Abstract
Genomic regions of nearly every species diverged into different haplotypes, mostly based on point mutations, small deletions, and insertions that do not affect the collinearity of genes within a species. However, the same genomic interval containing the z1C gene cluster of two inbred lines of Zea mays significantly lost their gene collinearity and also differed in the regulation of each remaining gene set. Furthermore, when inbreds were reciprocally crossed, hybrids exhibited an unexpected shift of expression patterns so that “overdominance” instead of “dominance complementation” of allelic and nonallelic gene expression occurred. The same interval also differed in length (360 vs. 263 kb). Segmental rearrangements led to sequence changes, which were further enhanced by the insertion of different transposable elements. Changes in gene order affected not only z1C genes but also three unrelated genes. However, the orthologous interval between two subspecies of rice (not rice cultivars) was conserved in length and gene order, whereas changes between two maize inbreds were as drastic as changes between maize and sorghum. Given that chromosomes could conceivably consist of intervals of haplotypes that are highly diverged, one could envision endless breeding opportunities because of their linear arrangement along a chromosome and their expression potential in hybrid combinations (“binary” systems). The implication of such a hypothesis for heterosis is discussed.
Keywords: z1C zein gene cluster, heterosis, genomic organization, overdominance, transposition
Corn or Zea mays is one of the most important crops in the world. Although wheat and rice surpass it as a staple and in acreage, no crop rivals its total grain yield and the diversity of its uses as processed food, for animal feed and sweetener in soft drinks, and its industrial applications such as fuel and adhesives. The preference of corn over other grains or legumes for such applications is because of the unusual properties of hybrids. When distant inbred lines are crossed, the resulting hybrids exhibit an unusual vigor (heterosis) that results in higher yields per acre than the parental lines would produce themselves (1). The premium exercised for the breeding of steadily improved maize inbreds customized for different geographic areas has generated a huge enterprise worldwide, particularly in the United States, yet the underlying molecular mechanism for hybrid vigor is not well understood. To decide between two models explaining “dominance complementation” or “overdominance” of heterozygotes, it will therefore be necessary to compare not only the genomic architecture of genes within an interval of several hundred kilobases but also the expression of these genes of two parental inbreds and their reciprocal hybrids.
The fact that some gene families are highly clustered within such lengthy intervals rather than spread over the entire genome is the springboard for our study. One such interval encodes the 22-kDa α-zein storage proteins. These proteins accumulate during maize seed development and provide an important nitrogen source for germinating seedlings in the next generation (2). They are also an important source for essential amino acids for human and animal nutrition. α-Zeins represent the major fraction of storage proteins in maize endosperm and are subdivided into four subfamilies: z1A, z1B, and z1D, each having a Mr of 19 kDa, and z1C, having a Mr of 22 kDa. In inbred B73, the three 19-kDa subfamilies are composed of 25 genes in total and occupy five different genomic locations (3). The genes for the z1C or 22-kDa subfamily have also been analyzed but only in inbred BSSS53, where they are located at two different sites of the short arm of chromosome 4 (4). The z1C-1 locus is composed of 22 gene copies arranged in a roughly tandem fashion within 170 kb. The z1C-2 locus is ≈20 centimorgans closer to the centromeric region and consists of a single copy. This locus is allelic to the floury2 allele, which is a mutation that interferes with the deposition of zeins in protein bodies (5). Of the 23 z1C copies in BSSS53, only seven produce detectable levels of mRNAs. Interestingly, a different inbred, IHO90, is lacking transcripts for three of the seven genes, whereas another inbred, Ohio43, is lacking transcripts for the two most recently amplified genes (4).
Earlier we compared the z1C-1 locus in maize to closely related grass species (sorghum and two subspecies of rice) at the DNA sequence level (6). The orthologous regions in maize, sorghum, and rice differed in gene content and intergenic structure, offering a mosaic organization of collinear and noncollinear genes. (The orthologous regions did not differ between the two rice subspecies.) However, when orthologous regions between other maize inbred lines were compared, the level of divergence resembled that between closely related species (7–9). To accurately assess the extent to which maize diverged from its close relatives, we needed to compare the same orthologous regions at the interspecies and intraspecies levels.
Accordingly, we surveyed the genomic organization of z1C gene loci among different common maize inbreds, using DNA probes specific to different sections of the region. Although the restriction patterns of these regions differed among common corn-belt inbred lines, they appeared to fit into a few major haplotypes, e.g., BSSS53 and B73. Having already sequenced the genomic regions of the z1C genes in BSSS53, we might expect analysis of the z1C loci in B73 to provide another haplotype. It should also provide the genomic organization of a complete set of α-zein genes within the same inbred. Cloning and sequencing the B73 z1C genomic regions gave us positional information for each gene in each inbred. We then analyzed gene expression in inbreds and reciprocal hybrid crosses. Inbreds differed in their gene content and also in their allelic and nonallelic interactions when crossed with each other. Whatever features emerged should help us explain the superior heterotic properties of maize when compared to closely related crops.
