, M., Borrmann, M., Lichtermann, D., Ertl, M. A., Maier, W. & Wildenauer, D. B. (1997) Mol. Psychiatry 2, 156-160. [DOI] [PubMed] [Google Scholar]
- 35.Crowe, R. R. & Vieland, V. (1999) Am. J. Med. Genet. 88, 229-232. [PubMed] [Google Scholar]
- 36.Paunio, T., Ekelund, J., Varilo, T., Parker, A., Hovatta, I., Turunen, J. A., Rinard, K., Foti, A., Terwilliger, J. D., Juvonen, H., et al. (2001) Hum. Mol. Genet. 10, 3037-3048. [DOI] [PubMed] [Google Scholar]
- 37.Badner, J. A. & Gershon, E. S. (2002) Mol. Psychiatry 7, 405-411. [DOI] [PubMed] [Google Scholar]
- 38.Lewontin, R. C. (1988) Genetics 120, 849-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muramatsu, T., Giri, P. R., Higuchi, S. & Kincaid, R. L. (1992) Proc. Natl. Acad. Sci. USA 89, 529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamagata, K., Kaufmann, W. E., Lanahan, A., Papapavlou, M., Barnes, C. A., Andreasson, K. I. & Worley, P. F. (1994) Learn. Mem. 1, 140-152. [PubMed] [Google Scholar]
- 41.Hippenmeyer, S., Shneider, N. A., Birchmeier, C., Burden, S. J., Jessell, T. M. & Arber, S. (2002) Neuron 36, 1035-1049. [DOI] [PubMed] [Google Scholar]
- 42.Mittelstadt, P. R. & Ashwell, J. D. (1998) Mol. Cell. Biol. 18, 3744-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stefansson, H., Sigurdsson, E., Steinthorsdottir, V., Bjornsdottir, S., Sigmundsson, T., Ghosh, S., Brynjolfsson, J., Gunnarsdottir, S., Ivarsson, O., Chou, T. T., et al. (2002) Am. J. Hum. Genet. 71, 877-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stefansson, H., Sarginson, J., Kong, A., Yates, P., Steinthorsdottir, V., Gudfinnsson, E., Gunnarsdottir, S., Walker, N., Petursson, H., Crombie, C., et al. (2003) Am. J. Hum. Genet. 72, 83-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greengard, P., Allen, P. B. & Nairn, A. C. (1999) Neuron 23, 435-447. [DOI] [PubMed] [Google Scholar]
- 46.Lai, M. M., Hong, J. J., Ruggiero, A. M., Burnett, P. E., Slepnev, V. I., De Camilli, P. & Snyder, S. H. (1999) J. Biol. Chem. 274, 25963-25966. [DOI] [PubMed] [Google Scholar]
- 47.Haucke, V. (2000) Nat. Neurosci. 3, 1230-1232. [DOI] [PubMed] [Google Scholar]
- 48.Cameron, A. M., Steiner, J. P., Roskams, A. J., Ali, S. M., Ronnett, G. V. & Snyder, S. H. (1995) Cell 83, 463-472. [DOI] [PubMed] [Google Scholar]
- 49.Genazzani, A. A., Carafoli, E. & Guerini, D. (1999) Proc. Natl. Acad. Sci. USA 96, 5797-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Day, M., Olson, P. A., Platzer, J., Striessnig, J. & Surmeier, D. J. (2002) J. Neurophysiol. 87, 2490-2504. [DOI] [PubMed] [Google Scholar]
- 51.Crabtree, G. R. & Olson, E. N. (2002) Cell 109 (Suppl.), S67-S79. [DOI] [PubMed] [Google Scholar]
- 52.Plyte, S., Boncristiano, M., Fattori, E., Galvagni, F., Paccani, S. R., Majolini, M. B., Oliviero, S., Ciliberto, G., Telford, J. L. & Baldari, C. T. (2001) J. Biol. Chem. 276, 14350-14358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.