Abstract
Prochlorophytes are a class of cyanobacteria that do not use phycobiliproteins as light-harvesting systems, but contain chlorophyll (Chl) a/b-binding Pcb proteins. Recently it was shown that Pcb proteins form an 18-subunit light-harvesting antenna ring around the photosystem I (PSI) trimeric reaction center complex of the prochlorophyte Prochlorococcus marinus SS120. Here we have investigated whether the symbiotic prochlorophyte Prochloron didemni also contains the same supermolecular complex. Using cells isolated directly from its ascidian host, we found no evidence for the presence of the Pcb–PSI supercomplex. Instead we have identified and characterized a supercomplex composed of photosystem II (PSII) and Pcb proteins. We show that 10-Pcb subunits associate with the PSII dimeric reaction center core to form a giant complex having an estimated Mr of 1,500 kDa with dimensions of 210 × 290 Å. Five-Pcb subunits flank each long side of the dimer and assuming each binds 13 Chl molecules, increase the antenna size of PSII by ≈200%. Fluorescence emission studies indicate that energy transfer occurs efficiently from the Pcb antenna. Modeling using the x-ray structure of cyanobacterial PSII suggests that energy transfer to the PSII reaction center is via the Chls bound to the CP47 and CP43 proteins.
Prochloron didemni, Prochlorothrix hollandica, and Prochlorococcus marinus are oxygenic photosynthetic prokaryotes known as prochlorophytes. They are closely related to cyanobacteria but differ in that they contain chlorophyll (Chl) a/b light-harvesting systems rather than phycobiliproteins (1). Their discovery has been discussed in terms of the endosymbiotic theory, which suggests that the Chl a/b containing chloroplasts of plants and green algae originate from free living oxygenic photosynthetic prokaryotes (2). Prochloron is symbiotic with didemnid ascidians of tropical waters (3, 4) and as yet there is no report of its existence as a free-living organism (5). In contrast, Prochlorothrix is a freshwater free-living filamentous prochlorophytes (6), whereas Prochlorococcus is a highly abundant, free-living marine phytoplankton (7).
Based on small subunit ribosomal (16S) RNA, all of these organisms fall within the clade of extant cyanobacteria (8); thus there must have been greater diversity of light-harvesting pigments within the cyanobacteria than previously conceived. This is reinforced by the recent discovery of a group of oxygenic photosynthetic bacteria containing Chl d as a major pigment (9), which also fall within the cyanobacterial clade, based on 16S rRNA analysis (H. Miyashita, personal communication).
In 1996 the sequence of the pcb genes encoding the Chl a/b-binding proteins of prochlorophytes was reported (10). Surprisingly, the sequence was found to be very different to that of the cab genes that encode the Chl a/b-binding proteins of chloroplasts (11). In fact, the pcb gene sequence was similar to that of the gene that encodes the Chl a-binding protein CP43 of photosystem II (PSII) and the IsiA protein induced in cyanobacteria under conditions of limiting iron levels (12, 13). Recent studies (14–16) have shown that the CP43-like IsiA protein forms a ring of 18 subunits around the cyanobacterial trimeric photosystem I (PSI) reaction center core complex. In so doing it increases the light-harvesting capacity of PSI by ≈80%. Following on from this, it was discovered that a similar light-harvesting antenna ring is present in a low light adapted strain SS120 of P. marinus (17). In this case, the 18 subunits were Pcb proteins containing both Chl a and Chl b, and the presence of this antenna ring did not depend on iron deficiency. In light of this finding, we set out to investigate whether the closely related organism P. didemni contained a similar Pcb–PSI supercomplex and to establish whether the 18 subunit antenna ring of PSI was a common feature of Chl b containing prochlorophytes. In so doing we have found that for Prochloron, isolated from its native environment, Chl a/b binding Pcb protein subunits associate with PSII rather than PSI. Here we report the structural analysis of this association.