Materials and Methods
Plant Materials. Our laboratory stocks maize inbred lines A188, A632, A636, A654, B37, B73, BSSS53, Mo17, Ill12E, W22, W23, and W64A, all are available from the National Plant Germplasm System (www.ars-grin.gov/npgs). Teosinte stocks (Zea mexicana, Zea parviglumis, and Zea luxurians) and the maize inbred line core collection (including 43 inbred lines) are also available from the National Plant Germplasm System. Genomic DNA samples from the inbred line core collection were a gift from Binzhang Shen (Harvard University, Boston).
Maize Bacterial Artificial Chromosome (BAC) Library Screening and BAC Clone Sequencing. A HindIII BAC library constructed from maize inbred line B73 was used for screening clones containing z1C gene sequences (10). High-density filter screening and BAC clone characterization were conducted as described (3). Two BAC clones (Z277I24 and Z190G16) were chosen for sequencing by the “shot-gun” strategy as described (3).
Sequence Analysis. The fgenesh program (Softberry, Mount Kisco, NY) was used for gene prediction analysis. The blastn program (www.ncbi.nlm.nih.gov/blast/) was used for sequence homology searches. The blast2 program (www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html) was used for sequence comparison between the B73 and BSSS53 z1C gene clusters.
The z1C Gene Expression Analysis. Maize endosperm tissue was dissected at 18 days after pollination (DAP), when z1C gene expression reaches its peak. Total RNA was extracted by using TRIzol reagent (Invitrogen). A Poly(A) Tract kit (Promega) was used to extract mRNA, which was transcribed with the reverse transcription kit of Promega. The reverse transcription mixtures were used as a template for the PCR with a universal z1C general primer set (5′ primer: ACACCATATGTTCATTATTCCACAATGCTCA; 3′ primer: TTAAGGATCCTATATAATCTAAAAGATGGCA) flanking the coding region of a z1C gene. PCR products (≈740 bp) were cloned into the pGEM-T vector (Promega). Two and three different sets of experiments were carried out for inbreds and hybrids, respectively. For each set, both ends of 96–192 random clones from RT-PCR libraries were sequenced by using universal primers and BigDye terminator chemistry (Applied Biosystems). Ten different sets of experiments yielded a total 2,689 high-quality sequence reads (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). Because zein genes do not have introns, RT-PCR sequences were directly aligned with genomic z1C sequences by using the megalign program of the Lasergene DNA sequence analysis package (DNASTAR, Madison, WI). Confidence intervals for each gene and each cross were calculated online (http://igs-server.cnrs-mrs.fr) by using the statistical method of Audic and Claverie (11). See Supporting Materials and Methods, which is published as supporting information on the PNAS web site, for additional methods.
Results
Haplotypes of the z1C Loci in Maize Inbreds and Teosinte. Is the variability of zein gene expression among maize inbred lines caused in part by the genomic organization of zein genes in different inbreds? From our previous analysis of the members of the z1C zein gene family, we chose a mix of sequences that could detect all of the z1C gene sequences in the genome, using them as a hybridization probe in Southern blot analysis of restriction fragment patterns of common maize inbred lines (Fig. 4, which is published as supporting information on the PNAS web site). From this analysis, many lines appear to share similar restriction patterns, indicating that the two loci for z1C genes, z1C-1 and z1C-2, might be composed of only a few haplotypes. According to the z1C-banding pattern, three major haplotypes (BSSS53, B73, and W64A) appear to be present in these inbred lines. We also included in this comparison some teosinte stocks to see whether other haplotypes existed in wild relatives of maize. Two different teosinte samples from the same stock showed differing z1C patterns. However, the extra z1C bands could simply reflect heterozygosity because teosinte stocks are highly heterogeneous and usually heavily contaminated by maize stocks.
Isolation of z1C Regions from Inbred Line B73. If we could determine contiguous genomic sequences comprising the loci of z1C zein in B73 that we had already determined from inbred line BSSS53 (4), we could make a high-resolution analysis of two haplotypes of a gene family and a gene-dense chromosomal region in maize. Using the same probes as described for the corn-belt inbred lines, we screened a maize HindIII BAC library constructed from inbred line B73 (10). We identified 15 positive BAC clones containing z1C gene sequences and/or the php200725 marker (Table 3, which is published as supporting information on the PNAS web site) and subjected them to DNA fingerprinting. The DNA fingerprints fell into two groups: six BAC clones contained a single copy of a z1C gene allelic to the floury2 locus (z1C-2 locus), and eight BAC clones contained the remaining z1C gene copies from the z1C gene cluster (z1C-1 locus). These noncontiguous genomic positions are consistent with the previously mapped loci in BSSS53. We also compared BAC clones to the maize-mapping project (12). Each clone set matched the corresponding finger printed contigs (FPC) (FPC no. 272 at map position 20.9 and FPC no. 227 at map position 51.8).
