Abstract
Two-component systems are the most common mechanism of transmembrane signal transduction in bacteria. A typical system consists of a histidine kinase and a partner response regulator. The histidine kinase senses an environmental signal, which it transmits to its partner response regulator via a series of autophosphorylation, phosphotransfer, and dephosphorylation reactions. Much work has been done on particular systems, including several systems with regulatory roles in cellular physiology, communication, development, and, in the case of bacterial pathogens, the expression of genes important for virulence. We used two methods to investigate two-component regulatory systems in Escherichia coli K-12. First, we systematically constructed mutants with deletions of all two-component systems by using a now-standard technique of gene disruption (K. A. Datsenko and B. L. Wanner, Proc. Natl. Acad. Sci. USA 97:6640-6645, 2000). We then analyzed these deletion mutants with a new technology called Phenotype MicroArrays, which permits assays of nearly 2,000 growth phenotypes simultaneously. In this study we tested 100 mutants, including mutants with individual deletions of all two-component systems and several related genes, including creBC-regulated genes (cbrA and cbrBC), phoBR-regulated genes (phoA, phoH, phnCDEFGHIJKLMNOP, psiE, and ugpBAECQ), csgD, luxS, and rpoS. The results of this battery of nearly 200,000 tests provided a wealth of new information concerning many of these systems. Of 37 different two-component mutants, 22 showed altered phenotypes. Many phenotypes were expected, and several new phenotypes were also revealed. The results are discussed in terms of the biological roles and other information concerning these systems, including DNA microarray data for a large number of the same mutants. Other mutational effects are also discussed.
Many gene families have now been identified by DNA sequencing. The so-called two-component regulatory genes of bacteria were among the earliest genes recognized in this manner (53). Computational analysis revealed that the NtrB and NtrC proteins of Bradyrhizobium sp. exhibit sequence similarities at the amino acid level with several other regulatory proteins. Extensive similarity was observed for the C-terminal domain of the NtrB protein and the C-terminal domains of the CpxA, EnvZ, and PhoR proteins and, to a lesser extent, the CheA protein of Escherichia coli (and Salmonella enterica serovar Typhimurium). Extensive similarity was also observed for the N-terminal domain of the NtrC protein and the N-terminal domains of the ArcA (then called SfrA and thought to interact with CpxA), OmpR, PhoB, CheB, and CheY proteins. Accordingly, Nixon et al. (53) proposed that the genes encode two-component regulatory systems and that these systems are involved in transduction of information about the environment from the C-terminal domain of a protein belonging to the first group of proteins to the N-terminal domain of the corresponding partner protein belonging to the second group.
Two-component regulatory systems are widespread in nature. Nearly all bacteria (mycoplasmas are exceptions) encode multiple systems of this type for diverse signaling processes. A typical two-component regulatory system is comprised of a signaling histidine kinase (HK) (also called a sensor kinase) that is usually membrane associated and a cytoplasmic response regulator (RR) that is usually a transcription factor (an activator or repressor). Similar systems control the expression of genes for nutrient acquisition, virulence, antibiotic resistance, and numerous other pathways in diverse bacteria. Due to the involvement of these two-component systems in so many cellular processes, several reviews of them have now been published. A monograph on two-component signal transduction has also been written (26). There are also analogous signaling systems in cells of lower eukaryotes, including fungi, amoebae, and plants (27, 42, 73, 78, 85).
Much work has been done on particular two-component systems, especially those with known roles in cell physiology, communication, development, and, in the case of bacterial pathogens, the expression of virulence genes. For example, the NtrB/NtrC and PhoR/PhoB systems were among the first such systems recognized; these systems control catabolic genes for nitrogen (N) and phosphorus acquisition, respectively (81, 89). Several two-component HKs were initially recognized because they can replace an HK of a nonpartner RR and thereby complement defects in the corresponding two-component HK mutants. For example, the HK CreC (originally called PhoM [83]) was originally discovered because it was found to replace the HK PhoR in activation of the response regulator PhoB in a phoR mutant. Such examples have led to the suggestion that cross-regulation (79) may be important for the integration of cellular processes involving multiple two-component systems (56).
E. coli is thought to encode 31 different two-component regulatory systems, based on experimental evidence and protein sequence similarities. The functions of many of these systems remain undefined. Also, even the most thoroughly studied systems may have functions other than those that are now known. Furthermore, if cross-regulation among different two-component systems has a fundamental biological role, then particular systems should have functions in common. In order to identify new roles of individual two-component systems and to uncover new interactions that can occur among them, we carried out an extensive, systematic phenotype analysis of E. coli mutants with deletions of all two-component systems and several related genes. Most mutants were constructed by using a recently developed gene disruption technique (10). The mutants included mutants with individual deletions of all two-component systems (L. Zhou, K. A. Datsenko, H. Aiba, K. Zhang, J. L. Masella, T. Mizuno, and B. L. Wanner, unpublished data).
The phenotype analysis was carried out by using a new tool, Phenotype MicroArrays (PMs). This technology can be used to find new functions of genes by testing mutants for a large number of phenotypes simultaneously (4a, 5, 61, 75). PM tests are performed in 96-well microplates containing different nutrients or inhibitors in which cell respiration is measured with a redox indicator. Here we report the results of PM tests performed with a large collection of mutants in which we examined nearly 2,000 cellular phenotypes by using a sensitive, highly controlled, reproducible format. Mutants were expected to display one or more altered phenotypes if the mutated gene has a role under the condition(s) tested. Of 37 different two-component mutants, 22 exhibited one or more phenotypic differences. Other mutants (including luxS and rpoS mutants) also exhibited altered phenotypes. Accordingly, PM technology provides a simple way to determine mutational effects on a genome-wide scale and adds to our understanding of intricate control systems.
MATERIALS AND METHODS
Bacteria.
All of the mutants used are derivatives of E. coli K-12 strain BW25113 (10) [lacIp4000(lacIq) rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1] or its rph+ derivatives BW28357 (86) and BW30383 (unpublished data). Most mutants were initially constructed by using a standard chromosomal gene disruption protocol (10). Briefly, PCR products were generated by using a pair of long (usually 56- to 60-nucleotide [nt]) primers and special template plasmids bearing a resistance marker flanked by FLP recombinase target sites. These oligonucleotides included 20 nt for priming on template plasmids and 36- to 40-nt homologous extensions for targeting recombination events to the corresponding loci on the E. coli chromosome. The PCR products were then introduced into cells expressing the phage λ Red recombinase, which permitted recombination in short homologous regions. Recombinants were then verified in a series of PCR tests in which locus-specific test primers were used, as described elsewhere (10). The test primers flanked the deleted region and usually were 200 nt or more from the homologous extension regions.
Two or more mutants of each type were constructed independently (Tables 1 and 2). Additional mutants were constructed by using long primers as described above or as follows. The simplest alternative method involved generating PCR products by using the corresponding flanking test primers for amplification and the mutants described above as templates. Accordingly, these PCR products had much longer regions of homologous sequences in common with the chromosome. As expected, they yielded huge numbers of recombinants when they were introduced into a new strain by using the Red system. Alternatively, the mutation was transferred by P1 transduction with P1kc lysates prepared with an initial mutant. All mutants were subsequently made antibiotic sensitive by using the FLP helper plasmid pCP20 to excise the resistance marker, as described elsewhere (10). Details concerning the construction and verification of these mutants will be described elsewhere (Zhou et al., unpublished). Some mutants were made in previous studies by using a standard two-step chromosomal gene deletion method (45).
TABLE 1.
