Abstract
SNARE [soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptor] proteins are essential for membrane fusion but their regulation is not yet fully understood. We have previously shown that the amino-terminal Longin domain of the v-SNARE TI-VAMP (tetanus neurotoxin-insensitive vesicle-associated membrane protein)/VAMP7 plays an inhibitory role in neurite outgrowth. The goal of this study was to investigate the regulation of TI-VAMP as a model of v-SNARE regulation. We show here that the Longin domain (LD) plays a dual role. First, it negatively regulates the ability of TI-VAMP and of a Longin/Synaptobrevin chimera to participate in SNARE complexes. Second, it interacts with the adaptor complex AP-3 and this interaction targets TI-VAMP to late endosomes. Accordingly, in mocha cells lacking AP-3δ, TI-VAMP is retained in an early endosomal compartment. Furthermore, TI-VAMPc, an isoform of TI-VAMP lacking part of the LD, does not interact with AP-3, and therefore is not targeted to late endosomes; however, this shorter LD still inhibits SNARE-complex formation. These findings support a mechanism controlling both localization and function of TI-VAMP through the LD and clathrin adaptors. Moreover, they point to the amino-terminal domains of SNARE proteins as multifunctional modules responsible for the fine tuning of SNARE function.
SNARE [soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptor] proteins are key mediators of membrane fusion as they are necessary for membrane fusion in vivo and are sufficient for fusion of liposomes in vitro (1, 2). However, the mechanisms ensuring proper targeting and regulation of SNAREs are still not well understood. All SNAREs share a 60- to 70-aa residue sequence called the SNARE motif, which mediates v-SNARE/t-SNARE interaction; additionally, several SNAREs contain extended amino-terminal regions, which might control their subcellular localization and protein interactions, and thus regulate their function (3, 4). In particular, the amino-terminal Longin domain (LD) of the v-SNARE TI-VAMP (tetanus neurotoxin-insensitive vesicle-associated membrane protein)/VAMP7/SYBL1, negatively regulates neurite outgrowth (5, 6). This domain is homologous to the profilin-like amino-terminal domains of sec22p and ykt6p (7–9), and defines a subfamily of SNAREs, called the Longins (9). The molecular mechanism underlying the function of LDs is not completely understood: the LD of ykt6 interacts directly with the SNARE motif (8), whereas this is not the case for Sec22 (7). In this article, we have analyzed the regulatory role of the LD of TI-VAMP as a model for v-SNARE regulation. Our data demonstrate a dual function for the LD, controlling both the ability of the v-SNARE to participate in SNARE complexes and its subcellular localization by an AP-3-mediated pathway. Therefore, our results reveal the multifunctional nature of the aminoterminal domains of SNAREs.
Materials and Methods
Antibodies and DNA Constructs. Mouse mAb clone 158.2 anti-TI-VAMP will be described elsewhere. Mouse mAbs anti-GFP (GFP; clones 7.1 and 13.1, Roche Diagnostics, Indianapolis), syntaxin 6 (clone 30, Transduction Laboratories, Lexington, KY), syntaxin 4 (clone 49, Transduction Laboratories), syntaxin 8 (clone 48, Transduction Laboratories), Vti1b (clone 7, Transduction Laboratories), and transferrin receptor (68.4, from I. Trowbridge, Salk Institute, La Jolla, CA) have been described. Rabbit sera anti-TI-VAMP (TG18), SNAP-23 (TG7) (10), syntaxin 3 (TG0) (10), VAMP8 (TG15) (11), and VAMP4 (TG19/ 20) (12) were purified by affinity chromatography. Rabbit polyclonal antibodies anti-syntaxin 10 and syntaxin 16 were generous gifts from Dr. W. Hong (Institute of Molecular and Cell Biology, Singapore). Rabbit polyclonal antibodies anti-AP-1 and AP-3 subunits and mouse mAb anti-AP-2 have been described elsewhere (13).
The cDNAs of human TI-VAMP (10) and the GFP-fusion constructs GFP-TIVAMP, GFP-ΔLongin-TIVAMP, and GFP-Longin have been reported (5). The cDNA of rat Syb2 (14) was cloned by PCR into the pEGFP-C3 vector (CLONTECH). For the production of the chimera GFP-Longin/Syb2, a PvuI site was introduced by site-directed mutagenesis in GFP-TIVAMP (after residue D124) and in GFP-Syb2 (after residue R30). PHLuorin from G. Miesenbock (Sloan–Kettering Memorial Hospital, New York) was used for the carboxyl-terminal GFP-fusion constructs TIVAMP-GFP and ΔLongin-TIVAMP-GFP. The cDNA of AP-3δ (13) was cloned into pCDNA3 by using the BamHI and XhoI sites. For the production of inducible clones, TI-VAMP and ΔLongin-TIVAMP were cloned in the tet off pBI-4 vector (15).