Examining BAC clone size and the coverage of different probes, we found that the z1C-1 region from B73 line was ≈250 kb long, much shorter than the 360-kb region in BSSS53. Such a drastic size difference suggests a significant loss of gene content in the B73 haplotype.
Sequence Analysis of the z1C-1 and z1C-2 Loci of Inbred Line B73. We sequenced two overlapping BAC clones (Z277I24 and Z190G16) that covered the whole B73 z1C-1 region (GenBank accession nos. AC144719 and AC144718). They yielded 263 kb of contiguous sequence. The z1C-1 locus, ≈112 kb, was in the middle of the sequence (Fig. 1). The restriction fragment length polymorphism reference marker php200725 was positioned ≈30 kb upstream of the z1C-1 locus, separated by various transposable elements. Approximately 100 kb of sequence downstream of the z1C-1 locus contained conserved genes (from CRb to CR4) also present in the BSSS53 genome (6). B73 has an array of only 15 z1C gene copies (Table 1), all of which, as in BSSS53, have the same polarity. Six have intact coding regions, six have premature stop codons in their coding regions, and three have large rearrangements either by transposon insertion (azs22.2-B73) or by large truncations (azs22.3-B73 and azs22.10-B73). In the z1C-2 locus, only a 14-kb fragment containing the single z1C gene was sequenced (GenBank accession no. AC144717). It has an intact gene and resembles the floury2 allele that was also found in BSSS53 as a single gene.
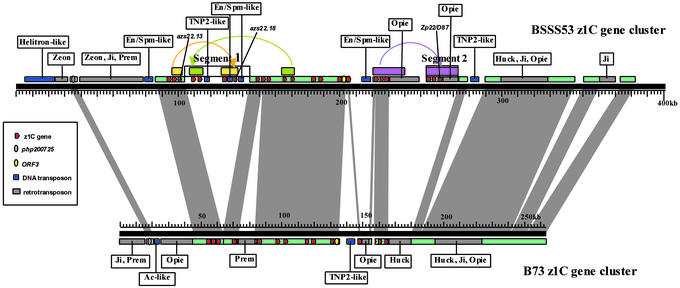
Fig. 1.
Sequence alignment of the two major haplotypes. The contiguous sequence of the B73 z1C-1 locus determined in this study was aligned with the previously sequenced orthologous region from BSSS53 by using orthologous markers. Conserved sequences are connected by vertical areas. Red arrows indicate polarity and position of zein gene copies. Zein gene copies that have been amplified, two duplications before and one triplication after formation of the haplotypes, are boxed in. Retrotransposons and transposons have been highlighted to illustrate noncollinear sequences and categorized (boxed-in names). Size markers for both contiguous sequences have been integrated to provide relative positions of sequence features.
Table 1. Summary of genes in BSSS53 and B73 z1C intervals.