Sources of two-component mutants
| Mutationa | Strain | Parent or controlb | Methodc |
|---|---|---|---|
| ΔarcA43 | BW27422 | BW25113 | a |
| ΔarcA45 | BW29409 | BW28357 | a |
| ΔarcB41 | BW26422 | BW25113 | a |
| ΔarcB41 | BW29859 | BW28357 | b |
| ΔatoSC571 | BW29009 | BW25113 | a |
| ΔatoSC573 | BW29010 | BW25113 | a |
| ΔbaeSR579 | BW27553 | BW25113 | b |
| ΔbaeSR610 | BW29744 | BW28357 | b |
| ΔbarA614 | BW27563 | BW25113 | b |
| ΔbarA1358 | BW29434 | BW25113 | a |
| ΔbasSR616 | BW27549 | BW25113 | b |
| ΔbasSR1287 | BW27848 | BW25113 | a |
| ΔbasSR620 | BW29371 | BW28357 | a |
| ΔcheZYBA1218d | BW28079 | BW25113 | a |
| ΔcheZYBA1218 | BW29661 | BW28357 | c |
| ΔcpxAR623 | BW27559 | BW25113 | b |
| ΔcpxAR623 | BW29849 | BW28357 | c |
| ΔcreABCD154 | BW26983 | BW25113e | d |
| ΔcreABCD176 | BW29135 | BW25113 | a |
| ΔcusSR1204 | BW30048 | BW28357 | c |
| ΔcusSR1204 | BW30049 | BW28357 | c |
| ΔdcuRS(yjdGH)1330 | BW27878 | BW25113 | a |
| ΔdcuRS(yjdGH)1330 | BW29657 | BW28357 | c |
| ΔdpiBA(citAB)1289 | BW27876 | BW25113 | a |
| ΔdpiBA(citAB)1289 | BW29656 | BW28357 | c |
| ΔevgAS1291 | BW27869 | BW25113 | a |
| ΔevgAS1291 | BW29663 | BW28357 | c |
| ΔfimZ1214 | BW28078 | BW25113 | a |
| ΔfimZ1214 | BW29659 | BW28357 | c |
| ΔkdpEDCBAF1224 | BW27564 | BW25113 | b |
| ΔkdpEDCBAF1224 | BW30166 | BW28357 | c |
| ΔnarLX1316 | BW27864 | BW25113 | a |
| ΔnarLX1316 | BW29658 | BW28357 | c |
| ΔnarP1312 | BW27873 | BW25113 | a |
| ΔnarP1312 | BW30265 | BW28357 | c |
| ΔnarQ1314 | BW27865 | BW25113 | a |
| ΔnarQ1314 | BW30008 | BW28357 | c |
| ΔntrCB(glnGL)1318 | BW27880 | BW25113 | a |
| ΔntrCB(glnGL)1318 | BW30011 | BW28357 | c |
| Δ(envZ ompR)520 | BW26424 | BW25113 | a |
| Δ(envZ ompR)520 | BW29655 | BW28357 | c |
| ΔphoBR580 | BW24476 | BW25113e | d |
| ΔphoBR758 | BW29134 | BW25113 | a |
| ΔphoBR758 | BW30046 | BW28357 | c |
| ΔphoQP1244 | BW27558 | BW25113 | b |
| ΔphoQP1244 | BW30007 | BW28357 | c |
| ΔqseBC(ygiXY)1302 | BW27551 | BW25113 | b |
| ΔqseBC(ygiXY)1304 | BW29747 | BW28357 | b |
| ΔrcsB1320 | BW27870 | BW25113 | a |
| ΔrcsB1320 | BW30009 | BW28357 | c |
| ΔrssB1275 | BW29745 | BW28357 | b |
| ΔrssB1273 | BW30418 | BW25113 | c |
| ΔrssB1273 | BW30419 | BW25113 | c |
| ΔrssB1273 | BW30420 | BW30383 | c |
| ΔrssB1273 | BW30421 | BW30383 | c |
| ΔrstAB1278 | BW27552 | BW25113 | b |
| ΔrstAB1280 | BW29806 | BW28357 | b |
| ΔtorSTRCAD518 | BW26423 | BW25113 | a |
| ΔtorSTRCAD518 | BW29855 | BW28357 | c |
| ΔuhpBA1322 | BW27871 | BW25113 | a |
| ΔuhpBA1322 | BW29852 | BW28357 | c |
| ΔuvrY1310 | BW29475 | BW25113 | a |
| ΔuvrY1362 | BW29476 | BW25113 | a |
| ΔyedVW1298 | BW27550 | BW25113 | b |
| ΔyedVW1300 | BW29746 | BW28357 | b |
| ΔyehTU1324 | BW27877 | BW25113 | a |
| ΔyehTU1324 | BW29858 | BW28357 | c |
| ΔyfhA1326 | BW27868 | BW25113 | a |
| ΔyfhA1326 | BW29851 | BW28357 | c |
| ΔyfhK1328 | BW27872 | BW25113 | a |
| ΔyfhK1328 | BW29857 | BW28357 | c |
| ΔyojN1332 | BW27866 | BW25113 | a |
| ΔyojN1332 | BW29856 | BW28357 | c |
| Δ(yojN rcsBC)1308 | BW27557 | BW25113 | b |
| Δ(yojN rcsBC)1308 | BW29861 | BW28357 | c |
| ΔypdAB1334 | BW27875 | BW25113 | a |
| ΔypdAB1334 | BW29860 | BW28357 | c |
| ΔzraSR(hydHG)1336 | BW27867 | BW25113 | a |
| ΔzraSR(hydHG)1336 | BW29660 | BW28357 | c |
Common alternative nomenclature is indicated in parentheses. Different alleles are used to identify mutations that differ with regard to the template or primer used for generation. The allele assignments for multiple gene deletions are in clockwise order in accordance with the K-12 map. Mutants are available from the Coli Genetic Stock Center (http://cgsc.biology.yale.edu/). Genes are designated according to their order within an operon.
E. coli K-12 strains BW28357 and BW30383 are independent rph+ derivatives of BW25113 that were made by using the Red system and P1 transduction, respectively (K. A. Datsenko and B. L. Wanner, unpublished data).
Mutants were made in one of four ways. In method a, mutants were isolated as kanamycin-resistant derivatives of the parent carrying pKD46 following introduction of a PCR fragment generated with long (56- to 60-nt) primers and pKD13 as the template, after which the resistance marker was eliminated with pCP20 as described elsewhere (10). In method b, mutants were isolated as kanamycin-resistant transductants by using P1kc grown on the corresponding kanamycin-resistant mutant, after which the resistance marker was eliminated with pCP20. In method c, mutants were isolated as kanamycin-resistant derivatives of the parent carrying pKD46 following introduction of a PCR fragment generated with locus-specific test primers and the corresponding kanamycin-resistant mutant as the template, after which the resistance marker was eliminated with pCP20. In method d, the mutation was constructed on a conditionally replicative plasmid by using standard cloning techniques and then recombined onto the chromosome by using our standard two-step allele replacement method (45), after which the mutation was transferred to a parent by cotransduction with nearby proC+ and thr+ markers for the phoBR and creABCD loci, respectively.
The ΔcheZYBA1218 mutation has a deletion of several nearby genes and corresponds to the Δ(flhEAB cheZYBR tap tar cheWA motBA flhCD IS1)1218 mutation.
BW24476 and BW26983 are descendents of strains like BW25113 and served as controls.
TABLE 2.
Sources of other mutants
| Mutationa | Strain | Parent | Methodb |
|---|---|---|---|
| ΔcbrA179 | BW29846 | BW28357 | a |
| ΔcbrA181 | BW29847 | BW28357 | a |
| ΔcbrBC183 | BW29869 | BW28357 | a |
| ΔcbrBC183 | BW30003 | BW28357 | a |
| ΔcbrBC185 | BW29848 | BW28357 | a |
| ΔcsgD1202 | BW28106 | BW25113 | a |
| ΔcsgD1202 | BW29850 | BW28357 | c |
| ΔluxS1368 | BW30044 | BW28357 | a |
| ΔluxS1368 | BW30045 | BW28357 | a |
| ΔphnP-C75c | BW29687 | BW28357 | a |
| ΔphnP-C75 | BW29688 | BW28357 | a |
| ΔphoA760 | BW29256 | BW28357 | a |
| ΔphoA760 | BW30047 | BW28357 | a |
| ΔphoH764 | BW29689 | BW28357 | a |
| ΔphoH766 | BW29690 | BW28357 | a |
| ΔpsiE768 | BW29844 | BW28357 | a |
| ΔpsiE770 | BW29845 | BW28357 | a |
| ΔrpoS1271 | BW28465 | BW28357 | a |
| ΔrpoS1271 | BW29923 | BW28357 | a |
| ΔugpQCEAB772 | BW29685 | BW28357 | a |
| ΔugpQCEAB774 | BW29686 | BW28357 | a |
Storage and handling of bacteria.
Bacteria are stored at −70°C in Luria-Bertani medium containing 8% dimethyl sulfoxide as described elsewhere (80). Cells were revived without thawing by scraping the surface with a toothpick and streaking the cells onto an appropriate agar medium. To minimize inadvertent selection of suppressor mutants, fresh colonies were routinely used to inoculate culture tubes.
PM tests.
Two or more independent mutants with mutations in each gene were examined. Mutants were compared pairwise with the otherwise isogenic control strain. In a few cases phenotype analysis of two independent mutants unexpectedly revealed substantial differences. Since such inconsistencies may have resulted from acquisition of unknown secondary mutations, additional mutants were sometimes constructed and assayed similarly. The results obtained with atypical strains are not described here.
With five exceptions, the mutants examined included mutants for which DNA microarray experiment results were recently reported (54). Even though all of these organisms are null mutants, it is still possible that some may contain unlinked secondary mutations. For example, during this study we found that the cusRS and rssB mutants (BW28077 and BW27555, respectively) used in DNA microarray experiments differed from other independently constructed cusRS mutants (BW30048, BW30049) and rssB mutants (BW29745, BW30416, BW30419, BW30420, BW30421), all of which were similar to other mutants of the same type. Also, as previously reported (54), the uvrY deletion mutant BW27874 that was used in the DNA microarray experiments has a growth defect. However, new uvrY mutants with in-frame (and presumably nonpolar) deletions did not have a growth defect. Accordingly, we report PM data for these new uvrY mutants (BW29475, BW29476) below. No data are reported for the atoSC and yfhA mutants (BW28878 and BW29430, respectively) that were used in DNA microarray experiments. However, this was due to an oversight. BW28878 and BW29430 were by chance not subjected to PM tests. Rather, two other independent atoSC and yfhA mutants were examined instead.