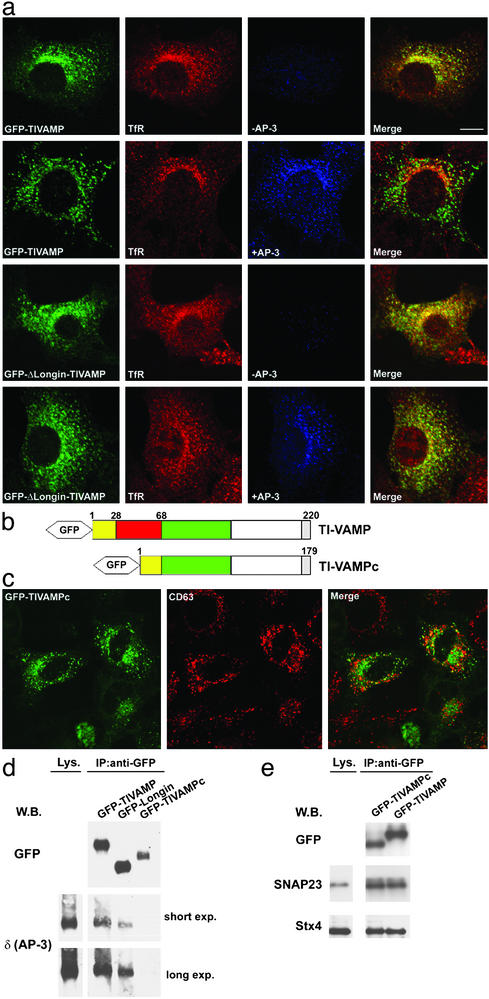
Identification of TI-VAMPc. Isoform c of TI-VAMP is the product of an alternative splicing variant of the gene SYBL1 (SYBL1c) that was isolated by RT-PCR, using human brain tissue as template. The existence of this isoform was subsequently confirmed by an RNase protection assay (unpublished observations). SYBL1c results from an intraexonic and in-frame splicing variant, which skips 123 nucleotides, lacking part of exon 2 and all of exon 3. The splicing is produced by using a cryptic GT donor splice site, within exon 2 at position 179 (position is in base pairs relative to GenBank accession no. X95804) and the known AG acceptor site, at the 3′-end of intron 3. Translation of this alternative transcript would produce a putative 179-aa protein that we named TI-VAMPc (GenBank accession no. AJ549301). This isoform differs from TI-VAMP (GenBank accession no. NP005629) because of a deletion from 28 to 68 aa (inclusive). For the GFP-fusion construct, the cDNA of TI-VAMPc was cloned into EcoRI and BamHI sites of pEGFP-C3 vector (CLONTECH).
In Vitro Interaction Assay. An Escherichia coli strain coexpressing GST-syntaxin 1 and His6-SNAP-25 was kindly provided by G. Schiavo (Cancer Research UK, London). Purification of the recombinant complex was as described (16). The GFP-fusion proteins GFP-TIVAMP, GFP-ΔLongin-TIVAMP, and GFP alone were produced in transiently transfected HeLa cells. Twenty-four hours after transfection, cells were lysed in RIPA buffer (50 mM Tris, pH 7.5/150 mM NaCl/10 mM EDTA/0.1% SDS/1% Triton X-100/0.5% deoxycholate) and the fluorescence of the extracts was quantified to include equal amounts in the in vitro interaction assay. After the indicated times of incubation at 4°C, the glutathione beads were washed three times in PBS plus 0.1% Triton X-100 and either quantified directly in a SpectraMax Gemini XS fluorometer (Molecular Devices; excitation wavelength, 488 nm, emission wavelength, 535 nm), or eluted in sample buffer for SDS/PAGE and Western Blot analysis of the bound GFP-fusion proteins. Nonlinear regression curves (one phase exponential association), statistics, rate constants and Bmax were calculated by using PRISM (GraphPad, San Diego).
Yeast Two-Hybrid Cloning and Analysis. Baits were PCR-amplified (Pfu, Stratagene) and cloned into the pB27 plasmid derived from the original pBTM116 (17). The Longin and cytosolic domains of human and Drosophila TI-VAMP comprised residues 1–120 and 1–188, respectively. Random-primed cDNA libraries from human placenta and Drosophila embryos (0–24 h) poly(A)+ RNA were constructed into the pP6 plasmid derived from the original pGADGH (18). The libraries were transformed into the Y187 yeast strain and 10 million independent yeast colonies were collected, pooled, and stored at -80°C. The mating protocol has been described elsewhere (19). Each screen was performed to ensure that a minimum of 50 million interactions were tested. The prey fragments of the positive clones were amplified by PCR and sequenced at their 5′ and 3′ junctions on a PE3700 sequencer. The resulting sequences were used to identify the corresponding gene in the GenBank database by using a fully automated procedure. Direct interaction assays were performed by mating, using the L40 and AMR70 strains (17).
Cell Culture and Transfection. HeLa and mocha cells were cultured in DMEM with 10% FCS and transfected by electroporation as described (5). For the sucrose treatment, cells were incubated with 0.05 M sucrose for 18 h, washed and chased for 5 h before fixation. Tet off Madin–Darby canine kidney (MDCK) cells (CLONTECH) cultured in DMEM with 7% FCS and 200 μg/ml G418 were cotransfected by electroporation with pBI-4-GFP-TIVAMP or pBI-4-GFP-ΔLongin-TIVAMP, and the pKT-Hyg vector conferring resistance to hygromycin in a ratio of 1:7. Clones were selected in medium containing 400 μg/ml hygromycin, 200 μg/ml G418, and 0.5 μg/ml doxycycline. Electron microscopy experiments were carried out at least 5 days after removal of doxycycline.
Immunocytochemistry. Cells were fixed with 4% paraformaldehyde and processed for immunofluorescence as described (20). Confocal laser scanning microscopy was performed by using either a TCS or an SP2 confocal microscope (Leica, Heidelberg, Germany). Images were assembled without modification by using PHOTOSHOP (Adobe Systems, San Jose, CA).
Immunoelectron Microscopy. MDCK cells were fixed with 2% paraformaldehyde in phosphate buffer 0.2 M, pH 7.4 (PB) for 2 h at room temperature. Fixed cells were processed for ultrathin cryosectioning, were ImmunoGold labeled, and contrasted as described (21). Anti-GFP polyclonal antibodies (Molecular Probes) were visualized with protein A-gold conjugates (PAGs) (Department of Cell Biology, Utrecht University, Utrecht, The Netherlands).
N-ethylmaleimide (NEM) Treatment, Chemical Crosslinking, and Immunoprecipitation. NEM treatment was as described by Galli et al. (10). Crosslinking was performed by incubating intact cells with 1 mM dithiobis(succinimidyl proprionate) (DSP) diluted from a freshly prepared stock at 25 mM in DMSO. After 30 min at room temperature, DSP was neutralized with 10 mM Tris, pH 7.6, for 15 min. After the NEM or DSP treatments, cells were lysed and processed for immunoprecipitation as described (5).