|
BSSS53 inbred
|
B73 inbred
|
|||
|---|---|---|---|---|
| Gene | Coding region | Status | Coding region | Status |
| php200725 | 36233-37925 | 18656-20346 | ||
| azs22.11 | 93984-94784 | Premature stop | 53874-54674 | Premature stop |
| azs22.12 | 97302-98102 | Intact | 57196-57996 | Intact |
| azs22.13 | 100397-101203 | Premature stop | 60291-61097 | Premature stop |
| azs22.14 | 108295-109095 | Intact | ||
| azs22.15 | 112406-113206 | Premature stop | ||
| azs22.17 | 127485-128292 | Premature stop | ||
| azs22.18 | 134925-135731 | Premature stop | ||
| azs22.1 | 147795-148594 | Premature stop | 68217-69017 | Premature stop |
| azs22.2 | 151904-152704 | Premature stop | 72327-84682 | Insertion, 11,426 bp |
| azs22.3 | 156010-156088 | 3′ truncated | 87988-88066 | Premature stop |
| azs22.4 | 164447-165247 | Intact | 96392-97192 | Intact |
| azs22.5 | 169654-170454 | Premature stop | 101599-102399 | Premature stop |
| azs22.6 | 183200-184006 | Premature stop | 115145-115951 | Premature stop |
| azs22.7 | 187312-188814 | Insertion, 714 bp | 119257-120060 | Intact |
| azs22.8 | 199338-200138 | Intact | 130584-131384 | Intact |
| ORF3 | 202981-203565 | 134226-134816 | ||
| Rous-associated virus-like protein | 138573-139271 | |||
| azs22.9 | 205153-205948 | Premature stop | 147608-148408 | Intact |
| azs22.10 | 220387-221187 | Intact | N/A* | 5′ truncation |
| azs22.19 | 224490-225289 | Premature stop | 160308-161108 | Intact |
| azs22.20 | 228600-229397 | Premature stop | 164425-165222 | Premature stop |
| Zp22/6 | 253535-254335 | Intact | ||
| azs22.21 | 257640-258440 | Premature stop | ||
| Zp22/D87 | 261759-262472 | Intact | ||
| Cytochrome p450 | 293515-295648 | 190126-192181 | ||
| Receptor kinase | 334124-336230 | 230630-232727 | ||
| Sugar transporter | 337394-341159 | 233866-237640 | ||
| Cytidine deaminase | 345917-346886 | |||
| 40S ribosomal protein | 347494-349825 | |||
| RNA-binding protein | 350586-358453 | 242081-249955 | ||
| Unknown protein 1 | 373678-376578 | 253600-256396 | ||
| Unknown protein 2 | 379420-382092 | 259155-261430 | ||
| azs22.16 (f/2) | † | Intact | † | Intact |
Coding region is completely truncated
Translocated z1C gene
Segmental Duplications/Deletions of the z1C-1 Loci of B73 and BSSS53. Using the orthologous flanking markers, we found that the 263-kb B73 z1C-1 zein gene region corresponds to a region of ≈360 kb in BSSS53 (Fig. 1). The sequences of the two inbred lines differed also in their gene content and intergenic regions (Table 1). Segmental duplications and deletions that affect the number of zein gene copies in each haplotype largely account for the length difference. The duplications and deletions were confined to two segments, also present in the BSSS53 z1C-1 locus. One segment was between the third gene and the eighth gene (segment 1), which contains four z1C genes. The other was at the 3′ end of z1C-1 locus (segment 2), which contains the last three copies of the z1C genes in BSSS53. Phylogenic analysis of the z1C gene copies in the gene cluster indicated that the four genes in segment 1 were derived from two independent gene duplications that both involved two linked gene copies (3). These duplications occurred before the separation of the two haplotypes. Therefore, the B73 haplotype resulted from a deletion of segment 1 and a loss of four gene copies. On the other hand, the BSSS53 haplotype gained genes by a segmental duplication involving the three z1C genes at the 3′ terminal (segment 2). Comparing all z1C genes in both inbreds, we find that genes in orthologous positions are more conserved than in paralogous positions.
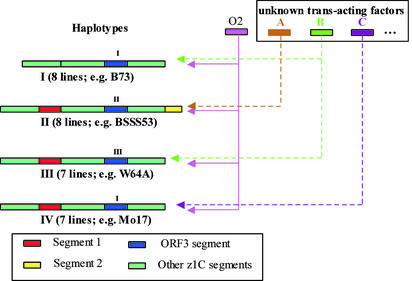
A Common Ancestral Interval of Maize Inbred Lines. On the basis of these sequence differences of the same interval for these two haplotypes, we could more extensively analyze haplotype variation among corn-belt inbred lines, including 43 inbred lines from a core collection. This analysis was based on Southern blots by using probes specific for different sections of the z1C gene cluster: segment 1 containing a duplicated azs22.13 copy (azs22.18) in the 5′ section, ORF3 in the middle section, and segment 2 containing Zp22/D87 in the 3′ section of the zein-gene cluster. Comparing the z1C-banding patterns, we find that the sampled inbreds exhibit four major haplotypes (types I to IV, Fig. 2), and two minor haplotypes (types V and VI, Table 4 and Fig. 5, which are published as supporting information on the PNAS web site). Recent deletion, insertion, or rearrangement events at different sections of the major haplotypes could explain the rest of the types and the sequence diversity among corn-belt inbred lines. One inbred line (B37) differed in the two analyses. Further analysis indicated that the B37 stock in our laboratory underwent sequence changes at the z1C-1 locus during seed propagation, switching from one haplotype to another (data not shown).
Fig. 2.
Known and hypothetical transacting interactions of the major haplotypes of the z1C-1 locus in maize. Major haplotypes of z1C-1 interval are represented by bars with different color-coded segments that are recognized by specific probes (Inset). The numbers above ORF3 segments indicate different subhaplotypes of this segment. The number of occurrences of a particular haplotype within the core collection (Table 4) and representative inbreds is marked below each bar. Colored bars to the right indicate genes that control the expression of all or subsets of zein genes within each haplotype. Arrows with dotted lines indicate hypothetical interactions to unknown transacting factors.