PM tests were performed essentially as described elsewhere (5), except that IF-0 inoculating fluid was used for PM1 to PM8 and IF-10 was used for PM9 to PM20. PM1 and PM2 are similar to ES. PM3 to PM8 are similar to EN, EPS, and EA, which used a defined medium containing 100 mM NaCl, 30 mM triethanolamine HCl (pH 7.1), 5.0 mM NH4Cl, 2.0 mM NaH2PO4, 0.25 mM Na2SO4, 0.05 mM MgCl2, 1.0 mM KCl, and 0.01% tetrazolium violet. PM3, PM4, and PM6 to PM8 contain various N, P, or S sources which are omitted from the defined medium. PM9 to PM20 are similar to ES1, ES2, and ES3, which used a rich medium containing 2.0 g of tryptone, 1.0 g of yeast extract, and 1.0 g of NaCl per liter. Strains were grown overnight at 35°C on BUG+B agar instead of R2A agar (64). All fluids, agar media, and PMs are commercially available from Biolog, Inc. (Hayward, Calif.), and all PMs were inoculated with cell suspensions at 100 μl per well. Cells were picked from the agar surface with a sterile cotton swab and suspended in 15 ml of IF-0, and the cell density was adjusted to 42% transmittance (T) on a Biolog turbidimeter. These suspensions were diluted sixfold by combining with 75 ml of IF-0 to give a density of 85% T (approximately an A420 of 0.12). PM1 and PM2, which measure C utilization phenotypes, were directly inoculated with 22 ml of the 85% T suspension. Six hundred sixty microliters of 2 M disodium succinate and 0.2 mM ferric citrate was then added to 66 ml of the 85% T suspension and used to inoculate PM3 to PM8, which measure N, P, and S utilization and auxotrophic phenotypes. Six hundred microliters of the 85% T suspension was diluted 200-fold into 120 ml of IF-10 and used to inoculate PM9 to PM20, which measure sensitivity to salt and pH stress, and to a wide variety of antibiotics, antimetabolites, and other inhibitors. All PMs were incubated at 36°C in an OmniLog and monitored for color change in the wells. Readings were recorded for 24 h for all PMs except PM3, PM4, and PM6 to PM8, for which readings were recorded for 48 h. Kinetic data were analyzed with OmniLog-PM software, release OL_PM_109M Jan. 14, 2002.
Additional tests were done to substantiate a subset of the PM data. To confirm phenotypes detected with the metabolic arrays (PM1 to PM8), tests were repeated once. To confirm phenotypes detected with the inhibitor sensitivity arrays (PM9 to PM20), 1.33-fold serial dilutions of some chemicals were used in 96-well microplates to retest particular mutants. BBL Sensi-Disc antimicrobial susceptibility test disks (Becton Dickinson and Co., Sparks, Md.) were also used to test some drug sensitivity phenotypes with the agar disk diffusion test procedure recommended by the manufacturer. These tests were carried out with cells which were grown in Difco Bacto Tryptic Soy broth (Becton Dickinson and Co.) and tested for antibiotic susceptibility on Difco Mueller-Hinton agar (Becton Dickinson and Co.), as recommended by the manufacturer.
RESULTS
Overview.
All phenotypic differences exhibited by two or more independent mutants of each type are briefly described below. Many results confirmed or expanded our knowledge of the two-component systems. Other results were unexpected and not easily understood in light of current knowledge. For clarity, two-component partner proteins are designated HK/RR below when the corresponding genes were deleted simultaneously. Otherwise, they are identified parenthetically as an HK, Hpt (histidine phosphotransfer domain only), hybrid (containing both HK and RR domains), or RR protein. Several mutants also had nearby related genes deleted (Tables 1 and 2). In general, no attempt was made to ascertain whether phenotypic differences were attributable to the loss of the two-component system per se or to the loss of the function of a nearby gene(s).
We first describe results for the two-component systems (ArcB/ArcA, PhoR/PhoB, UhpB/UhpA, and NtrB/NtrC) that gave phenotypes which were largely in agreement with expectations. These results are especially valuable for validation purposes. We then discuss other systems alphabetically. No significant differences were observed for 15 different two-component mutants [BasS/BasR, CheA (HK)/CheB (RR); CheY (RR), CreC/CreB, DpiB/DpiA, EvgS/EvgA, FimZ (RR), NarX/NarL, NarQ (HK), TorS/TorR, YedV/YedW, YehU/YehT, YfhK (HK), YfhA (RR), YpdA/YpdB, and ZraS/ZraR]. The CsgD mutants also exhibited no phenotypic difference. The effects of mutations in phoBR- and creBC-regulated genes are described in the context of the PhoR/PhoB and CreC/CreB systems, respectively. Data for the luxS and rpoS mutants are also summarized below.
ArcB/ArcA.
The two-component system consisting of ArcB (HK) and ArcA (RR) modulates expression of numerous operons and regulons involved in respiratory and fermentative metabolism in response to oxygen deficiency or redox potential (28, 29). The oxidized forms of quinones are thought to be ArcB-specific signals that silence, rather than stimulate, ArcB kinase activity during aerobiosis (19), thus showing that there is a direct connection between the control of gene expression by the Arc system and the electron transport chain. The Arc system has also been shown to inhibit E. coli chromosomal initiation in vitro (40).
The arcA and arcB genes are not linked, so each gene was deleted individually and the corresponding mutants were tested independently. The arcA and arcB mutants were both highly pleiotropic, displaying more than 50 phenotypes (Table 3). There was a high degree of overlap of the phenotypic changes, confirming their relationship. Both arcA and arcB mutants exhibited increased resistance to β-chloro-l-alanine and hypersensitivity to more than 40 chemicals, most of which affect membrane or membrane-associated functions, such as respiration. Phospho-ArcA acts as a transcriptional activator of several genes and as a repressor of other genes. Accordingly, the discordance for phenotypes not in common may have been due to different effects resulting from the loss of ArcA or phospho-ArcA on gene expression. Alternatively, they may have been due to an additional specific role(s) for one partner protein. For example, ArcB controls not only ArcA-dependent gene expression but also OmpR-dependent gene expression in vivo (44). While the arcA and arcB mutants had two phenotypes (thioridazine and cobalt chloride sensitivity) in common with EnvZ/OmpR mutants, neither phenotype was specific for arcA or arcB mutants.
TABLE 3.
arcA and arcB mutant phenotypes
| Testa | Differenceb
|
Mode of action | |
|---|---|---|---|
| arcA | arcB | ||
| β-Methyl-d-glucuronate | 60 | —d | C source |
| β-Chloro-l-alanine | 120 | 88 | Amino acid analog |
| Potassium tellurite | — | 78 | Transport, toxic anion |
| Atropine | −150 | −180 | Acetylcholine antagonist |
| pH 8 to pH 10c | −64 | −68 | Alkaline pH |
| Myricetin | −110 | −170 | Antimicrobial |
| Sanguinarine | −130 | — | ATPase inhibitor |
| Aztreonam | −140 | −200 | Cell wall, lactam |
| EGTA | — | −210 | Chelator, Ca2+ |
| EDTA | −180 | −250 | Chelator, hydrophilic |
| Sodium pyrophosphate | −210 | −140 | Chelator, hydrophilic |
| 1,10-Phenanthroline | −120 | −83 | Chelator, lipophilic |
| 2,2′-Dipyridyl | — | −110 | Chelator, lipophilic |
| 5-Chloro-7-iodo-8-hydroxyquinoline | — | −130 | Chelator, lipophilic |
| 8-Hydroxyquinoline | — | −140 | Chelator, lipophilic |
| Fusaric acid | −71 | −94 | Chelator, lipophilic |
| Caffeine | — | −71 | Cyclic AMP phosphodiesterase |
| Benserazide | −72 | −62 | Fungicide |
| Dichlofluanid | −240 | −220 | Fungicide |
| Nordihydroguaiaretate | — | −94 | Fungicide, lipoxygenase |
| Harmane | — | −76 | Imidazoline binding sites, agonist |
| Trifluoperazine | −120 | −110 | Ion channel, Ca2+ |
| Dequalinium | −200 | −220 | Ion channel, K+ |
| 2-Phenylphenol | −110 | — | Membrane agent |
| m-Cresol | — | −100 | Membrane agent |
| o-Cresol | −70 | — | Membrane agent |
| p-Cresol | −150 | −150 | Membrane agent |
| Phenylethanol | −120 | −160 | Membrane agent |
| Alexidine | −90 | — | Membrane, biguanide, electron transport |
| Benzethonium chloride | — | −63 | Membrane, cationic detergent |
| Methyltrioctylammonium chloride | −150 | −91 | Membrane, cationic detergent |
| Guanidine hydrochloride | −85 | −75 | Membrane, chaotropic agent |
| Lauryl sulfobetaine | — | −120 | Membrane, zwitterionic detergent |
| Amitriptyline | — | −94 | Membrane, transport |
| Glycyl-l-cysteine | −120 | −57 | N source |
| 5% Sodium sulfate | −320 | −400 | Osmolarity |
| Lawsone | −360 | −340 | Oxidizing agent |
| Plumbagin | −160 | −220 | Oxidizing agent |
| Potassium superoxide | −92 | −100 | Oxidizing agent |
| Chlorpromazine | — | −79 | Phenothiazine |
| Compound 48/80 | −110 | — | Phospholipase C, ADP ribosylation |
| Chloramphenicol | — | −120 | Protein synthesis |
| Puromycin | — | −160 | Protein synthesis |
| Tylosin | — | −110 | Protein synthesis |
| Geneticin (G418) | −110 | — | Aminoglycoside |
| Paromomycin | −130 | — | Aminoglycoside |
| Troleandomycin | — | −77 | Macrolide |
| Thioglycerol | −210 | −170 | Reducing agent |
| Tetrazolium violet | −110 | −180 | Respiration |
| Thioridazine | −110 | −170 | Respiration |
| Pentachlorophenol | −150 | −130 | Respiration, H+ ionophore |
| Menadione | — | −120 | Respiration, uncoupler |
| Sodium azide | −180 | — | Respiration, uncoupler |
| 100 mM ammonium sulfate (pH 8) | −200 | — | Toxicity, ammonia |
| 200 mM sodium benzoate (pH 5.2) | — | −67 | Toxicity, benzoate |
| 100 mM sodium nitrite | −650 | −840 | Toxicity, nitrite |
| 200 mM sodium phosphate (pH 7) | −430 | −500 | Toxicity, phosphate |
| Sodium cyanate | −110 | −210 | Transport, toxic anion |
| Sodium cyanide | −150 | −170 | Transport, toxic anion |
| Sodium metaborate | −61 | −79 | Transport, toxic anion |
| Sodium metasilicate | −120 | −150 | Transport, toxic anion |
| Sodium nitrite | — | −200 | Transport, toxic anion |
| Sodium tungstate | — | −160 | Transport, toxic molybdate analog |
| Cobalt chloride | −120 | −190 | Transport, toxic cation |
| Manganese(II) chloride | — | −160 | Transport, toxic cation |
| Thallium(I) acetate | −170 | −220 | Transport, toxic cation |
| Zinc chloride | −250 | −220 | Transport, toxic cation |
Chemicals were tested in 96-well PMs.