Materials and Methods
Organism. At present P. didemni cannot be cultured (5, 18). Therefore colonies of the ascidian Lissoclimum patella containing the Prochloron cells as symbionts were collected from a depth of 2–4 m in the “Blue pools” area of the Heron Island Reef (Great Barrier Reef, Australia). Prochloron cells were squeezed out of cut ascidian colonies into buffered seawater (0.1 M Tris, pH 9.2). The seawater was taken directly from the Heron Island Reef and passed through a sterile filter before use. The cells were collected by low-speed centrifugation (1,000 × g) and washed with buffered seawater (50 mM Mops, pH 7.5). The concentrated cells were then transferred within 24 h on ice to the University of Sydney for the extraction of thylakoids.
Isolation of Thylakoid Membranes. The procedure used was as described (19). Freshly harvested cells were suspended in seawater (50 mM Hepes, pH 7.8) or aqueous buffer (50 mM Hepes, pH 7.8/15 mM NaCl/5 mM MgCl2/20% glycerol) and passed twice through a prechilled French press at a pressure of ≈100 MPa. The resulting unbroken cells and debris were removed from the French press extracts by centrifugation with a JA-10 Beckman rotor at 1,000 × g for 10 min at 4°C. By using a Ti70 Beckman rotor, thylakoid membranes were pelleted at 10,000 × g and stored in 50 mM Mes, pH 6.0 at -70°C. For experiments samples were rapidly thawed and washed twice with an aqueous buffer (50 mM Mes-NaOH, pH 6.0/10 mM CaCl2/5 mM MgCl2/20% glycerol/1 mM PMSF).
Isolation and Purification of Supercomplexes. The thylakoid membrane fraction of Prochloron was solubilized for 10 min in the dark at 4°C by using 1% β-dodecyl maltoside. The solubilized complexes were separated on continuous sucrose density gradients generated by the freeze-thawing technique (20) containing 50 mM Mes-NaOH (pH 6.0), 500 mM betaine, 20 mM CaCl2, 2.5 mM MgCl2, and 0.03% β-dodecyl maltoside and ultracentrifuged at 90,000 × g for 16 h at 4°C (SW28 Beckman rotor). The procedure was similar to that used previously for isolating PSI supercomplexes from Synechocystis PCC 6803 (14, 15) and Prochlorococcus SS120 (17). The green pigment-containing fractions were removed carefully by using a syringe.
Spectral Analysis. Room temperature absorption spectra were performed by using a Shimadzu UV-1601 spectrophotometer. The room and low temperature (77 K) fluorescence spectra were recorded by using a Perkin–Elmer LS50 luminescence spectrometer (5-nm slit width for emission) with an excitation wavelength of 430 nm.
Chl Analysis. HPLC was used to determine Chl a/b ratios following a published procedure (21). Pigment was extracted into 100% acetone and subjected to HPLC analysis by using a Kontron Spherisorb ODS-1 C18 reverse-phase column (Zurich) and calibrated with purified Chl a and Chl b obtained from Sigma.
SDS/PAGE Analysis. The polypeptide composition of supercomplexes isolated by sucrose density gradient centrifugation was resolved by Tricine-SDS/PAGE (10%) with 6 M urea (22). Before the samples were loaded onto the gels, they were denatured by incubating with 50 mM Tris·HCl (pH 6.8), 4% (wt/vol) SDS, 12% sucrose, and 100 mM DTT at room temperature for 1 h. Low molecular weight makers (Bio-Rad) were used to estimate polypeptide size.
Immunoblotting and N-Terminal Sequencing. Western blotting was conducted by using a published procedure (20) with Abs raised to the PSII reaction center PsbA (D1) protein of Synechocystis 6803 and to the reaction center proteins of PSI (PsaA) of Chlamydomonas reinhardtii (kindly provided by P. Nixon, Imperial College). Polypeptides separated by SDS/PAGE were transferred to poly(vinylidene difluoride) membrane before identification by N-terminal sequencing conducted by J. Keen (University of Leeds).