Other Gene Insertions/Deletions in z1C Loci. Although gene density between haplotypes remains nearly the same (1/11 kb in BSSS53; 1/10.8 kb in B73), differences other than deletions/insertions of z1C gene sequences occur. B73 possesses one additional single gene that interrupts the zein gene cluster. The gene is also absent in the orthologous position of sorghum and rice (6), and it has a high homology (e-144) to a Rous-associated virus-like B3 domain DNA-binding protein. BSSS53 has two additional linked single genes, the segment containing them being downstream of the zein gene cluster. One gene encodes a cytidine deaminase-like protein, and the other a ribosomal protein. These two genes are missing in the orthologous regions of sorghum and rice (6). These three single genes, together with seven zein genes in segments 1 and 2, exhibit a mosaic organization of gene order between the two maize inbred lines. This is more typical for interspecies comparisons than for comparisons within the same species (6). For instance, comparing the same interval of japonica and indica rice subspecies shows no change in gene order for the same region. Except for the seven zein genes and the additional three single genes, all other genes form orthologous pairs, conserving their positions and sequences (Table 1).
Transpositions After Haplotype Formation. A comparison of orthologous pairs indicated that z1C genes underwent extensive changes after the two haplotypes were separated. These changes included insertions, truncation, or premature stop codons formed by point mutations in the coding regions. Only a few z1C genes (7/22 in BSSS53; 6/15 in B73) retained an intact coding region. Intergenic spacing also changed after haplotype formation. Only ≈180 kb of sequence (≈66% of the B73 region, but only 50% of the BSSS53 region) was conserved between the two maize inbreds. Insertions occurred in both BSSS53 and B73 in different positions by different transposable and retrotransposable elements (Fig. 1), which also have been found at the bz and adh1 loci except for the Helitron at the 5′ end of the BSSS53 sequence (9, 13, 14). However, the largest expansion of intergenic space occurred upstream of the z1C-1 locus, which was formed by different retrotransposition events. Because transposition events affect both the length of an interval and the amount of conserved sequences, the same intervals of two maize inbreds apparently underwent more drastic genomic rearrangements than intraspecies rearrangements in rice.
Expression of z1C Genes in B73, BSSS53, and Their Hybrids. How do the changes that have occurred in the same interval of two maize inbred lines affect the expression of genes? The precise sequence and genomic locations of all z1C genes in both inbred lines enabled us to measure the expression of each individual gene in each inbred and their crosses. We selected endosperm tissue at 18 DAP. Although seed maturity varies between different maize lines and their hybrids, 18 DAP is within the window of maximal zein gene expression. Using a universal z1C primer set, z1C cDNAs were cloned by RT-PCR. To reconstruct the pool of z1C transcripts in endosperm tissue, random z1C cDNA clones were sequenced for each of the two inbred lines and their reciprocal crosses (see Materials and Methods). Because zein genes do not have introns, we could match each RT-PCR sequence with its gene, thus permitting us to determine its relative level of mRNA accumulation (Table 2). Two sets of experiments were compiled for each inbred, and three sets of experiments were conducted for each hybrid. Fig. 3 shows expression levels with confidence intervals for all z1C genes in B73 and BSSS53 and their reciprocal hybrids at a significance level of 0.05 (11).
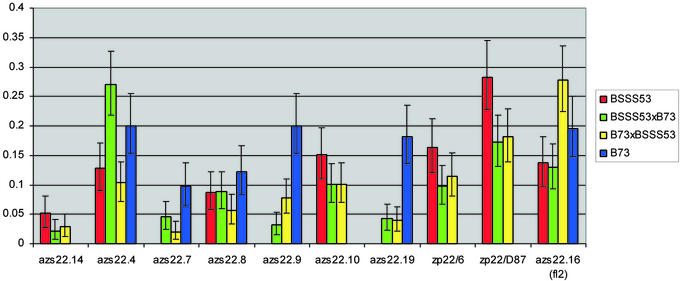
Fig. 3.
Relative z1C gene expression in inbreds and hybrids. Different sets of samples from immature, 18 DAP, endosperm tissue were processed for the measurement of steady-state mRNA levels as described in Materials and Methods. Relative expression levels were determined for each set as shown in Table 2. Bars, color-coded for inbreds and their reciprocal hybrids as shown in the key, represent the average expression levels in percentage of the total 22-kDa zein mRNA mixture with confidence intervals for each gene (Table 2).