Antibody Binding Assay. HeLa cells transfected with TIVAMP-GFP or with ΔLongin-TIVAMP-GFP were incubated in the presence of 5 μg/ml anti-GFP antibody in culture medium for 1 h at 4°C, washed extensively with PBS, fixed with 4% PFA and processed for immunofluorescence.
Results
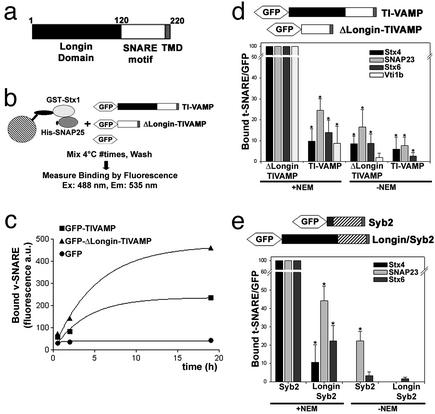
The LD of TI-VAMP Inhibits SNARE-Complex Formation in Vitro and in Vivo. To better understand the molecular mechanisms underlying the regulatory function of the amino-terminal extensions of SNARE proteins, we first set up an assay to quantitatively measure the binding of v-SNAREs to t-SNAREs in vitro (Fig. 1b). Recombinant t-SNARE composed of GST-syntaxin 1 and His6-SNAP-25 was immobilized on beads (see Fig. 6a, which is published as supporting information on the PNAS web site, www.pnas.org) and incubated with diluted detergent extracts of HeLa cells expressing GFP, GFP fused to full-length TI-VAMP (GFP-TIVAMP), or GFP fused to TI-VAMP lacking the LD (GFP-ΔLongin-TIVAMP). As measured by fluorescence, deletion of the LD induced a more efficient interaction of TI-VAMP with the immobilized t-SNARE (Figs. 1c and 6b). Analysis of the kinetics of the v-SNARE/t-SNARE interaction revealed a two-fold increase in the Bmax (bound v-SNARE) of GFP-ΔLongin-TIVAMP compared with GFP-TIVAMP (470 ± 37 versus 235 ± 33 fluorescence arbitrary units respectively; two-tailed P value is 0.04), with no significant difference in the rate constant (0.25 ± 0.12 h-1 versus 0.19 ± 0.05 h-1; two-tailed P value is 0.70), reflecting the fact that both proteins have the same SNARE motif.
Fig. 1.
The LD inhibits SNARE-complex formation in vitro and in vivo. (a) Scheme of the structure of TI-VAMP. TMD, transmembrane domain. (b and c) Quantitative in vitro assay to measure the interaction between recombinant syntaxin 1 and SNAP25 with TI-VAMP or ΔLongin-TIVAMP. The graphic shows one representative experiment of three independent experiment. (d and e) HeLa cells transfected with GFP-TIVAMP or GFP-ΔLongin-TIVAMP (d) or with GFP-Syb2 or GFP-Longin/Syb2 (e), and treated as described in Fig. 6c, were lysed and immunoprecipitated with an anti-GFP antibody. Coimmunoprecipitated SNAREs were detected by Western blot and quantified by densitometry. The histograms show the statistical analyses of at least three independent experiments. *, P < 0.05, Student's t test.
To understand the relevance of this autoinhibition in vivo, we compared the ability of GFP-TIVAMP and GFP-ΔLongin-TIVAMP to form SNARE complexes in transfected HeLa cells. We found that the LD inhibited the formation of SNARE complexes with cognate plasma membrane and endosomal t-SNAREs of TI-VAMP (Figs. 1d and 6 c and d). Indeed, compared with GFP-TIVAMP, GFP-ΔLongin-TIVAMP made between 4- and 10-fold more SNARE complexes with SNAP-23, syntaxin 4, syntaxin 6, and Vti1b (Figs. 1d and 6d). The specificity of the SNARE interactions was not affected by the presence of the LD because the repertoire of t-SNAREs coimmunoprecipitated by endogenous TI-VAMP (Fig. 7c, which is published as supporting information on the PNAS web site), GFP-TIVAMP, and GFP-ΔLongin-TIVAMP was identical (Figs. 1d and 6 c and d), thus suggesting that TI-VAMP and ΔLongin-TIVAMP mediated fusion with the same target membranes. We then asked whether the LD could function in the context of another v-SNARE. Therefore we produced a GFP-fusion of synaptobrevin 2 (GFP-Syb2) and a chimeric protein between TI-VAMP and Syb2 by exchanging the first 30 aa of Syb2 with the LD of TI-VAMP (GFP-Longin/Syb2). We found that the LD also inhibited the formation of SNARE complexes in the context of Syb2 (Figs. 1e and 6d). Indeed, GFP-Longin/Syb2 made between 2- and 10-fold fewer SNARE complexes than GFP-Syb2 (Fig. 1e) with known partners of Syb2, i.e., syntaxin 4, SNAP-23 (11), and syntaxin 6 (22). These results suggest that the LD could prevent the accessibility of the SNARE motif of the v-SNARE to its cognate t-SNAREs as suggested for syntaxin 7 (23) and ykt6p (8). In the case of ykt6p, a direct interaction of the aminoterminal domain and the SNARE motif was detected in a yeast two-hybrid assay (8). In our case, however, we did not detect any interaction between the LD of TI-VAMP and either its SNARE motif or its full cytoplasmic domain in a yeast two-hybrid assay (Fig. 6e). The lack of high-affinity interaction between the LD and the SNARE motif of TI-VAMP is further supported by the fact that TI-VAMP was not recovered in multiple yeast two-hybrid screens using the LD as bait (see below, Fig. 4) and also by the following further evidence (unpublished observations): (i) in vitro assays with recombinant proteins in which there was no interaction between the LD and the SNARE motif of TI-VAMP; (ii) the lack of coimmunoprecipitation from cells transfected with both domains of the protein; (iii) the observation that addition of recombinant LD had no effect on the binding of GFP-TIVAMP and GFP-ΔLongin-TIVAMP to the immobilized t-SNARE as described above; and (iv) the autoinhibition of the LD when fused to Syb2 (Figs. 1e and 6d). These results also suggest that the Longin domain does not mediate homooligomerization of the protein. We cannot exclude that the LD-mediated autoinhibition may result from a low-affinity intramolecular interaction, but an intramolecular steric hindrance would be a more likely mechanism.