Electron Microscopy and Image Processing. Electron microscopy analyses were carried out by negatively staining the samples with 2% (wt/vol) uranyl acetate and imaging at room temperature by using a Philips CM100 electron microscope set to a calibrated magnification of ×51,500. The 14 best micrographs for each sample, displaying minimal or no discernible drift or astigmatism, were digitized on a Leafscan 45 densitometer at a step size of 10 μm. The resultant sampling frequency of 1.94 Å per pixel on the specimen scale was coarsened to 3.88 Å per pixel to aid in the speed of subsequent processing by using the imagic-5 software environment (23, 24). On calculation of the contrast transfer function, the first zero-crossing for each micrograph was consistently found to be in the range of 18.6–21.3 Å. We identified and analyzed populations of particles in the heaviest sucrose gradient fractions. Application of classification procedures (25) enabled a number of subpopulations within each dataset to be identified, differing in both size and orientation. Each subpopulation was treated de novo and iteratively refined, leading to improved 2D class averages.
Modeling Studies. All studies were conducted by using the O software package (26), with structural coordinates derived from the 3.8-Å resolved model of PSII (27) designated under Protein Data Bank ID code 1FE1 (www.rcsb.org/) elucidated from the x-ray diffraction of crystals of PSII isolated from Synechococcus elongatus. Subunits attributed to Pcb protein were modeled by using the coordinates assigned to the six transmembrane helices of CP43 in 1FE1 (27), given the high structural homology known to exist between these two protein families (15).
Results
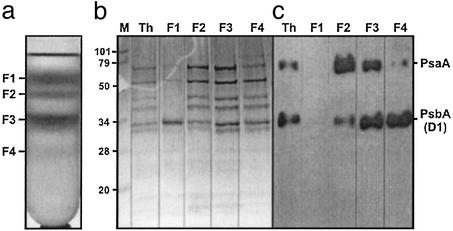
Biochemistry. Thylakoid membranes solubilized with β-dodecyl maltoside were subjected to sucrose density centrifugation with the view of isolating high molecular weight supercomplexes containing Pcb protein. Fig. 1a shows that four fractions (F1-F4) were resolved. SDS/PAGE analyses (Fig. 1b) suggested that PSI and PSII proteins were present in fractions F2, F3, and F4 as confirmed by immunological blotting analyses (Fig. 1c) by using Abs raised to the reaction center proteins of PSI (PsaA) and PSII (PsbA/D1 protein). The immunological blotting, however, indicated that F2 was enriched in PSI, whereas F4 was highly enriched in PSII. The F1 fraction was dominated by a protein having an apparent Mr of ≈34 kDa, and N-terminal sequencing identified this as a Pcb protein. This Pcb protein was also present in the other three fractions.
Fig. 1.
Sucrose density gradient and characterization of proteins in the green fractions. (a) Sucrose density gradient showing separation of four clear fractions, F1–F4. (b) Coomassie blue-stained SDS/PAGE of the thylakoid membrane and fractions obtained by sucrose density gradient centrifugation. (c) Immunoblotting with polyclonal Abs raised to PsaA PSI proteins of Chlamydomonas and PsbA (D1) PSII protein of Synechocystis 6803. Lanes 1–4 are for fractions F1–F4 as identified by sucrose density centrifugation. Th, thylakoids; M, molecular mass markers (in kDa).