Whereas the z1C gene copies vary dramatically between the two inbred lines, the number of genes expressed at detectable level is about the same (six in B73; seven in BSSS53). In both inbreds, the translocated z1C-2 allele (floury2) is expressed. Only two additional copies of z1C genes, alleles of azs22.4 and azs22.8, are expressed in both inbred lines. Therefore, nearly 50% of the expressed gene copies differ between the two inbred lines (3/6 in B73; 4/7 in BSSS53). Within the same inbred line, the active genes are not uniformly expressed at the same level. In inbred line BSSS53, the highest expressed gene is Zp22/D87, which accounts for nearly 25% of the z1C transcripts in the endosperm; whereas the lowest expressed gene is azs22.14-BSSS53, only ≈25% of the amount of Zp22/D87. Other expressed genes in BSSS53 produce mRNA at similar levels. Two uniquely expressed genes in BSSS53 arose from a segmental duplication event (Zp22/6 and Zp22/D87) and contribute nearly 40% of the z1C transcripts. Therefore, this most recent amplification (<1 million years ago) shifts z1C transcripts nearly 65% toward the new genes, a shift that might be partially caused by the change of their transcriptional regulation (Fig. 2). Unlike all of the other z1C gene copies, these genes no longer require the transcriptional activator O2 (4). However, in inbred line B73 expression levels of different genes are quite uniform. The only gene that is expressed at relatively lower levels is azs22.7-B73.
In reciprocal crosses between B73 and BSSS53, all active copies remained expressed, although at altered levels. The triploid nature of endosperm tissue (two maternal copies and one paternal copy) leads us to expect mRNA levels that reflect the gene dosage of the parental allele: three doses in the inbred, two doses when the inbred is the maternal parent, and one dose when the inbred is the paternal parent. But in hybrids between B73 and BSSS53, this is rarely the case for z1C gene expression. Only azs22.9-B73, one of the nonallelic-expressed gene, appears to exhibit a dosage effect. In all other cases, expression of the z1C genes seems to deviate from the dosage of their genomic copies. Except for azs22.9-B73, five of the other six nonallelic-expressed genes (except azs22.7-B73) show similar levels of gene expression in reciprocal hybrids, regardless of their dosage between the hybrids. One nonallelic gene (azs22.7-B73) exhibits an unusual expressional pattern that is reverse of its gene dosage in hybrids. Three allelic-expressed genes show a significant shift of gene expression level between reciprocal hybrids. Both azs22.4 and azs22.16 (floury 2) exhibit a level of gene expression that is significantly more elevated in one reciprocal hybrid than in the other. Even considering the confidence intervals for each gene and each cross, one of the hybrids surpasses the expression level in each of the parents (Fig. 3). In contrast, azs22.8 exhibits a decreased expression level in one reciprocal hybrid that is also lower than that of either parent.
Discussion
Interval Differences Because of Segmental Indels and Transposable Elements. Our study presents an analysis that compares the same genome interval between and within species for closely related cereals. This interval also contains a small, but highly clustered, gene family in maize and sorghum that is missing in rice.
Two aspects of the study are significant: the diversity of the maize germplasm within a small chromosomal region, and the impact of such diversity on gene expression in hybrids. Because maize was domesticated only 10,000 years ago, it is quite unusual for the sequence differences between the two major haplotypes of the interval common to BSSS53 and B73 to be so extensive given that allelic 22-kDa zein sequences are conserved between 97% and 100%. Although haplotypes of loci in other species consist primarily of single nucleotide polymorphisms or small insertions and deletions (indels), the same genome interval of the two maize inbreds differs substantially in size and content. Genes are missing or added as whole sequence segments that contain more than one gene. Our previous phylogenetic analysis of the z1C-1 cluster in BSSS53 showed that the cluster did not arise by unequal crossing over between genes but by the amplification of segments containing at least two genes (4), a finding confirmed by comparing the inbred lines as well. The two extra genes downstream of the BSSS53 z1C gene cluster also represent a segmental indel. In a recent study comparing an interval containing the bz gene between two different maize lines, four extra genes in one inbred line are also clustered within a single segment (9). Conceivably, therefore, segmental indels could dramatically change gene content and size of the same interval in maize inbred lines.
In the same way, transposable elements, particularly DNA transposons and retrotransposons of relatively large size, affect sequence content and size of the same interval of different inbred lines (Fig. 1). Although DNA transposable elements contribute to sequence divergence, retrotransposons also contribute to the size variation of the same interval. Because active retrotransposons reach a length between 8 and 12 kb and tend to insert on top of each other (“hotspot” effect) (6), reiterative retrotranspositions result in the large retrotransposon blocks observed in the maize genome (13). Retrotransposons also seem to “hitchhike” with segmental duplications. The recent segmental amplification of the three downstream zein genes (segment 2) also duplicates an opie retrotransposon. Therefore, roughly half of the extra DNA in the BSSS53 interval consists of retrotransposable elements and the other half consists of extra segments containing genes.