The OmniLog-PM software generates time course curves for respiration (tetrazolium color formation) and calculates differences in the areas for mutant and control cells. The units are arbitrary. Positive values indicate that the mutant showed greater rates of respiration than the control. Negative values indicate that the control showed greater rates of respiration than the mutant. The differences are averages of values reported for two or more mutants of each type compared with the corresponding control strains.
Minimal differences are given for alkaline pH sensitivities.
—, software reported no significant difference.
PhoR/PhoB.
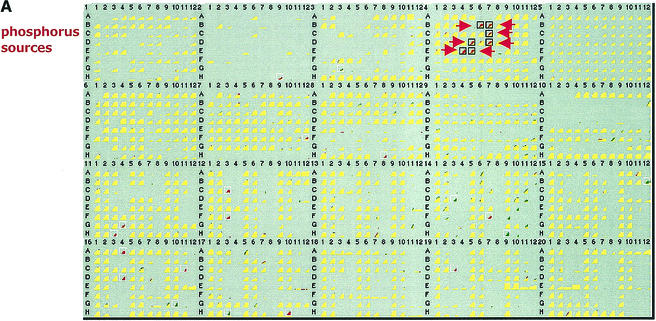
The PhoR/PhoB two-component system controls genes of the phosphate (Pho) regulon for assimilation of alternative P sources (81). Representative PM results for a phoBR mutant showed that the principal phenotypic changes were in the top five rows of PM4 (Fig. 1), which measured P metabolism. All three phoBR mutants showed decreased use of several organophosphates as P sources, which was expected.
FIG. 1.
Phenotypic changes in PM assays. Significant changes are enclosed in boxes and indicated by arrows. Yellow indicates that growth of the wild type and growth of the mutant were similar. Red indicates faster growth of the wild type. Green indicates faster growth of the mutant. The quantitative difference values are shown in Table 4. (A) Comparison of BW29134 (ΔphoBR) with BW25113. Well B6, d-2-phosphoglyceric acid; well B7, d-3-phosphoglyceric acid; well C7, 6-phosphogluconic acid; well D5, O-phospho-d-serine; well D7, O-phospho-l-threonine; well E4, phosphorylcholine; well E5, ο-phosphorylethanolamine. (B) Comparison of BW27880 (ΔntrBC) with BW25113.
We also tested the effects of mutations of Pho regulon genes (Table 2), including deletions of the phoA gene (which encodes the bacterial alkaline phosphatase Bap), the ugpBAECQ operon (which encodes an ABC transporter for uptake of sn-glycerol-3-phosphate and a glycerophosphoryl diester hydrolase), the phnC-phnP operon (which codes for breakdown of phosphonates), and two genes of unknown function (phoH and psiE). The phoA and phoBR mutants showed decreased use of the same six organophosphates (Table 4). Hence, the phenotypic changes for the phoBR mutants in PM4 were due to loss of Bap, a nonspecific phosphohydrolase. Accordingly, Bap is required for use of these six organophosphates as sole phosphorus sources. The failure to detect changes in the ugpBAECQ mutants is also understandable. An effect on use of glycerol 3-phosphate as a P source is likely to be masked in ugpBAECQ mutants that are both phoA+ and glpT+. Bap can hydrolyze glycerol 3-phosphate, whereas GlpT is an alternative glycerol 3-phosphate transporter.
TABLE 4.
Mutant phenotypes
| Mutants | Testa | Differenceb | Mode of action |
|---|---|---|---|
| Two-component mutants | |||
| atoSC | Glucuronamide | 64 | C source |
| Polymyxin B | −180c | Membrane agent, outer | |
| Methyltrioctylammonium chloride | −150c | Membrane, detergent, cationic | |
| 6% NaCl | −51 | Osmolarity | |
| Dihydrostreptomycin | −140c | Aminoglycoside | |
| Iodonitrotetrazolium violet | −130c | Respiration | |
| baeSR | Myricetin | −140 | Antimicrobial |
| Gallic acid | −220 | Antimicrobial | |
| Nickel chloride | −76 | Transport, toxic cation | |
| Sodium tungstate | −390d | Transport, toxic molybdate analog | |
| barA | Melibionic acid | 59 | C source |
| d-Melibiose | 77e | C source | |
| cpxRA | pH 9.5 + anthranilate, glycine, l-alanine, l-arginine, l-asparagine, l-aspartate, l-glutamate, l-isoleucine, l-leucine, l-methionine, l-phenylalanine, or l-threonine | −70 to −110e | Alkaline pH, deaminase |
| Leu-Trp | −87 | N source | |
| 15 or 20% Ethylene glycol | −230 | Osmolarity | |
| Amikacin | −140d,f | Aminoglycoside | |
| Dihydrostreptomycin | −150d | Aminoglycoside | |
| Geneticin (G418) | −170d | Aminoglycoside | |
| Hygromycin B | −84d | Aminoglycoside | |
| Kanamycin | −110d,f | Aminoglycoside | |
| Tobramycin | −140d,f | Aminoglycoside | |
| Methane sulfonate | −72 | S source | |
| Lithium chloride | −79 | Transport, toxic cation | |
| cusRS | 1,10-Phenanthroline | −110d | Chelator, lipophilic |
| dcuSRg | Bromosuccinate | −65 | C source |
| dl-Malate or d-Malate | −130 | C source | |
| Fumarate | −110 | C source | |
| l-Asparagine | −71 | C source | |
| l-Aspartate | −140 | C source | |
| l-Malate | −130 | C source | |
| m-Tartarate | −73 | C source | |
| Succinate | −57 | C source | |
| kdpFABCDE | Novobiocin | 95d | DNA topoisomerase |
| Hygromycin B | −75c | Aminoglycoside | |
| narP | 5-Chloro-7-iodo-8-hydroxyquinoline | −110d | Chelator, lipophilic |
| ntrBC | β-Chloro-l-alanine | 120 | Amino acid analog |
| d-Cycloserine | 85d | Cell wall, sphingolipid synthesis | |
| pH 9.5 + l-alanine | −76 | Alkaline pH, deaminase | |
| Nitrofurazone | −150 | DNA synthesis | |
| d-Serine | −240d | Inhibitor, 3-phosphoglycerate dehydrogenase | |
| Poly-l-lysine | −110 | Membrane, detergent, cationic | |
| Arg-Arg | −75 | N source | |
| Arg-Lys | −66 | N source | |
| Arg-Phe | −39 | N source | |
| Arg-Tyr | −43 | N source | |
| d-Lysine | −36 | N source | |
| l-Argininc | −50 | N source | |
| l-Lysine | −44 | N source | |
| l-Ornithine | −45 | N source | |
| Lys-Arg | −68 | N source | |
| Met-Arg | −32 | N source | |
| δ-Amino-N-valerate | −82 | N source | |
| Amikacin | −100d,f | Aminoglycoside | |
| Geneticin (G418) | −120d | Aminoglycoside | |
| Gentamicin | −120f | Aminoglycoside | |
| Neomycin | −100d | Aminoglycoside | |
| Paromomycin | −110d | Aminoglycoside | |
| Tobramycin | −120d,f | Aminoglycoside | |
| Thioridazine | −102 | Respiration | |
| Capreomycin | −62 | Respiration, Na+-K+ ATPase | |
| ompR-envZ | β-d-Allose | 56 | C source |
| d-Fructose | 64e | C source | |
| d-Mannitol | 65 | C source/PICK> | |
| N-Acetyl-d-glucosamine | 87 | C source | |
| α-d-Glucose | 100 | C source | |
| Cefamandole | 250d | Cell wall, cephalosporin | |
| Cephalothin | 260d,f | Cell wall, cephalosporin | |
| Aztreonam | 140 | Cell wall, lactam | |
| Norfloxacin | 120 | DNA topoisomerase | |
| Sulfachloropyridazine | 89 | Folate antagonist | |
| Ata-Ser | 61 | N source | |
| Lys-Ser | 46 | N source | |
| Oxacillin | −110 | Cell wall, lactam | |
| 15% Ethylene glycol | −97 | Osmolarity | |
| Thioridazine | −130 | Respiration | |
| Cobalt chloride | −120 | Transport, toxic cation | |
| Sodium selenite | −253 | Toxic anion | |
| Potassium chromate | −74 | Transport, toxic anion | |
| Lithium chloride | −71 | Transport, toxic cation | |
| Sodium dichromate | −83 | Transport, toxic SO4 analog | |
| phoBRh | d-2-Phosphoglycerate, d-3-phosphoglycerate, O-phospho-d-serine, O-phospho-l-threonine, O-phosphorylethanolamine, phosphorylcholine | −53 to −82 | P source |
| Tobramycin | −140c,i | Aminoglycoside | |
| Paromomycin | −130c | Aminoglycoside | |
| phoPQ | d-Fructose | 54e | C source |
| d-Mannitol | 52 | C source | |
| qseBC (ygiXY) | Ruthenium red | −73 | Respiration, mitochondrial Ca2+ porter |
| Cesium chloride | −75 | Transport, toxic cation | |
| Cobalt chloride | −75 | Transport, toxic cation | |
| Cupric chloride | −150c | Transport, toxic cation | |