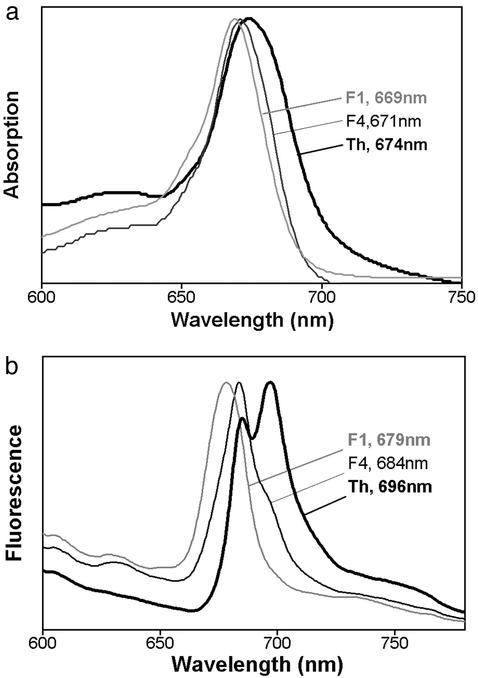
Spectroscopic Analyses. Pigment extraction and HPLC analysis gave a Chl a/b ratio of ≈5 for the isolated thylakoids and therefore the long-wavelength room temperature (RT) absorption peak was at 674 nm and dominated by Chl a (Fig. 2a). This agrees with the findings of Christen et al. (19), but higher Chl b levels can be found in this organism depending on the light intensity to which the cells are exposed in the ascidian host (31). The Chl a/b ratio for the Pcb-rich F1 fraction was 3 and the absorption peak was blue-shifted to 669 nm (Fig. 2a). This free Pcb fraction had an emission peak at 676 nm at RT (data not shown) and at 679 nm at 77 K (see Fig. 2b). In contrast, the fluorescence emission from thylakoids was maximum at 682 nm at RT (data not shown) and 684 and 696 nm at 77 K (see Fig. 2b). The low temperature peaks are typical of PSII emission, 685 nm from CP43 and 697 nm from CP47 (32). As noted (19), no significant low-temperature emission band for PSI was present, contrasting with cyanobacterial and higher plant PSI. The absence of significant 679-nm Pcb fluorescence at 77 K from thylakoids suggests that the Pcb proteins transfer energy efficiently to reaction centers in line with their role as a light-harvesting system.
Fig. 2.
Spectral properties of Prochloron thylakoids (Th) and sucrose density fractions (F1 and F4). (a) RT absorption spectra giving long wavelength maxima. (b) A 77-K fluorescence emission spectra giving wavelengths of the maximum peaks.
The heavy fraction (F4) had a red absorption maximum at 671 nm (Fig. 2a) and a RT emission maximum of 679 nm (data not shown). Importantly the low-temperature fluorescence spectrum of this heavy fraction peaked at 684 nm with no significant peak or shoulder at 679 nm due to free or excitonically uncoupled Pcb proteins (Fig. 2b). Moreover a shoulder at 697 nm confirms the presence of PSII in this fraction. The long wave-length absorption maxima of F2 and F3 were at 671 and 672 nm, respectively, and their RT and 77 K emission spectra were dominated by fluorescence from free Pcb protein or Pcb proteins not fully functionally associated with reaction centers.
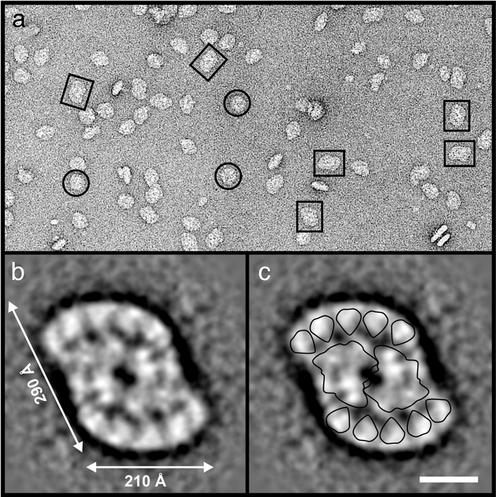
Electron Microscopy. The Pcb–PSI supercomplex isolated from Prochlorococcus SS120 with its 18-Pcb subunits is a giant circular particle having a diameter of 320 Å (17). Despite using the same solubilization and isolation procedures for Prochloron as used for Prochlorococcus SS120, we observed no such particle in the heaviest fractions, F4 or F3, or any particle that could have been derived from a partial degradation of this PSI supercomplex. We did, however, identify a circular particle having a diameter of ≈220 Å (circled in Fig. 3a), which was at low frequency in the F4 fraction but common in F3. We assign this particle to a PSI trimer based on its similarity to the PSI trimer of cyanobacteria (28) (see below and Fig. 4 a and b). However, an oblong-shaped particle having dimensions of 210 × 290 Å was observed in F4 (boxed in Fig. 3a). This represented the largest and most common particle in F4. Based on past experience of viewing PSII core dimers (29) and PSI supercomplexes containing Pcb proteins (17) in the electron microscope, we assign the large Prochloron structures boxed in Fig. 3a as being PSII supercomplexes. We interpret these supercomplexes to consist of a central PSII dimer and associated Pcb proteins (Fig. 3b). By overlaying the outlines for the PSII dimer of cyanobacterial PSII and for CP43 based on x-ray models [the latter as an analog of a Pcb protein (15, 30)], the large particles are seen to consist of a core dimer with a flanking region accommodating five Pcb proteins along each edge, making a total of 10 Pcb subunits per PSII dimer (Fig. 3c). Assuming a Mr of 850 kDa for the PSII dimer and 46 kDa for the Pcb protein, to include the bound Chls, then the estimated Mr of this large particle is ≈1,500 kDa.