Reference Intervals for Intraspecies Comparisons in Maize. We know that conserved sequences are flanked by nonconserved sequences between different maize inbred lines. In one case all sequences flanking a unique sequence of 2,773 bp were completely different (7). In another case, two lines differed by a 7.3-kb insertion and a 14-kb deletion that included an active gene (8). Recent work on the maize bz region further extended this observation, providing a more complete picture of such sequence divergence of the same interval (9). However, in all of these previous studies intraspecies changes could not be compared to interspecies changes. Being able to compare the same interval in sorghum and rice, close relatives of maize, would be an essential reference for assessing the uniqueness of these features for maize breeding. The same interval on chromosome 11 of two rice subspecies (≈50 kb), a japonica variety (Lemont) and an indica variety (Teqing), is nearly completely conserved (6). On the same rice chromosome, the gene order of the interval containing the Adh1–Adh2 region is also completely conserved >340 kb between two japonica cultivars (Nipponbare; GenBank accession nos. AC123518, AC123523, AC123522, AC123521, and AC123515) and a hybrid (Yashiro-mochi X Tsuyuake) (15). Infrequent transposition events and other intergenic sequences were the only intraspecies differences detectable. It is therefore striking that maize inbreds differ significantly from rice in haplotype differences.
Furthermore, the z1C-1 interval in maize and sorghum appears different as between maize inbreds alone, whereas sorghum and rice appear to be much more conserved downstream of the z1C gene cluster (5). When compared to rice or sorghum, maize seemed to be much more active in segmental rearrangements and transpositions of its strains. By accumulating just a few “large unit” mutations, maize lines rapidly yield divergent intervals from a common ancestral chromosomal region.
Molecular Basis of Heterosis in Maize. Reciprocal crosses of two inbred lines containing two major haplotypes provide a pattern of gene expression based on the presence and absence of gene copies (“binary” systems). Because this type of noncollinearity within the same species has been observed mainly in maize, this feature might contribute to heterosis in maize. However, genes missing in one inbred could have moved to another genomic location of the same genome (9), a change that does not necessarily provide a selective advantage. However, if a gene is completely deleted from one inbred line, its gene product is lost as well. For instance, the single copy gene ORF3 in the z1C-1 interval, which is potentially involved in disease response, was missing from several inbred lines. In a gene family clustered at a single genomic location, the copy number could change dramatically, as shown for the z1C-1 locus. Breeding haplotype combinations through improved inbreds could conceivably lead to the production of hybrids that perform better. We would expect that expression patterns between inbreds and hybrids would be additive, as in the case of 27-kDa γ-zein, where reciprocal crosses between the S and Ra haplotypes seemed to follow a simple gene dosage effect (16, 17). However, such “dominance complementation” fits the expression pattern only of the azs22.9-B73 gene in the z1C-1 locus. Five of six other nonallelic-expressed genes (not azs22.7-B73), show a nearly uniform level of expression between the two reciprocal hybrids, regardless of their dosage difference. Even more surprising, azs22.7-B73 exhibits an expressional pattern that reverses its gene dosage. The allelic-expressed gene that is present in both parents should exhibit no gene dosage, but in the average a nearly 2-fold difference in expression levels occurs in azs22.4 and azs22.16 (fl2), depending on the direction of the cross (Fig. 3). The more highly expressed gene also surpasses the expression level of any of the parents, typical of the heterosis phenomenon. Such “overdominance” clearly differs from the dominance complementation model favored by evidence from quantitative genetics (1). A less pronounced difference can be observed for another allelic-expressed gene azs22.8, but here the level of gene expression is repressed in one reciprocal hybrid. Expression levels also seem to drop where inactive alleles are present and not simply deleted as in asz22.7-B73 and azs22.19-B73 (Fig. 3 and Table 1).
Changes in gene functions can differ substantially in two other ways: functional genes can drop out, or transacting mechanisms can diverge. For example, 15 of 22 z1C genes in BSSS53 and 9 of 15 z1C genes in B73 were deprived of function by point mutations, insertions of transposable elements (which may have deactivated azs22.2-B73 and azs22.7-BSSS53), or by truncation (which affected azs22.3 and azs22.10-B73) (Table 1). Only a small portion of coding regions of z1C genes remained intact and expressed, most of them nonallelic between the two haplotypes. The last segmental duplication in the BSSS53 haplotype exemplifies the second process: the Zp22/D87 and Zp22/6 genes no longer require the transcriptional activator O2 for their expression (4). Here, the trans-acting mechanisms that regulate the expressed genes have diverged (Fig. 2).