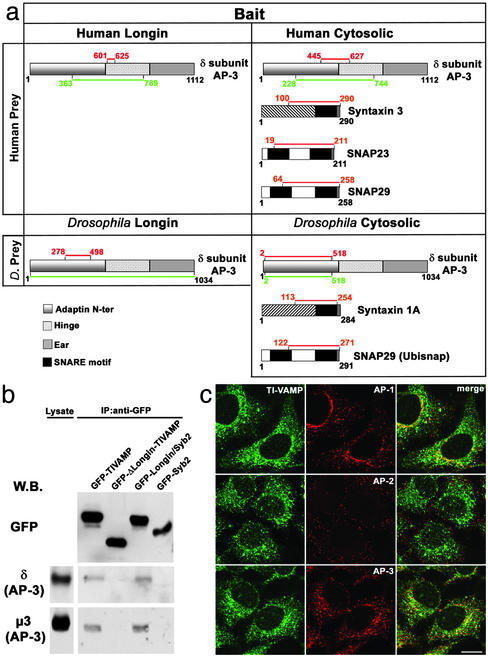
Fig. 4.
Direct interaction between AP-3δ and the LD. (a) Results of a yeast two-hybrid screening using either the Longin or the cytoplasmic domain of Drosophila or human TI-VAMP as baits. SNAP-29, a yet unknown partner of TI-VAMP, was also detected both in the human and in the Drosophila screening, and biochemical experiments validated this interaction (unpublished observations). The red and green lines highlight the intersection and total coverage of all prey clones identified in the yeast two-hybrid screen, respectively. (b) HeLa cells transfected with the GFP-fusion constructs indicated were crosslinked, lysed, and immunoprecipitated with an anti-GFP antibody. Coimmunoprecipitated proteins were detected by Western blot. (c) HeLa cells fixed and double stained for endogenous TI-VAMP and AP-1, AP-2, and AP-3 are shown. (Bar, 15 μm.)
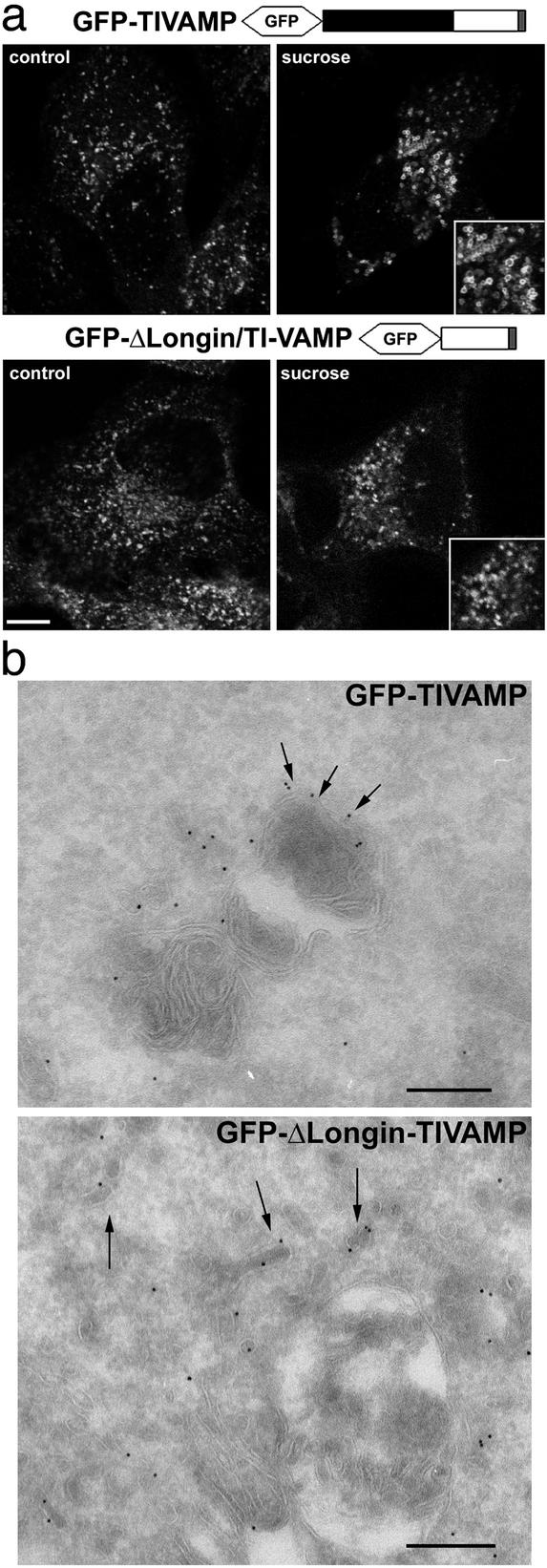
The LD Controls the Subcellular Localization of the v-SNARE. We then asked whether the form of TI-VAMP lacking the LD would show a different subcellular distribution. As we have seen before in PC12 cells, the GFP-TIVAMP staining was identical to that of endogenous TI-VAMP in HeLa cells (unpublished observations). Treatment of cells with sucrose induced the osmotic swelling of the GFP-TIVAMP-positive structures (Fig. 2a), suggesting a late endosomal/lysosomal nature (24). In contrast, GFP-ΔLongin-TIVAMP-positive structures were not affected by the sucrose treatment (Fig. 2a), indicating a different intracellular distribution. The subcellular localization of both proteins was also investigated at the ultrastructural level (Fig. 2b). Immunogold labeling on ultrathin cryosections revealed that the bulk of GFP-TIVAMP localized to compartments similar to lysosomes on the basis of their morphological appearance (i.e., electron dense membranous content). Occasionally, membrane vesicles were also labeled with the anti-GFP antibodies. In contrast, the subcellular distribution of GFP-ΔLongin-TIVAMP was restricted to tubulovesicular elements.