Fig. 3.
Electron micrographs and image analysis of a total of 2,590 particle data set contained in fraction 4 (F4) of the sucrose density gradient. (a) Negatively stained particles are identified as large Pcb–PSII supercomplexes (boxed) and PSI trimers (circled). (b) Top view of Pcb–PSII supercomplex obtained by image processing of 713 particles of the type boxed in a. (c) Pcb–PSII supercomplex with outlines of the core dimer and CP43 (for each Pcb subunit) derived from x-ray analysis of PSII isolated from the cyanobacterium S. elongatus (27). (Bar, 100 Å.)
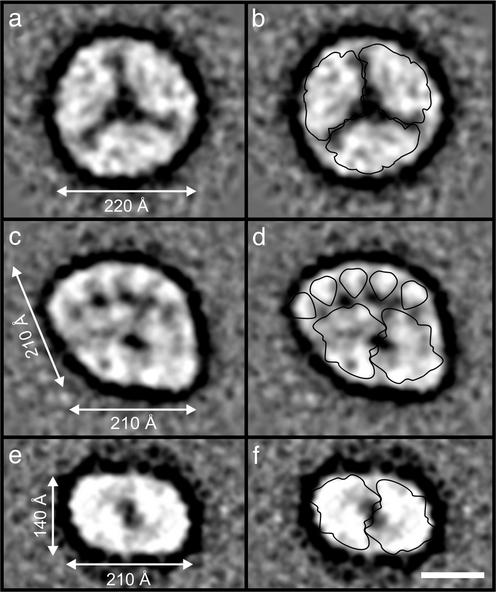
Fig. 4.
Averaged projection maps of particles present in F3 fraction of the sucrose density gradient (see Fig. 1). (a) Top view of the PSI trimer after image-processing 600 particles. (b) Overlay of outline of the PSI trimer derived from x-ray data of Jordan et al. (28). (c) Top view of a partially degraded Pcb–PSII supercomplex after processing 681 particles. (d) Overlay of outlines of the core dimer and CP43 (for each Pcb subunit) derived from x-ray analysis of PSII isolated from the cyanobacterium S. elongatus (27). (e) Top view of PSII core dimers after processing 231 particles. (f) Overlay of outline of PSII core dimer from ref. 27. (Bar, 100 Å.)
Although the structure described above is representative of the majority of particles in fraction F4 of the sucrose density gradient, there were other PSII particles within the F3 band that seemed to have lost some or all of their flanking Pcb subunits. We interpret the particle shown in Fig. 4c, e.g., as a PSII dimer with 5-Pcb subunits missing from one side, having dimensions of 210 × 210 Å. This interpretation is supported by overlaying the outline of the x-ray structures (Fig. 4d) and is consistent with the assignments within the complete Pcb–PSII supercomplex as shown in Fig. 3c. Fig. 4 e and f shows the top view of a PSII core dimer present in F3 with no Pcb subunits attached. Also contained within the F3 fraction, as mentioned above, were PSI trimers observed at a higher frequency than in F4, shown as a processed image in Fig. 4a with an overlay of the outline of the x-ray structure of a PSI trimer from S. elongatus (28) in Fig. 4b. When viewed in the electron microscope, F2 seems to consist mainly of monomers of PSI although some dimeric PSII particles were present in this fraction.