The significance of these two processes lies in their dramatic impact on changes within maize. Because intraspecies changes would tend to be deleterious for most gene copies, cumulative loss of gene products would result from inbreeding, leading in time to inbreeding depression. However, if two maize lines would differ dramatically in their gene losses, crosses between inbreds could rescue the predictable depression by complementing different functional genes. Instead of exhibiting complementation (e.g., dosage), the expression patterns of individual z1C genes in hybrid crosses frequently exhibit overdominance. Because multiple interactions can occur between different products unique to each inbred, different layers of interactions are synergistic. In particular, the combination of diverged z1C genes from two parental lines can draw on alternative regulatory factors that differ between the two lines as illustrated in Fig. 2. Although hybrid combinations of different functional genes might provide a selective advantage and be favored in subsequent generations, the resulting differing inbreds might produce thousands of new hybrid combinations. If any of those events are reiteratively selfed, the inbreeding depression process begins again, because not only would thousands of useful combinations be segregated but the accumulation of deleterious mutations would also be renewed. However, the outcrossing nature of maize itself could intervene before inbreeding depression. Maize seeds have large embryos that determine in the seeds the cell fate of the adult plant's six-leaf stage (almost half of the plant) (18). If any disfavored combination would be catastrophic for plant development, selection would already have occurred at the seed stage by seed abortion or by germination failure.
Supplementary Material
Acknowledgments
We thank S. Kavchok, S. Young, A. Bronzino, and G. Keizer for technical assistance; Dr. B. Shen for providing maize DNA samples; and Dr. Z. Swigonova for helping with the statistical analysis of the expression profiles. Department of Energy Grant DE-FG05-95ER20194 and National Science Foundation Grant 9975618 (to J.M.) supported this work.
Abbreviations: DAP, days after pollination; BAC, bacterial artificial chromosome.
References
- 1.Hallauer, A. R., Russell, W. A. & Lamkey, K. R. (1988) in Corn and Corn Improvement, eds. Sprague, G. F. & Dudley, J. W. (Am. Soc. Agron., Madison, WI), pp. 463-564.
- 2.Ueda, T. & Messing, J. (1993) in Genetic Engineering, ed. Setlow, J. K. (Plenum, New York), Vol. 15, pp. 109-130. [DOI] [PubMed] [Google Scholar]
- 3.Song, R. & Messing, J. (2002) Plant Physiol. 130, 1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song, R., Llaca, V., Linton, E. & Messing, J. (2001) Genome Res. 11, 1817-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman, C. E., Clore, A. M., Higgins, R., Lopes, M. A. & Larkins, B. A. (1997) Proc. Natl. Acad. Sci. USA 94, 7094-7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song, R., Llaca, V. & Messing, J. (2002) Genome Res. 12, 1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timmermans, M. C. P., Das, O. P. & Messing, J. (1996) Genetics 143, 1771-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu, W., Timmermans, M. C. P. & Messing, J. (1998) Genetics 150, 1229-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu, H. & Dooner, H. K. (2002) Proc. Natl. Acad. Sci. USA 99, 9573-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yim, Y.-S., Davis, G., Duru, N., Musket, T., Linton, E., Messing, J., McMullen, M., Soderlund, C., Polacco, M., Gardiner, J., et al. (2002) Plant Physiol. 130, 1686-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Audic, S. & Claverie, J.-M., (1997) Genome Res. 7, 986-995. [DOI] [PubMed] [Google Scholar]
- 12.Cone, K., McMullen, M., Bi, I., Davis, G., Yim, Y., Gardiner, J., Polacco, M., Sanchez-Villeda, H., Fang, Z., Schroeder, S., et al. (2002) Plant Physiol. 130, 1598-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.SanMiguel, P., Tikhonov, A., Jin, Y., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P., Edwards, K., Lee, M., Avramova, Z., et al. (1996) Science 274, 765-768. [DOI] [PubMed] [Google Scholar]
- 14.Lal, S. K., Giroux, M. J., Brendel, V., Vallejos, C. E. & Hannah, L. C. (2003) Plant Cell 15, 381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarchini, R., Biddle, P., Wineland, R., Tingy, S. & Rafalski, A. (2000) Plant Cell 12, 381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das, P. & Messing, J. (1987) Mol. Cell. Biol. 7, 4490-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geetha, K. B., Lending, C. R., Lopes, M. A., Wallace, J. C. & Larkins, B. A. (1991) Plant Cell 3, 1207-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiesselbach, T. A. (1949) The Structure and Reproduction of Corn (Cold Spring Harbor Lab. Press, Plainview, NY), 50th Ed.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.