Fig. 2.
The LD controls the intracellular localization of TI-VAMP. (a) HeLa cells transfected with GFP-TIVAMP or GFP-ΔLongin/TIVAMP were either directly fixed (control) or fixed after sucrose treatment (sucrose). Note the swelling of GFP-TIVAMP-positive vesicles, but not that of GFP-ΔLongin-TIVAMP-positive vesicles (Insets). (Bar, 10 μm; bar in Insets,7 μm.) (b) Ultrathin cryosections of MDCK cells expressing GFP-TIVAMP and GFP-Δ Longin-TIVAMP were ImmunoGold-labeled with anti-GFP antibodies and PAG10. TI-VAMP localizes to the limiting membrane of lysosomal compartments (arrows). Occasionally, TI-VAMP is detected in small closely apposed vesicles (arrowheads). GFP-Δ Longin-TIVAMP localizes to tubular vesicular membrane structures (arrows). (Bars, 200 nm.)
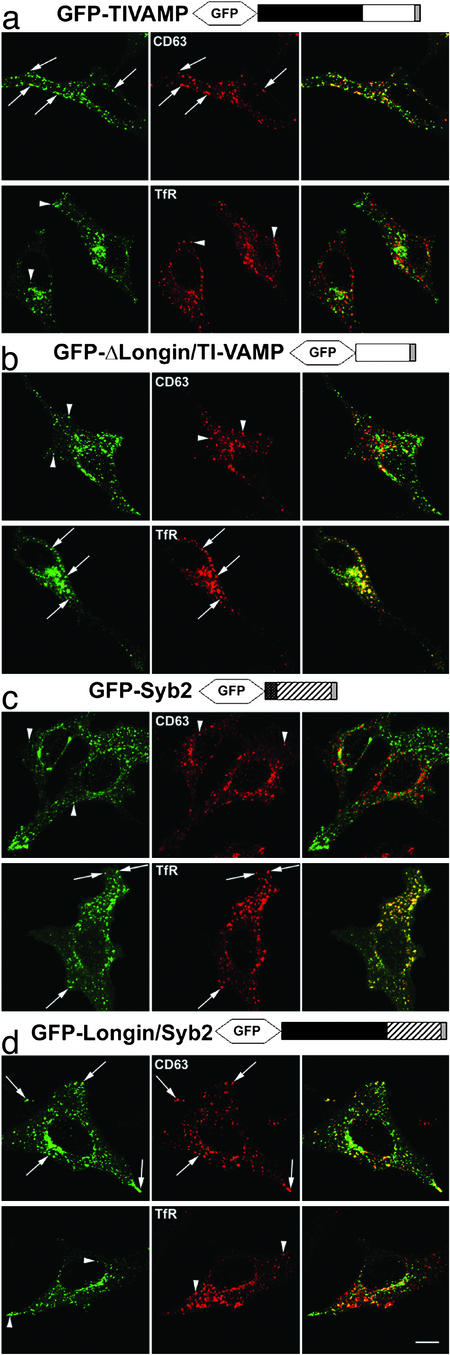
When compared with endosomal markers, GFP-TIVAMP colocalized significantly with the tetraspanin protein CD63, but not with the transferrin receptor (TfR) (Fig. 3a), supporting a late endosomal/lysosomal localization in these cells, as seen in 3T3 cells (25). In contrast, GFP-ΔLongin-TIVAMP, while still localized in vesicles distributed all over the cytoplasm, did not colocalize with CD63, but did colocalize with the TfR (Fig. 3b). Conversely, GFP-Syb2 distributed mostly to early/recycling endosomes, as shown by extensive colocalization with the TfR, but not with CD63 (Fig. 3c), whereas the chimera GFP-Longin/Syb2 displayed the inverse pattern, with some codistribution with CD63 and virtually no colocalization with the TfR (Fig. 3d). These effects were not due to the absence of the short aminoterminal domain of Syb2, but to the presence of the LD of TI-VAMP, because a form of Syb2 lacking the first 30 residues behaved like full-length Syb2 (unpublished observations). These results suggested that the LD had a second role in the targeting of TI-VAMP to late endosomes/lysosomes.
Fig. 3.
The LD controls the intracellular localization of v-SNAREs. HeLa cells transfected with the constructs indicated were fixed and stained for the endosomal markers CD63 and TfR. Arrows point to structures labeled by both the GFP-fusion protein and either CD63 or TfR. Arrowheads point to structures positive for only one of the proteins. (Bar, 13 μm.)