| Nickel chloride | −74 | Transport, toxic cation | |
| rcsB | Nitrofurazone | −68d | DNA synthesis |
| Trimethoprim | −110i | Folate antagonist | |
| 5 to 6% NaCl | −25 to −62d | Osmolarity | |
| Iodonitrotetrazolium violet | −210d | Respiration | |
| rssBg | pH 4.5 + l-lysine | −85 | Acidic pH, decarboxylase |
| α-Ketoglutarate | −110 | C source | |
| α-Methyl-d-galactoside | −91 | C source | |
| β-Hydroxypyruvate | −62 | C source | |
| Bromosuccinate | −84 | C source | |
| dl-α-Glycerol phosphate | −94 | C source | |
| dl-Malate | −100 | C source | |
| d-Alanine | −120 | C source | |
| d-Glucuronate | −69 | C source | |
| d-Malate | −68 | C source | |
| Fumarate | −90 | C source | |
| Glyoxylate | −98 | C source | |
| l-Asparagine | −100 | C source | |
| l-Aspartate | −130 | C source | |
| l-Malate | −89 | C source | |
| m-Tartarate | −90 | C source | |
| Succinate | −88 | C source | |
| Ala-Asn | −62 | N source | |
| Ala-Leu | −65 | N source | |
| Ala-Ser | −87 | N source | |
| Arg-Tyr | −60 | N source | |
| Asp-Leu | −92 | N source | |
| Asp-Phe | −60 | N source | |
| d-Ala-Gly | −95 | N source | |
| d-Ala-Gly-Gly | −66 | N source | |
| δ-Amino-N-valerate | −130 | N source | |
| d-Asparagine | −77 | N source | |
| Glu-Trp | −60 | N source | |
| Gly-D-Ala | −68 | N source | |
| Gly-Gly-d-Leu | −78 | N source | |
| Gly-Gly-Leu | −60 | N source | |
| Gly-Phe-Phe | −73 | N source | |
| Gly-Tyr | −79 | N source | |
| His-Trp | −74 | N source | |
| l-Cysteine | −160 | N source | |
| Leu-Ala | −86 | N source | |
| Leu-Glu | −82 | N source | |
| Leu-Gly | −85 | N source | |
| Leu-Trp | −89 | N source | |
| l-Proline | −89 | N source | |
| Met-Trp | −67 | N source | |
| Phe-Trp | −130 | N source | |
| Pro-Leu | −71 | N source | |
| Putrescine | −75 | N source | |
| Ser-Leu | −75 | N source | |
| Trp-Leu | −120 | N source | |
| Trp-Phe | −90 | N source | |
| Trp-Trp | −92 | N source | |
| Trp-Tyr | −100 | N source | |
| l-Cysteate | −80 | S source | |
| Methane sulfonate | −77 | S source | |
| Sodium nitrite | −76 | Transport, toxic anion | |
| Sodium tungstate | −76 | Transport, toxic molybdate analog | |
| rstAB | Ketoprofen | −100 | Anticapsule, anti-inflammatory |
| Pridinol | −72 | Cholinergic antagonist | |
| Troleandomycin | −140 | Macrolide | |
| uhpAB | d-Glucose 6-phosphate | −69 | C source |
| d-Fructose 6-phosphate | −73 | C source | |
| uvrY | d-Melibiose | 73e | C source |
| Hydroxylamine | 89 | DNA damage, antifolate | |
| Nitrofurazone | −70d | DNA synthesis | |
| Methyltrioctylammonium chloride | −130c | Membrane, cationic detergent | |
| Polymyxin B | −150c | Membrane agent, outer | |
| Dihydrostreptomycin | −120c | Aminoglycoside | |
| Iodonitrotetrazolium violet | −67c | Respiration | |
| yojN | 20% Ethylene glycol | −130 | Osmolarity |
| 3 to 6% NaCl | −13 to −83 | Osmolarity | |
| yoJN rcsBC | Trimethoprim | −110i | Folate antagonist |
| 3 to 6% NaCl | −21 to −100 | Osmolarity | |
| Other mutants | |||
| cbrA (yidS) | Hydroxylamine | 94 | DNA damage, mutagen, antifolate |
| Ofloxacin | −81 | DNA topoisomerase, quinolone | |
| 5,7-Dichloro-8-hydroxyquinaldine | −86 | Chelator, lipophilic | |
| 18-Crown-6 ether | −87 | Respiration, ionophore | |
| cbrBC (yieIJ)j | Nitrofurazone | −68 | DNA synthesis |
| luxS | Caffeine | 110 | Cyclic AMP phosphodiesterase |
| Oxycarboxin | 75 | Fungicide, respiratory enzymes | |
| pH 10 | −57 | Alkaline pH | |
| pH 9.5 + l-alanine, l-histidine, or l-lysine | −39 to −47 | Alkaline pH, deaminase | |
| 1,10-Phenanthroline | −110 | Chelator, lipophilic | |
| 5,7-Dichloro-8-hydroxyquinaldine | −98 | Chelator, lipophilic | |
| Sulfamethoxazole | −92 | Folate antagonist | |
| Sulfadiazine | −94 | Folate antagonist | |
| Sulfanilamide | −74 | Folate antagonist | |
| Sulfathiazole | −70 | Folate antagonist | |
| d-Valine | −41 | N source | |
| l-Homoserine | −21 | N source | |
| phoA | Phosphorylcholine, d-2-phosphoglycerate, O-phospho-l- threonine, O-phosphorylethanolamine, O-phospho-d-serine, d-3-phosphoglycerate | −54 to −90 | P source |
| phoH | Pridinol | 73 | Cholinergic antagonist |
| Troleandomycin | 94 | Macrolide | |
| Cefoxitin | −100 | Cell wall, cephalosporin | |
| Spiramycin | −61 | Macrolide | |
| phn | Troleandomycin | 95 | Macrolide |
| pH 10 | −70 | Alkaline pH | |
| Cefoxitin | −69 | Cell wall, cephalosporin | |
| 5,7-Dichloro-8-hydroxyquinaldine | −79 | Chelator, lipophilic | |
| psiE | 8-Hydroxyquinoline | −90 | Chelator, lipophilic |
| rpoS | l-Threonine | 68 | C source |
| β-Methyl-d-glucuronate | 60 | C source | |
| l-Threonine | 75 | N source | |
| Tyr-Tyr | 65 | N source | |
| Guanosine | 75 | N source | |
| Tyr-Phe | 62 | N source | |
| l-Tyrosine | 67 | N source | |
| Ile-Tyr | 74 | N source | |
| Methylene diphosphonate | 58 | P source | |
| α-Hydroxybutyrate | −98 | C source | |
| Glycyl-l-aspartate | −86 | C source | |
| α-Ketobutyrate | −80 | C source | |
| Hygromycin B | −64 | Aminoglycoside |
Not confirmed by serial dilution (see Materials and Methods).
Confirmed qualitatively by serial dilution (see Materials and Methods).
Confirmed by repeating the PM tests (see Materials and Methods).
Confirmed qualitatively with BBL Sensi-Disc tests (see Materials and Methods).
The dcuSR and rssB mutants were defective in the use of succinate as a C source and showed many defects with the normal PM3 to PM8 plates. These mutants were therefore reassayed by using PM3 to PM8 with glycerol as the C source, on which the dcuSR mutants showed no phenotypic differences. Results obtained for PM3 to PM8 with glycerol as the C source are shown for the rssB mutants. See text.
Unlike BW29134 and BW30046, BW24476 did not show increased aminoglycoside sensitivity. See text.
Not confirmed with BBL Sensi-Disc tests (see Materials and Methods).
Unlike BW29869 and BW30003, BW29848 did not show increased nitrofurazone sensitivity. See text.
The phn mutants showed defective growth at pH 10 and hypersensitivity to a few inhibitors, as did the psiE and phoH mutants (Table 4). It is interesting, although not surprising, that mutants in which genes activated by the PhoR/PhoB system were deleted displayed phenotypes that were not observed in phoBR mutants. Basal-level expression of individual Pho regulon genes or operons may be sufficient for functioning of the corresponding gene products. Other controls also act on these genes (81). The phn operon encodes 14 gene products, including an ABC transporter, a C-P lyase, and two apparent transcriptional regulators. No tests were done to determine which phn gene(s) is responsible for these phenotypes. The phoH gene product is an ATP-binding protein, and the psiE product is a highly conserved, putative membrane protein. New studies are needed to define more precisely the genetic basis, as well as the biochemical and physiological basis, of these phenotypic changes.
UhpB/UhpA.
The UhpB/UhpA system controls synthesis of the hexose phosphate transporter UhpT (84). As expected, we found that the uhpAB mutants were specifically defective in the use of glucose 6-phosphate and fructose 6-phosphate as carbon sources. Since the uhpAB mutants were phoA+, they were able to use these compounds as P sources.
NtrB/NtrC.