Discussion
Although a Pcb–PSI supercomplex consisting of 18 subunits of Chl a/b-binding Pcb protein surrounding a trimeric PSI reaction center core has been found to occur in the prochlorophytes P. marinus SS120 (17), we were unable to detect a similar PSI supercomplex in Prochloron, despite adopting the same solubilization and isolation procedures. It should be noted, however, that Prochloron contains an isiA-like gene (10) and perhaps under iron stress conditions or other environmental variables, such a PSI supercomplex assembles, as in the case of cyanobacteria (14–16). At present it is not possible to test this hypothesis by using Prochloron because there is no reported culturing procedure. However, we have identified a large supercomplex that has Pcb proteins associated with PSII. This supercomplex in Prochloron is the largest particle present anywhere in the sucrose density gradient, and we interpret it to be a single PSII core dimer with five Pcb proteins attached to each side. Consistent with this conclusion is the fact that the particle is found in a sucrose density fraction, F4, which is enriched in PSII relative to PSI. Both room and low temperature fluorescence characteristics of the fraction containing the Pcb–PSII supercomplex suggest that energy is transferred efficiently from the Pcb proteins to PSII, indicating it is a functional photosynthetic unit within the thylakoid membrane.
Assuming the PSII core monomer binds 35 Chl and Pcb proteins are like CP43 and bind 13 Chls (33), then the Pcb antenna system of the supercomplex detected in F4 increases the light-harvesting capacity of PSII by almost 200%. This is a substantial increase in the absorption cross section of PSII and presumably compensates for the lack of phycobiliproteins in this organism. Thus the situation in Prochloron anticipates very closely that found in green plastids of higher plants, where the major light-harvesting Chl-binding proteins are attached to PSII (11), as predicted from previous studies of Prochloron (19, 34).
Interestingly, in no case was a complete ring of Pcb proteins observed around a PSII core dimer, which contrasts with the arrangement of Pcb proteins around PSI in Prochlorococcus SS120 (17). The fact that some PSII core dimers had <10 Pcb proteins associated with them is probably the consequence of the detergent solubilization procedure used, which, although very mild, did bring about the release of some Pcb proteins, which were clearly detected in the F1 fraction. This partial and in some cases complete stripping down of the Pcb–PSII supercomplex gave rise to subpopulations, particularly in the F2 and F3 fractions.
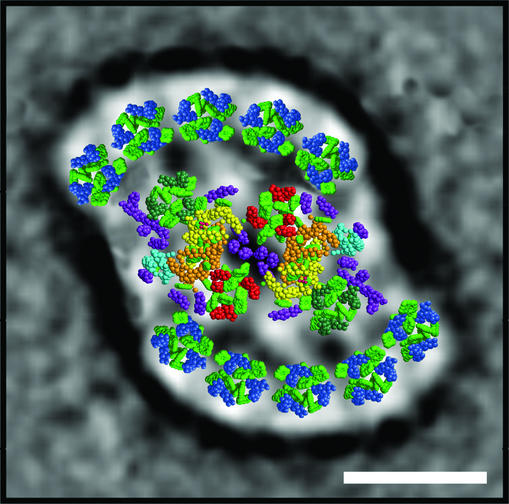
The work presented here represents the structural description of the association of Pcb proteins with PSII core dimers and potentially may be representative of Pcb–PSII complexes in other related prochlorophytes. The model shown in Fig. 5, in which the x-ray data of Zouni et al. (27) has been incorporated, is a representation of the largest Pcb–PSII supercomplex that we detected in Prochloron, but we cannot be certain that it is the largest complex in vivo. However, this model shows that the Pcb subunits closest to PSII are adjacent to CP47 and CP43. This arrangement shows that the minimum Chl-Chl distances between the Pcb antenna and CP47 and CP43 subunits are 10–15 and ≈20 Å, respectively. Chl molecules in the other Pcb subunits are distant from those within the PSII core, suggesting that energy migration occurs between the Pcb antenna proteins before being transferred to the PSII reaction center. Note that the position of the Pcb proteins within the density of the projection map in Fig. 5 has allowed for a detergent layer of ≈10 Å, which is typical for negatively stained membrane protein complexes (35).
Fig. 5.