TI-VAMP Interacts with the AP-3 Adaptor Complex Through the LD. To understand the LD-dependent effect on subcellular distribution of v-SNAREs, we performed a yeast two-hybrid screen by using either the LD or the cytoplasmic domain, comprising both the LD and the SNARE motif, of Drosophila or human TI-VAMP as bait. The t-SNARE proteins syntaxin 1A, syntaxin 3, and SNAP-23, known partners of TI-VAMP (5, 10), were identified as partners of the cytoplasmic domain, thereby validating the screen. Importantly, this screen revealed an interaction between the LD and the δ subunit of the AP-3 complex (Fig. 4a). This adaptor complex has been shown to mediate traffic to late endocytic compartments (13, 26–29) and to the plasma membrane (30). The interaction between TI-VAMP and the δ subunit of AP-3 was confirmed by transfection, followed by crosslinking and coimmunoprecipitation experiments (Fig. 4b). The endogenous δ subunit of AP-3 was recovered in an immunoprecipitation from cells transfected with GFP-TIVAMP, but not from cells transfected with GFP-Syb2. The μ3 subunit of AP-3 was also recovered, demonstrating that TI-VAMP interacts with the whole AP-3 complex. In agreement with the two-hybrid results, when the immunoprecipitation was done from cells transfected with GFP-ΔLongin-TIVAMP, neither the δ subunit nor the μ3 subunit were recovered, whereas both were recovered from cells transfected with GFP-Longin/Syb2 (Fig. 4b). The AP-1 and AP-2 complexes were not coimmunoprecipitated in these conditions (Fig. 7 and unpublished observations). We then compared the distribution of endogenous TI-VAMP with that of the different clathrin adaptor complexes (Fig. 4c). TI-VAMP did not colocalize with the plasma membrane adaptor AP-2, whereas it colocalized partially with AP-1 in the perinuclear area and more evidently with AP-3. Altogether these results suggest a strong biochemical link between TI-VAMP and AP-3.
In particular, our data indicate that the LD of TI-VAMP interacts directly with AP-3δ and suggest that this interaction would mediate TI-VAMP localization to a late endosomal/ lysosomal compartment. To test this hypothesis, we made use of mocha cells, which lack the δ subunit and thus a functional AP-3 complex. We reasoned that if AP-3 was implicated in the targeting of TI-VAMP to a CD63-positive late endosomal compartment, then the distribution of TI-VAMP in mocha cells should be affected. This was indeed the case: in mocha cells, GFP-TIVAMP colocalized with early/recycling endosomes labeled with the TfR (Fig. 5a). When these cells were cotransfected with the δ subunit, thus allowing formation of functional AP-3 complex, GFP-TIVAMP recovered its distribution to late endosomes/lysosomes and did not colocalize with the TfR (Fig. 5a). Furthermore, if the AP-3-mediated targeting of TI-VAMP depended on the interaction between its LD and the δ subunit, there should be no difference in the distribution of GFP-ΔLongin-TIVAMP, regardless of the presence or the absence of the AP-3 complex. Again, this was the case: in mocha cells, cotransfected or not with the δ subunit, GFP-ΔLongin-TIVAMP colocalized with the TfR (Fig. 5a), as seen before in HeLa cells (Fig. 3b).
Fig. 5.
Localization and function of TI-VAMP depends on its interaction with functional AP-3 complex. (a) Mocha cells were transfected with either GFP-TIVAMP or GFP-ΔLongin-TIVAMP alone, or were cotransfected with AP-3δ. GFP-TIVAMP and GFP-ΔLongin-TIVAMP were detected by direct GFP fluorescence (green), whereas AP-3δ (blue) and TfR (red) were detected by coimmunolabeling. Merge, the level of colocalization between the TfR and GFP-TIVAMP or GFP-ΔLongin-TIVAMP. (b) Scheme of the structure of TI-VAMP compared with TI-VAMPc. Isoform c lacks residues 28–68 (red box). (c) HeLa cells transfected with GFP-TIVAMPc were fixed and stained for endogenous CD63. (d) HeLa cells transfected with the GFP-fusion constructs indicated were crosslinked, lysed, and immunoprecipitated with an anti-GFP antibody. Coimmunoprecipitated proteins were detected by Western blot. (e) HeLa cells transfected with the GFP-fusion constructs indicated were incubated with NEM as in Fig. 6c, lysed, and immunoprecipitated with a mouse monoclonal anti-GFP antibody. Coimmunoprecipitated SNAREs were detected by Western blot. (Bar in a, 10 μm; bar in c, 13 μm.)
The Truncated LD of an Isoform of TI-VAMP, TI-VAMPc, Does Not Interact with AP-3 But Still Inhibits SNARE-Complex Formation in Vivo. SNARE proteins could be regulated by mRNA splicing (31). We have recently identified a splicing isoform of TI-VAMP, TI-VAMPc (M.V., F.F., and M.D., unpublished observations), which lacks approximately one-third of the LD (Fig. 5b). Coimmunoprecipitation experiments showed that GFP-TIVAMPc did not interact with AP-3δ (Fig. 5d), whereas AP-3δ was coimmunoprecipitated with GFP-TIVAMP and with the LD of TI-VAMP fused to GFP (GFP-Longin) (Fig. 5d), a cytosolic protein (5). Interestingly, and by contrast to GFP-TIVAMP, GFP-TIVAMPc did not colocalize with CD63 (Fig. 5c), thus confirming the direct link between AP-3δ binding and late endosomal localization and indicating that a crucial domain for AP-3δ binding resides between residues 28–68 of TI-VAMP. However, TI-VAMPc behaved like TI-VAMP, regarding its ability to participate in SNARE complexes with cognate t-SNAREs (Fig. 5e). This latter result indicates that the shorter LD of TI-VAMPc retains the autoinhibitory function, but not the AP-3 binding function of the full LD.