The NtrB/NtrC system controls the expression of genes for N assimilation. These include the glnALG operon (glnA encodes glutamine synthetase, glnL encodes NtrB, and glnG encodes NtrC), the glnHPQ operon (which encodes glutamine permease), the glnK-amtB operon (glnK encodes a PII homolog, and amtB encodes an ammonium uptake protein), the nac gene (which encodes a nitrogen assimilation control protein), the astCADBE operon (which encodes an arginine catabolic system), and other transporters and enzymes important in N assimilation (89). Approximately 2% of the E. coli genome (∼75 genes) appears to be under NtrC control, and nearly two-thirds of the genes encode systems for scavenging N-containing compounds, including those released into the periplasmic space during cell growth and division (e.g., d-alanine and the d-alanyl-d-alanine dipeptide).
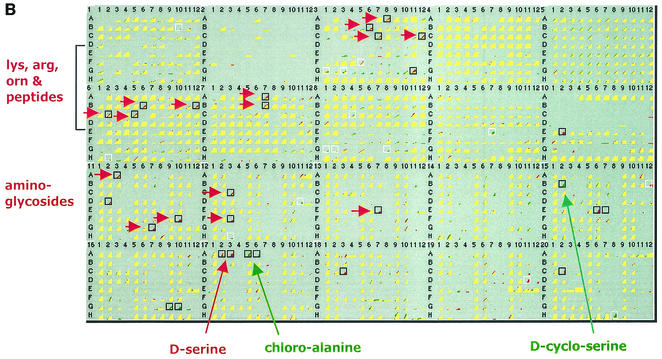
PM analysis of ntrBC mutants provided the best example for comparing PM results (Fig. 1B and Table 4) directly with a detailed gene expression analysis (89). In general, the agreement was very good. As expected, the mutants grew normally on minimal medium and did not require glutamine (Fig. 1B, PM5, well A1) (68). Defects were clearly detected in N catabolic pathways (Fig. 1B, PM3, PM6, and PM7), but these defects were specific for utilization of arginine, lysine, and ornithine or peptides of these amino acids. This was presumably due to Ntr regulation of the ast operon (arginine catabolism) and/or the argT (basic amino acid transport) and ygjG (probable ornithine aminotransferase) genes (89).
The ntrBC mutants were resistant to β-chloro-l-alanine and d-cycloserine and hypersensitive to d-serine, which suggests that there was altered regulation of the nac, cycA, dadA, and/or metC genes (38). The cycA gene (uptake of d-alanine, d-serine, and glycine) and the nac gene (nitrogen assimilation control) are already known to be involved in Ntr control (89). The cycA protein is controlled by the Nac protein (89), which is controlled by NtrC and also connects control by NtrC to σ70-dependent genes (50). The toxicity of d-serine is elevated if it is not efficiently deaminated, leading to inhibition of 3-phosphoglycerate dehydrogenase (9).
In addition to being resistant to the l-alanine analog β-chloro-l-alanine, the ntrBC mutants were also defective in l-alanine deamination (PM10, well E2). This could have resulted from an effect on expression of an unknown gene(s), a gene(s) encoding a shared Ntr-regulated alanine transport system, or a gene encoding a deaminase (or transaminase) required for detoxifying the analog. A defect was also seen in the use of δ-amino-N-valeric acid as an N source. This is a novel phenotype, and no genes are known to be involved. All of these phenotypic changes observed in the ntrBC mutants are reasonably attributable to the NtrB/NtrC function in the regulation of amino acid catabolism as an N source.
Unexpectedly, the ntrBC mutants were hypersensitive to several aminoglycosides, including tobramycin, paromomycin, gentamicin, Geneticin, neomycin, and amikacin, and to nitrofurazone, thioridazine, and capreomycin. Aminoglycoside sensitivity was verified by additional tests (Table 4). The biochemical or physiological basis of these susceptibilities is unknown and remains to be explained. Some possible candidate genes are ubiF, topA, rplF, and especially metC, which also mediates β-chloro-l-alanine resistance (38).
AtoS/AtoC.
The AtoS/AtoC system regulates expression of the atoDAEB operon for acetoacetate metabolism (31). The only metabolic phenotype detected in the atoSC mutants was increased use of glucuronamide as a carbon source. The atoSC mutants also showed increased sensitivity to sodium chloride but not to potassium chloride. In addition, these mutants showed greater susceptibilities to membrane agents (polymyxin B and methyltrioctylammonium chloride), the aminoglycoside dihydrostreptomycin, and the respiration inhibitor iodonitrotetrazolium violet (Table 4). However, we were unable to confirm these susceptibilities by using serial dilution tests as described in Materials and Methods. The basis of these phenotypes is not known.
BaeS/BaeR.
The BaeS/BaeR system is now thought to control genes for an efflux pump (mdtABC) (2, 51) and a third envelope stress pathway (62). This system was originally revealed by the ability of BaeS to suppress the loss of the HKs CreC, EnvZ, and PhoR in the corresponding mutants (52). The baeSR mutants showed increased sensitivity to myricetin, gallic acid, and nickel chloride and especially to sodium tungstate (Table 4). Because tungstate is a molybdate analog, the BaeS/BaeR system may also regulate a gene(s) whose product has a molybdate cofactor. The increased susceptibilities of the baeSR mutants support the hypothesis that the BaeS/BaeR system controls an efflux pump.
BarA/UvrY.
The barA and uvrY genes are not linked. BarA and UvrY were shown to be two-component partner proteins by finding that barA and uvrY mutants are both hypersensitive to hydrogen peroxide and that phospho-BarA efficiently transfers its phosphoryl group to UvrY (58). The hydrogen peroxide sensitivity is attributed to an effect on RpoS, which in turn controls synthesis of the major catalase (KatE) in E. coli. BarA (also called AirS) is a hybrid HK. Homologous BarA/UvrY systems have been widely studied due to their roles in animal and plant pathogenesis. BarA homologs include ExpS in Erwinia and GacS (also called LemA or PheN) in Pseudomonas species; UvrY homologs include ExpA in Erwinia, GacA in Pseudomonas, SirA in Salmonella, and VarA in Vibrio species. Many effects attributed to these systems are likely to be indirect. So far, the only direct target of UvrY is csrB, which encodes a small regulatory RNA and whose homologs include rsmZ in Pseudomonas and rsmB in Erwinia species (72). The BarA/UvrY system has a role in biofilm formation (30).
PM analysis revealed that both barA and uvrY mutants gave a stronger carbon utilization response with d-melibiose, providing further evidence that there is an association between BarA and UvrY. Recently, this system was found to be required for efficient switching between glycolytic and gluconeogenic carbon sources in uropathogenic E. coli (57). It is therefore a bit surprising that PM tests failed to reveal additional carbon source effects for these mutants. The uvrY mutant, but not the barA mutant, also showed increased resistance to hydroxylamine and increased sensitivity to several unrelated inhibitors (Table 4).
CpxA/CpxR.
The CpxA/CpxR system controls expression of genes involved in relieving envelope protein stress, biofilm formation, motility and chemotaxis, cell proliferation, adaptation to or recovery from the stationary phase, and pathogenesis. The Cpx system also plays a critical role in the regulation of adhesion-induced gene expression (55). Based on a combination of computational and experimental approaches, it has been estimated that about 100 promoters (including the ompC promoter) are controlled by the CpxA/CpxR system in E. coli (14).
The cpxRA mutants had two dominant phenotypes. They were hypersensitive to several amino acids at an alkaline pH and to aminoglycosides (Table 4). These results are in agreement with accounts describing, among several other seemingly unrelated phenotypes, effects of a cpxA* mutation on resistance to amikacin and growth at high pH, which were shown to be CpxR dependent (13, 63). The cpxRA mutants also showed defects in the use of Leu-Trp as an N source, the use of methane sulfonate as an S source, and sensitivity to lithium chloride and the nonionic osmolyte ethylene glycol.
CreC/CreB.
Neither the role nor the signal for the CreC/CreB system is known, although this system appears to be connected to carbon and energy metabolism (82). Elsewhere, genes controlled by this system that were identified by searching for promoter-lacZ fusions regulated by CreB have been described (86). This search revealed open reading frames of unknown function, which were designated cbrA, cbrB, and cbrC (creB-regulated genes A, B, and C, respectively). No phenotypic changes were observed for the creABCD mutants, yet differences were apparent in the cbrA and cbrBC mutants. The cbrA mutants displayed greater resistance to hydroxylamine and hypersensitivity to ofloxacin, a lipophilic chelator, and an ionophore (Table 4). Two cbrBC mutants showed hypersensitivity to nitrofurzone; however, a third mutant did not (Table 4). The identification of phenotypes for the cbrA and cbrBC mutants but no differences for the creABCD mutants is consistent with the hypothesis that the cbrA and cbrBC genes are subject to an additional control(s). New studies are needed to understand the basis of these phenotypes.
CusS/CusR.
The CusS/CusR system is required for expression of at least one chromosomal gene, cusC, which probably encodes a copper ion efflux system (49). The CusS/CusR system also controls copper-induced expression of the promoter for pcoE in the plasmid-borne copper resistance pco operon.
The cusRS mutants displayed a single phenotypic change, hypersensitivity to 1,10-phenanthroline, which was also displayed by arcA, arcB, and luxS mutants (Table 4). 1,10-Phenanthroline is a chelator with high affinities for copper and other metals, including iron and zinc (69). These data support the hypothesis that the CusS/CusR system plays a role in control of a copper efflux system.
DcuS/DcuR.
The DcuS/DcuR system is closely related to a subgroup of two-component systems, including the citrate-responsive CitA/CitB system of Klebsiella (47). These systems activate expression of dcuB (which encodes the anaerobic fumarate-succinate antiporter), frdABCD (which encodes fumarate reductase), and dctA (which encodes the aerobic succinate carrier) in response to the C4 dicarboxylates fumarate, succinate, malate, aspartate, tartrate, and maleate (20, 88).