The structure of the largest Pcb–PSII supercomplex isolated from Prochloron, incorporating the x-ray structures of the PSII core dimer and CP43 of S. elongatus (27), emphasizing the positioning of the Chl molecules (shown in light green) and transmembrane helices. CP43 is used as a model for the Pcb subunits with transmembrane helices shown in blue and with helices 5 and 6 facing the PSII core dimer. In the case of the PSII core dimer, the color coding for the transmembrane helices is: yellow, D1 protein; orange, D2 protein; red, CP47; dark green, CP43; cyan, cytochrome b559; purple, low molecular weight subunits. (Bar, 100 Å.)
Whether the lack of a complete Pcb ring around PSII is a general feature required to facilitate quinone diffusion from the QB site of PSII to the cytochrome b6f complex has yet to be determined, but this idea has often been argued for the possible lack of a complete light harvesting one antenna ring around the purple bacterial reaction center (36). Interestingly the thylakoid membranes of Prochloron are organized into stacked and unstacked regions (37) reminiscent of the grana and stromal lamellae of higher plant chloroplasts. It is possible that the Pcb–PSII supercomplex is located in the stacked regions because there is some evidence that there is lateral separation between PSI and PSII in the thylakoid membranes of prochlorophytes equivalent to that found in higher plant chloroplasts (38).
Acknowledgments
We thank James Duncan for helping with the Chl analyses. J.B. and A.W.D.L. acknowledge the Biotechnology and Biological Research Council and Australian Research Council, respectively, for financial support. J.N. holds a Royal Society University fellowship.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PSI and PSII, photosystems I and II, respectively; Chl, chlorophyll; RT, room temperature.
References
- 1.Matthijs, H. C. P., van der Staay, G. W. M. & Mur, L. R. (1994) in The Molecular Biology of Cyanobacteria, ed. Bryant, D. A. (Kluwer, Dordrecht, The Netherlands), pp. 49-64.
- 2.Margulis, L. (1981) Symbiosis in Cell Evolution (Freeman, San Francisco).
- 3.Lewin, R. A. (1976) Nature 261, 697-698. [DOI] [PubMed] [Google Scholar]
- 4.Newcomb, E. H. & Pugh, T. D. (1975) Nature 253, 533-534. [Google Scholar]
- 5.Kühl, M. & Larkum, A. W. D. (2002) in Cellular Origin and Life in Extreme Habitats, ed. Seckbach, J. (Kluwer, Dordrecht, The Netherlands), Vol. 3, pp. 273-290. [Google Scholar]
- 6.Burger-Wiersma, T., Voenhuis, M., Korthals, H. J., van de Wiel, C. C. M. & Mur, L. R. (1986) Nature 320, 262-264. [Google Scholar]
- 7.Chisholm, S. N., Olson, R. J., Zettler, E. R., Goericke, R., Waterbury, J. B. & Welschmeyer, N. A. (1988) Nature 344, 340-343. [Google Scholar]
- 8.Turner, S. (1997) Plant Syst. Evol. Suppl. 11, 43-52. [Google Scholar]
- 9.Miyashita, H., Ikemoto, H., Kurano, N., Adachi, K., Chihara, M. & Miyachi, S. (1996) Nature 383, 402. [Google Scholar]
- 10.La Roche, J., van der Staay, G. W. M., Partensky, F., Durret, A., Aebersold, R., Li, R., Golden, S. S., Hiller, R. G., Wrench, P. M., Larkum, A. W. D., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 15244-15248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green, B. R. & Durnfold, D. G. (1996) Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 685-714. [DOI] [PubMed] [Google Scholar]
- 12.Pakrasi, M. B., Goldenberg, A. & Sherman, L. A. (1985) Plant Physiol. 79, 290-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Straus, N. A. (1994) in The Molecular Biology of Cyanobacteria, ed. Bryant, D. A. (Kluwer, Dordrecht, The Netherlands), pp. 731-750.