The finding that GFP-ΔLongin-TIVAMP interacts more efficiently with plasma membrane and endosomal t-SNAREs (Fig. 1), together with its early endosomal localization (Figs. 2 and 3b), suggests that the LD may regulate the exocytic function of TI-VAMP, i.e., the fusion of TI-VAMP's vesicles with the plasma membrane. Therefore, we produced GFP-fusion proteins with the GFP tag fused to the carboxyl terminus of TI-VAMP (TIVAMP-GFP) or of ΔLongin-TI-VAMP (ΔLongin-TI-VAMP-GFP), and measured their expression at the plasma membrane by incubating living cells with an anti-GFP antibody. Fusion of the GFP tag at the carboxyl terminus of TI-VAMP or ΔLongin-TIVAMP did not affect their steady-state localization. However, cells transfected with ΔLongin-TIVAMP-GFP bound the anti-GFP antibody more efficiently than cells transfected with TIVAMP-GFP (Fig. 8, which is published as supporting information on the PNAS web site). Whereas these results are consistent with the trafficking of other AP-3-dependent cargo proteins such as CD63 (32), the fact that TI-VAMP is a v-SNARE-mediating fusion at the plasma membrane (5), together with the autoinhibitory role of the LD in plasma membrane SNARE-complex formation, suggest that the presence of the LD correlates with the low exocytic activity of TI-VAMP.
Discussion
The extended amino-terminal domains of certain SNAREs have been proposed to be responsible for their regulation, and in some cases, like for syntaxin 1 and 7 and ykt6p, a role in modulating SNARE-complex formation has been shown (8, 23, 33). Our data point to the LD of TI-VAMP as, to our knowledge, the first example of a SNARE amino-terminal domain playing a dual role by modulating both the fusogenic activity and the subcellular localization of the v-SNARE. Altogether our results show that the LD of TI-VAMP plays two distinct functions: (i) it controls the capacity of this v-SNARE to interact with its endosomal and plasma membrane t-SNARE partners (Fig. 1), and (ii) by interacting with the δ subunit of AP-3, it targets TI-VAMP to late endosomes/lysosomes (Figs. 2, 3, 4, 5). The first role of the LD unraveled by our in vitro assay can function independently from the second because (i) the interaction between AP-3 and TI-VAMP is labile and could only be observed after in vivo crosslinking, and (ii) the autoinhibition but not the AP-3-dependent targeting is conserved in the shorter form of the LD present in TI-VAMPc. Interaction between SNAREs and coat proteins have been described (34, 35), but this is an example of a SNARE domain playing a dual role. Hence, our findings suggest that the amino-terminal domains of SNAREs can regulate at least two independent biochemical properties, one directly linked to their fusogenic activity, and one linked to their targeting to a specific subcellular compartment. Furthermore, the multifunctional properties of the LD point to an important coordination between localization and function, a crucial aspect of cell homeostasis. In addition, our data show the involvement of AP-3 in transport from early/recycling to late endosomes. Given our demonstration that TI-VAMP mediates neurite outgrowth (5) and apical transport in epithelial cells (10, 36), and its role in the endocytic pathway (25), the important regulation unraveled here suggests that the neurological defects seen in mocha mice (37), and the defects in renal function seen in mocha and pearl mice (38, 39), may be linked to a defect in the proper targeting of TI-VAMP. TI-VAMP is likely to interact with as yet unknown membrane proteins, the trafficking of which would also be affected when this pathway is perturbed. In the light of the link between the AP-3 and TI-VAMP pathways, it will now be important to further identify the regulators and cargo of the TI-VAMP/AP-3-mediated pathway in both nonneuronal and neuronal cells.
Supplementary Material
Acknowledgments
We thank all Hybrigenics staff for their contributions; Margaret Robinson for reagents and helpful comments; and Fabienne Paumet, Christophe Lamaze, Ludger Johannes, and Anne Schmidt for discussion of the results. This work was supported in part by Human Frontier Science Program Grant RGY0027/2001-B101; European Community Grant QLG3-CT-2001–02430_Retrograde Signaling; Association pour la Recherche sur le Cancer Grant ARC 5873 (to T.G.); GenHomme Network Grant 02490-6088 to Hybrigenics and Institut Curie; Telethon Grant GGP02308 Ministero dell'Universitá e della Ricerca Scientifica e Tecnologica Cluster 02 (to M.D.); and Ministero dell'Universitá e della Ricerca Scientifica e Tecnologica 60% (to F.F.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NEM, N-ethylmaleimide; SNARE, soluble NSF (NEM-sensitive factor) attachment protein receptor; LD, Longin domain; VAMP, vesicle-associated membrane protein; TI-VAMP, tetanus neurotoxin-insensitive VAMP; TfR, transferrin receptor.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ549301).