The dcuSR mutants gave many expected findings. In particular, defects in the use of d- and l-malic acids, fumaric acid, m-tartaric acid, bromosuccinic acid, l-asparagine, and l-aspartic acid as C sources were expected. The dcuSR mutants also had a slight defect in the use of succinate, in agreement with a previous report (20). Many additional defects were also seen for use of various N, P, and S sources. These results are not shown in Table 4 because they were secondary to the defect in metabolism of succinate, which was used as the C source in the assays. When the tests were repeated with glycerol as the C source, no phenotypic differences were seen.
EnvZ/OmpR.
The EnvZ/OmpR system controls expression of the ompF and ompC porin genes and many other genes in response to the medium osmolarity (1). The amounts of OmpF and OmpC vary differently. OmpC synthesis is favored under high-osmolarity conditions, and OmpF synthesis is dominant under low-osmolarity conditions (59). OmpF and OmpC synthesis is also subject to other controls, which involve both OmpR-dependent and OmpR-independent effects of adenylate cyclase, RpoS, and acetyl phosphate synthesis, pH, nutrient limitation, and other factors (41, 59, 60).
The ompR-envZ mutants showed diverse phenotypic changes (Table 4). Their greater resistance to several antibiotics (including cephalosporins, β-lactam, topoisomerase inhibitor, and folate antagonist) was likely a consequence of defects in porin synthesis that prevented access through the outer membrane. Membrane defects were also probably the cause of hypersensitivity to other agents, including thioridazine, cobalt chloride, and sodium dichromate. These mutants showed increased use of several hexoses as C sources (allose, fructose, mannitol, N-acetyl-d-glucosamine, and glucose), which was unexpected. They also displayed increased use of two serine-containing dipeptides as N sources. The ompR-envZ mutants were hypersensitive to ethylene glycol (an osmotic agent), but not to other osmotic agents, such as sodium chloride, sodium sulfate, and sodium lactate. These results support the hypothesis that the EnvZ/OmpR system plays a role in the control of porin synthesis and provide new, unexpected data concerning phenotypic differences as well.
KdpD/KdpE.
The KdpD/KdpE system controls expression of the kdpFABCDE operon, which encodes the high-affinity K+ transporter, as well as KdpD and KdpE (18, 77). The expression of this operon is inducible by both NaCl and CsCl via different mechanisms (33). The entire kdpFABCDE operon was deleted in the KdpD/KdpE mutants. These mutants displayed increased resistance to novobiocin and increased sensitivity to hygromycin (Table 4). Whether these differences resulted from a direct role of the Kdp transporter in uptake or efflux of these antibiotics or were an effect of the KdpD/KdpE system on expression of unlinked genes is not known. Our failure to detect phenotypic changes directly attributable to a K+ transport defect was probably a consequence of the presence of multiple K+ transporters in E. coli (16).
NarP (RR).
The Nar system is comprised of two HKs (NarQ and NarX) and two RRs (NarL and NarP), which control genes for anaerobic respiration and fermentation in response to the electron acceptors nitrate and nitrite (3, 39, 71). The genes regulated by the Nar system include adhE (which encodes alcohol dehydrogenase), dmsABC (which encodes dimethyl sulfoxide/trimethylamine-N-oxide reductase), fdnGHI (which encodes formate dehydrogenase), frdABCD (which encodes fumarate reductase), modABCD (which encodes components of a molybdate uptake system), narK (which encodes a nitrite exporter), narGHJI and napA (which encode cellular nitrate reductases), nirBDC and nrfABCDEFG (which encode nitrite reductases), and pfl (which encodes pyruvate-formate lyase).
Three different nar mutants were examined, as narL and narX are adjacent while narQ and narP are at different loci. No phenotypic differences were observed for narQ and narXL mutants. The narP mutants displayed a single difference, increased sensitivity to the lipophilic chelator 5-chloro-7-iodo-8-hydroxyquinoline (Table 4). Such sensitivity could be indicative of an essential metal cofactor. It is reasonable to suppose that finding additional phenotypic differences would require testing of the nar mutants under anaerobic or microaerophilic conditions. This has not been done, however.
PhoQ/PhoP.
The PhoQ/PhoP system has been studied primarily in S. enterica, in which it was first recognized as a regulatory system that controls synthesis of an acid phosphatase (36) and was later shown to be required for pathogenesis (17, 22, 48). The finding that the PhoQ/PhoP system controls synthesis of an Mg2+ transporter led to the discovery that PhoQ senses Mg2+ in several gram-negative bacteria (21). Many genes are activated or repressed by this system in S. enterica (23). The genes regulated by the PhoQ/PhoP system in response to Mg2+ in E. coli include phoPQ, mgtA, and mgrB (35). No growth phenotype has ever been reported for phoPQ mutants.
The phoPQ mutants showed increased use of fructose and mannitol as C sources. Curiously, these phenotypes were also seen in ompR-envZ mutants (Table 4). The PM assays were done under conditions of excess Mg2+. Other phenotypes may be specific to low-magnesium conditions.
QseC/QseB.
The QseC/QseB system has recently been shown to be involved in quorum sensing and transcriptional regulation of flhDC, the master regulator operon for the flagellar and motility genes in E. coli (70). Quorum sensing in E. coli is thought to respond to a universal signal (called autoinducer-2) which is produced by LuxS in a large number of bacteria and which has been shown to be a furanosyl borate diester (7). Whether QseC is the only sensor kinase that responds to autoinducer-2 is unclear.
The qseBC mutants displayed hypersensitivity to several toxic cations (cesium, cobalt, copper, nickel, and ruthenium) (Table 4). Accordingly, the QseC/QseB system may have an undetermined role in metal metabolism.
RcsC (HK)/YojN (Hpt)/RcsB (RR).
The RcsC/YojN/RcsB system is a three-component phosphorelay system, in which the Hpt protein YojN accepts a phosphoryl group from phospho-RcsC and then transfers it to RcsB (74). This system regulates expression of the capsular polysaccharide synthesis cps genes, the cell division ftsAZ genes, an osmoregulated osmC gene, and the small RNA gene rprA, as well as genes involved in motility and chemotaxis (11, 43). The RcsC sensor appears to respond to stress that affects the cell membrane; however, the precise signal is unknown (8).
The rcsB, yojN, and rcsC genes are adjacent, but they are not in an operon. Three kinds of mutants were examined: mutants with individual deletions of rcsB and yojN and mutants with all three genes deleted. The rcsB mutants showed hypersensitivity to sodium chloride as an osmotic agent and to nitrofurazone, iodonitrotetrazolium violet, and trimethoprim. Likewise, the yojN mutant was hypersensitive to sodium chloride. However, it was also hypersensitive to ethylene glycol as an osmotic agent. The yojN mutant did not show greater sensitivity to other inhibitors. The triple yojN-rcsBC mutant showed hypersensitivity to sodium chloride as an osmotic agent and to trimethoprim but no other phenotypic differences. Although variations were also apparent, all RcsC/YojN/RcsB system mutants were hypersensitive to sodium chloride. Accordingly, this system may have a role in osmotic protection.
RssB (RR).
RssB is an orphan RR for which no partner HK has been identified. Unlike most RRs, RssB is also not a transcription factor. Rather, RssB regulates the stability of the sigma factor RpoS and is essential for RpoS proteolysis (25). Phospho-RssB catalyzes the delivery of RpoS to the protease ClpXP for degradation (87).
PM analysis showed that the rssB mutants were highly pleiotropic. The rssB mutants also resembled the dcuSR mutants in that both types of mutants showed defects in the use of several carboxylic acids, including succinic acid, l- and d-malic acids, fumaric acid, l-aspartic acid, l-asparagine, d-alanine, and other acids. Accordingly, like the dcuSR mutants, the rssB mutants were retested for effects on the use of N, P, and S sources with glycerol as a C source. However, whereas defects in the use of N, P, and S sources were eliminated in the dcuSR mutants under these conditions (see above), the rssB mutants still showed defects in the use of several N sources (including leucine dipeptides, aspartic acid dipeptides, tryptophan dipeptides, and others) (Table 4). The rssB mutants also showed defects in the use of l-cysteate and methane sulfonate as S sources and hypersensitivity to sodium nitrite and tungstate. It is reasonable to suspect that many of these phenotypic changes are attributable to the role of RssB in proteolysis; however, this cannot be ascertained.
RstB/RstA.
No role has yet been reported for the RstB/RstA system. The rstAB mutants were hypersensitive to ketoprofen, pridinol, and troleandomycin. The basis for these sensitivities is unknown.
LuxS.
As noted above, LuxS synthesizes a furanosyl borate diester that is believed to be a universal autoinducer for cell-to-cell communication among diverse bacteria (7, 12). We examined luxS mutants because autoinducer-2 has been proposed to act as a signaling molecule for a two-component system(s), including the QseC/QseB system (70).
The luxS mutants showed hypersensitivity to two chelators,1,10-phenanthroline and 5,7-dichloro-8-hydroxyquinaldine(Table 4). 1,10-Phenanthroline has a high affinity for copper, as well as other metals, such as iron and zinc (15). Hypersensitivity to 1,10-phenanthroline was also observed for the cusRS mutants, which affect copper transport (49). The luxS mutants were also more sensitive to several folate antagonists, including sulfamethoxazole, sulfadiazine, sulfathiazole, and sulfanilamide, and were more resistant to oxycarboxin and caffeine. In addition, the luxS mutants showed defects in the use of d-valine or l-homoserine as an N source, a growth defect at pH 10, and sensitivities to l-alanine, l-histidine, and l-lysine at basic pH values (which are indicators of a deaminase defect[s]). Further studies are required to unravel the basis of these phenotypes.