- 14.Bibby, T. S., Nield, J. & Barber, J. (2001) Nature 412, 743-745. [DOI] [PubMed] [Google Scholar]
- 15.Bibby, T. S., Nield, J. & Barber, J. (2001) J. Biol. Chem. 276, 43246-43252. [DOI] [PubMed] [Google Scholar]
- 16.Boekema, E. L., Hifneg, A., Yakushevska, A. E., Piotrowski, M., Keegstra, W., Berry, S., Michel, K. P., Pistoius, E. K. & Kruip, J. (2001) Nature 412, 745-748. [DOI] [PubMed] [Google Scholar]
- 17.Bibby, T. S., Nield, J., Partensky, F. & Barber, J. (2001) Nature 413, 590. [DOI] [PubMed] [Google Scholar]
- 18.Lewin, R. A. & Cheng, L. (1989) Prochloron: A Microbiological Enigma (Chapman & Hall, New York).
- 19.Christen, G., Stevens, G., Lukins, P. B., Renger, G. & Larkum, A. W. D. (1999) FEBS Lett. 449, 264-268. [DOI] [PubMed] [Google Scholar]
- 20.Hankamer, B., Nield, J., Zheleva, D., Boekema, E. J., Jansson, S. & Barber, J. (1997) Eur. J. Biochem. 243, 422-429. [DOI] [PubMed] [Google Scholar]
- 21.Zheleva, D., Hankamer, B. & Barber, J. (1996) Biochemistry 35, 15074-15079. [DOI] [PubMed] [Google Scholar]
- 22.Schagger, H. & von Jagow, G. (1987) Anal. Biochem. 166, 368-379. [DOI] [PubMed] [Google Scholar]
- 23.van Heel, M., Schatz, M. & Orlova, E. (1992) Ultramicroscopy 46, 309-316. [Google Scholar]
- 24.van Heel, M., Gowen, B., Matadeen, R., Orlova, E. V., Finn, R., Pape, T., Cohen, D., Stark, H., Schmidt, R., Schatz, M. & Patwardhan, A. (2000) Q. Rev. Biophys. 33, 307-369. [DOI] [PubMed] [Google Scholar]
- 25.Ruprecht, J. & Nield, J. (2001) Prog. Biophys. Mol. Biol. 75, 121-164. [DOI] [PubMed] [Google Scholar]
- 26.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 27.Zouni, A., Witt, H. T., Kern, J., Fromme, P., Krauss, N., Saenger, W. & Orth, P. (2001) Nature 409, 739-743. [DOI] [PubMed] [Google Scholar]
- 28.Jordan, P., Fromme, P., Witt, H. T., Klukas, O., Saenger, W. & Krauss, N. (2001) Nature 411, 909-917. [DOI] [PubMed] [Google Scholar]
- 29.Nield, J., Kruse, O., Ruprecht, J., da Fonseca, P., Büchel, C. & Barber, J. (2000) J. Biol. Chem. 275, 27940-27946. [DOI] [PubMed] [Google Scholar]
- 30.Hiller, R. G. & Larkum, A. W. D. (1985) Biochim. Biophys. Acta 806, 107-115. [Google Scholar]
- 31.Alberte, R. S., Cheng, L. & Lewin, R. A. (1986) Mar. Biol. (Berlin) 90, 575-587. [Google Scholar]
- 32.Bricker, T. M. & Frankel, L. K. (2002) Photosynth. Res. 72, 131-146. [DOI] [PubMed] [Google Scholar]
- 33.Vasil'ev, S., Orth, P., Zouni, A., Owens, T. G. & Bruce, D. (2001) Proc. Natl. Acad. Sci. USA 98, 8602-8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schreiber, U., Ralph, P., Gademann, R. & Larkum, A. W. D. (1997) Plant Cell Physiol. 38, 945-951. [Google Scholar]
- 35.Boekema, E. J., Hankamer, B., Bald, D., Kruip, J., Nield, J., Boonstra, A. F., Barber, J. & Rögner, M. (1995) Proc. Natl. Acad. Sci. USA 92, 175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jungas, C., Ranck, J. L., Rigauld, J. L., Joliot, P. & Vermeglio, A. (1999) EMBO J. 18, 534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giddings, T. R., Jr., Withers, N. W. & Staehelin, A. L. (1980) Proc. Natl. Acad. Sci. USA 77, 352-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Staay, G. W. M. & Staehelin, A. L. (1994) J. Biol. Chem. 269, 24834-24844. [PubMed] [Google Scholar]