References
- 1.Rothman, J. E. (2002) Nat. Med. 8, 1059-1062. [DOI] [PubMed] [Google Scholar]
- 2.Galli, T. & Haucke, V. (2001) Sci. STKE 2001, RE1. [DOI] [PubMed] [Google Scholar]
- 3.Jahn, R., Lang, T. & Sudhof, T. C. (2003) Cell 112, 519-533. [DOI] [PubMed] [Google Scholar]
- 4.Hasegawa, H., Zinsser, S., Rhee, Y., Vik-Mo, E. O., Davanger, S. & Hay, J. C. (2003) Mol. Biol. Cell 14, 698-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Arca, S., Alberts, P., Zahraoui, A., Louvard, D. & Galli, T. (2000) J. Cell Biol. 149, 889-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Arca, S., Coco, S., Mainguy, G., Schenk, U., Alberts, P., Bouille, P., Mezzina, M., Prochiantz, A., Matteoli, M., Louvard, D. & Galli, T. (2001) J. Neurosci. 21, 3830-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez, L. C., Weis, W. I. & Scheller, R. H. (2001) J. Biol. Chem. 276, 24203-24211. [DOI] [PubMed] [Google Scholar]
- 8.Tochio, H., Tsui, M. M., Banfield, D. K. & Zhang, M. J. (2001) Science 293, 698-702. [DOI] [PubMed] [Google Scholar]
- 9.Filippini, F., Rossi, V., Galli, T., Budillon, A., D'Urso, M. & D'Esposito, M. (2001) Trends Biochem. Sci. 26, 407-409. [DOI] [PubMed] [Google Scholar]
- 10.Galli, T., Zahraoui, A., Vaidyanathan, V. V., Raposo, G., Tian, J. M., Karin, M., Niemann, H. & Louvard, D. (1998) Mol. Biol. Cell 9, 1437-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paumet, F., Le Mao, J., Martin, S., Galli, T., David, B., Blank, U. & Roa, M. (2000) J. Immunol. 164, 5850-5857. [DOI] [PubMed] [Google Scholar]
- 12.Mallard, F., Tang, B. L., Galli, T., Tenza, D., Saint-Pol, A., Yue, X., Antony, C., Hong, W., Goud, B. & Johannes, L. (2002) J. Cell Biol. 156, 653-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simpson, F., Peden, A. A., Christopoulou, L. & Robinson, M. S. (1997) J. Cell Biol. 137, 835-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elferink, L. A., Trimble, W. S. & Scheller, R. H. (1989) J. Biol. Chem. 264, 11061-11064. [PubMed] [Google Scholar]
- 15.Baron, U., Freundlieb, S., Gossen, M. & Bujard, H. (1995) Nucleic Acids Res. 23, 3605-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber, T., Zemelman, B. V., McNew, J. A., Westermann, B., Gmachl, M., Parlati, F., Sollner, T. H. & Rothman, J. E. (1998) Cell 92, 759-772. [DOI] [PubMed] [Google Scholar]
- 17.Vojtek, A. B. & Hollenberg, S. M. (1995) Methods Enzymol. 255, 331-342. [DOI] [PubMed] [Google Scholar]
- 18.Bartel, P. L. (1993) in Cellular Interactions in Development: A Practical Approach, ed. Hartley, D. A. (Oxford Univ. Press, Oxford), pp. 153-179.
- 19.Rain, J. C., Selig, L., De Reuse, H., Battaglia, V., Reverdy, C., Simon, S., Lenzen, G., Petel, F., Wojcik, J., Schachter, V., et al. (2001) Nature 409, 211-215. [DOI] [PubMed] [Google Scholar]
- 20.Coco, S., Raposo, G., Martinez, S., Fontaine, J. J., Takamori, S., Zahraoui, A., Jahn, R., Matteoli, M., Louvard, D. & Galli, T. (1999) J. Neurosci. 19, 9803-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raposo, G., Kleijmer, K. J., Posthuma, G., Slot, J. W. & Geuze, H. J. (1997) in Handbook of Experimental Immunology, eds. Herzenberg, L. A., Weir, D. M. & Blackwell, C. (Blackwell, Malden, MA), pp. 1-11.
- 22.Davanger, S., Bock, J. B., Klumperman, J. & Scheller, R. H. (1997) Mol. Biol. Cell 8, 1261-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antonin, W., Dulubova, I., Arac, D., Pabst, S., Plitzner, J., Rizo, J. & Jahn, R. (2002) J. Biol. Chem. 277, 36449-36456. [DOI] [PubMed] [Google Scholar]
- 24.Astarie-Dequeker, C., Carreno, S., Cougoule, C. & Maridonneau-Parini, I. (2002) J. Cell Sci. 115, 81-89. [DOI] [PubMed] [Google Scholar]
- 25.Advani, R. J., Yang, B., Prekeris, R., Lee, K. C., Klumperman, J. & Scheller, R. H. (1999) J. Cell Biol. 146, 765-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dell'Angelica, E. C., Ohno, H., Ooi, C. E., Rabinovich, E., Roche, K. W. & Bonifacino, J. S. (1997) EMBO J. 16, 917-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darsow, T., Burd, C. G. & Emr, S. D. (1998) J. Cell Biol. 142, 913-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blumstein, J., Faundez, V., Nakatsu, F., Saito, T., Ohno, H. & Kelly, R. B. (2001) J. Neurosci. 21, 8034-8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reusch, U., Bernhard, O., Koszinowski, U. & Schu, P. (2002) Traffic 3, 752-761. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura, N., Plutner, H., Hahn, K. & Balch, W. E. (2002) Proc. Natl. Acad. Sci. USA 99, 6755-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersson, J., Fried, G., Lilja, L., Meister, B. & Bark, C. (2000) Eur. J. Cell Biol. 79, 781-789. [DOI] [PubMed] [Google Scholar]
- 32.Dell'Angelica, E. C., Shotelersuk, V., Aguilar, R. C., Gahl, W. A. & Bonifacino, J. S. (1999) Mol. Cell 3, 11-21. [DOI] [PubMed] [Google Scholar]
- 33.Parlati, F., Weber, T., McNew, J. A., Westermann, B., Sollner, T. H. & Rothman, J. E. (1999) Proc. Natl. Acad. Sci. USA 96, 12565-12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peden, A. A., Park, G. Y. & Scheller, R. H. (2001) J. Biol. Chem. 276, 49183-49187. [DOI] [PubMed] [Google Scholar]
- 35.Springer, S. & Schekman, R. (1998) Science 281, 698-700. [DOI] [PubMed] [Google Scholar]
- 36.Lafont, F., Verkade, P., Galli, T., Wimmer, C., Louvard, D. & Simons, K. (1999) Proc. Natl. Acad. Sci. USA 96, 3734-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kantheti, P., Qiao, X., Diaz, M. E., Peden, A. A., Meyer, G. E., Carskadon, S. L., Kapfhamer, D., Sufalko, D., Robinson, M. S., Noebels, J. L. & Burmeister, M. (1998) Neuron 21, 111-122. [DOI] [PubMed] [Google Scholar]
- 38.Swank, R. T., Reddington, M., Howlett, O. & Novak, E. K. (1991) Blood 78, 2036-2044. [PubMed] [Google Scholar]
- 39.Novak, E. K. & Swank, R. T. (1979) Genetics 92, 189-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.