RpoS.
RpoS is the stationary-phase sigma factor (also called σ38). We examined rpoS mutants because the RR RssB is involved in its turnover (25). The rpoS mutants were pleiotropic. They showed increased use of l-threonine and β-methyl-d-glucuronic acid as C sources, increased use of l-threonine, guanosine, l-tyrosine, and dipeptides such as Tyr-Tyr, Tyr-Phe, and Ile-Tyr as N sources, and increased use of methylene diphosphonic acid as a P source. The rpoS mutants also exhibited defects in the use of α-hydroxybutyric acid, glycyl-l-aspartic acid, and α-ketobutyrate as C sources and sensitivity to hygromycin B (Table 4). None of these phenotypic differences was exhibited by the rssB mutants. Both rpoS and rssB mutations affected l-threonine metabolism, but they did so in opposite ways. Two rssB mutants were defective in the use of l-threonine as an N source, while the rpoS mutants showed improved growth.
DISCUSSION
We surveyed mutants with mutations in all two-component systems and several related genes in E. coli by determining the phenotypes of a large set of well-defined deletion mutants that are otherwise isogenic. We found phenotypic changes for 22 different two-component-system mutants and not for 15 other systems (Tables 3 and 4). Several two-component system mutants (arcA, arcB, cpxRA, ompR-envZ, ntrBC, rssB) were highly pleiotropic. We discovered new phenotypes or functions for 14 systems, including the AtoS/AtoC, BaeS/BaeR, BarA/UvrY, CpxA/CpxR, CusS/CusR, EnvZ/OmpR, KdpD/KdpE, NarP, NtrB/NtrC, PhoQ/PhoP, QseC/QseB, RcsC/YojN/RcsB, RssB, and RstB/RstA systems (Table 5). We also identified phenotypic changes for 8 of 10 related non-two-component mutants surveyed (Table 4).
TABLE 5.
Summary of PM data for two-component mutantsa
| System (HK/RR) | Function(s) | No. of phenotypesb
|
New phenotype(s) or function(s) | |
|---|---|---|---|---|
| Gained | Lost | |||
| ArcA (RR) | Respiration control | 2 | 47 | Nonec |
| ArcB (hybrid) | Respiration control | 2 | 59 | Nonec |
| AtoS/AtoC | Acetoacetate metabolism | 1 | 5 | Glucuronamide use and NaCl sensitivity |
| BaeS/BaeR | Unknown | 0 | 4 | Sodium tungstate sensitivity |
| BarA (hybrid) | Hydrogen peroxide sensitivity | 2 | 0 | Melibiose metabolism |
| UvrY (RR) | Hydrogen peroxide sensitivity | 2 | 5 | Melibiose metabolism |
| CpxA/CpxR | Cell envelope stress | 0 | 23 | Ethylene glycol sensitivity |
| CusS/CusR | Response to copper | 0 | 1 | 1,10-Phenanthroline sensitivity |
| DcuS/DcuR | C4 dicarboxylate utilization | 0 | 9 | None |
| EnvZ/OmpR | Osmotic regulation | 12 | 8 | Increased use of carbohydrates, cephalosporin resistance |
| KdpD/KdpE | Potassium transport | 1 | 1 | Novobiocin resistance |
| NarP (RR) | Nitrate regulation | 0 | 1 | Chelator sensitivity |
| NtrB/NtrC | Nitrogen regulation | 2 | 23 | Aminoglycoside sensitivity |
| PhoQ/PhoP | Response to magnesium | 2 | 0 | Increased use of fructose and mannitol |
| PhoR/PhoB | Phosphate regulation | 0 | 8 | Noned |
| QseC/QseB | Quorum sensing | 0 | 5 | Metal sensitivity |
| RcsB (RR) | Capsule synthesis | 0 | 6 | NaCl and trimethoprim sensitivity |
| RcsC/YojN(Hpt)/RcsB | Capsule synthesis | 0 | 6 | NaCl and trimethoprim sensitivity |
| YojN(Hpt) | 0 | 6 | NaCl sensitivity | |
| RssB (RR) | σS Stability | 0 | 53 | Decreased use of C4 di- and monocarboxylates and amino acid N sources |
| RstB/RstA | Unknown | 0 | 3 | Ketoprofen, pridinol, and troleandomycin sensitivity |
| UhpB/UhpA | Hexose phosphate uptake | 0 | 2 | None |
The following 15 other two-component mutants did not have altered phenotypes: BasS/BasR, CheA (HK)/CheB (RR)/CheY (RR), DpiB/DpiA, CreC/CreB, EvgS/EvgA, FimZ (RR), ZraS/ZraR, NarX/NarL, NarQ (HK), TorS/TorR, YedV/YedW, YehU/YehT, YfhK (HK), YfhA (RR), and YpdA/YpdB.
Phenotypes gained means PM assays showing increased growth or respiration (positive values in Table 4). Phenotypes lost means PM assays showing decreased growth or respiration (negative values in Table 4).
The arcA and arcB mutants had many phenotypes that were attributed to membrane-associated functions, which may be considered new.
As mentioned in the text, two of the three phoBR mutants showed increased sensitivity to aminoglycoside.
This study was enabled by two technologies. One was a method for one-step inactivation of chromosomal genes in E. coli with PCR products (10), which permits easy deletion or modification of a target gene(s). The other was the PM technology (5), which permits examination of cellular phenotypes in a high-throughput format.
Altogether, we constructed over 100 deletion mutants, including two or more independent mutants with mutations in each gene or gene cluster. The mutations included 65 different deletions (50 distinguishable two-component mutations and 15 different other function mutations) (Tables 1 and 2). Many of these mutations were obtained by using new special template plasmids for creation of multiple mutations in the same strain. Details on the methodology, construction, and verification of these mutants will be described elsewhere (Zhou et al., unpublished).
The PM technology used in this study is an extension of previous work (5). Previously, assays were developed for measuring about 700 different phenotypes in seven 96-well microplates. This system was validated by using a small collection of E. coli mutants having lesions in genes whose functions are known. While most results were confirmatory, surprises were also encountered. For example, xylA and ynjB mutants, which had been thought to have single mutations, were shown to carry secondary lesions, including a linked lesion (5). Accordingly, phenotypic effects can be uncovered by detailed comparisons of mutant and parental strains by using standardized PM assays. High-throughput assays based on 300 growth phenotypes have also been developed for Saccharomyces cerevisiae (65-67).
PMs have now been expanded to include nearly 2,000 assays in a set of 20 96-well microplates. One-half of these assays measure basic cellular metabolism and stress functions. The others measure susceptibility to about 240 inhibitors at four different concentrations. Because the PM technology is new, it was important to evaluate and validate the data. We did this in three ways: (i) we carefully compared mutants with mutations in several systems for which many phenotypic changes were predictable; (ii) we conducted additional phenotypic tests to confirm numerous changes; and (iii) we always examined two or more independent mutants and looked for phenotypic changes that they had in common in order to reduce the chance of finding effects of secondary mutations.
We detected most, if not all, of the expected phenotypes for mutants with mutations in five systems (ArcB/ArcA, DcuS/DcuR, NtrB/NtrC, PhoR/PhoB, and UhpB/UhpA). We also found many expected phenotypes, as well as new phenotypes for several systems and, in particular, for the rssB and rpoS mutants (Table 5). Many mutants had phenotypes that are also reasonable based on current knowledge. For example, we found that the baeSR mutants exhibited antibiotic and toxic metabolite susceptibilities. This finding supports the recent discovery that the BaeS/BaeR system controls synthesis of an efflux pump (2, 51). We found that the cusRS mutants are hypersensitive to 1,10-phenanthroline, a high-affinity copper chelator (69). This provides further evidence that this system plays an important role in copper homeostasis (49). These results therefore provide confidence in the validity of the PM assays.
In a few cases we were unable to confirm phenotypic changes by additional tests. Since it was not feasible to conduct additional tests for all mutants, we are not certain about the cause of such discrepancies. It is notable that we were unable to confirm most changes for the atoSC and uvrY mutants by serial dilution tests (Table 4). We do not know whether this reflects variability with regard to these mutants or whether particular PM tests are less reliable. In contrast, most other confirmation tests substantiated the PM assays. Perhaps, these mutants are phenotypically less stable than other mutants. Without more detailed knowledge of the basis of these phenotypes, it is difficult to be certain about the validity of the results of particular PM assays, such as these assays.
PM analysis permits a “big picture” perspective at the biological level. Not only does it reveal phenotypes that change, but in revealing phenotypes that do not change, it eliminates possibilities and helps focus the direction of future investigations. However, it is important to also consider the limitations of the technology. There are a number of reasons why PM technology does not uncover all phenotypes and all members of a regulon. First, the phenotyping set is, of course, not all-inclusive. Certainly, it is likely to miss phenotypes involving surface structures and functions such as flagella, attachment, biofilm formation, motility, and chemotaxis, as well as functions turned on only under anaerobic conditions. In this study, we were unable to detect motility and chemotaxis phenotypes (e.g., in cheABYZ mutants) and anaerobic phenotypes (e.g., in narXL, narP, narQ, and torRS mutants). Second, deletion of genes requiring other special conditions for expression would also not result in a mutant phenotype. And third, deletion of genes may not reveal mutant phenotypes if they involve redundant cellular functions.
Conclusions about gene function drawn from this study rely on the validity of comparing isogenic strains. Even genetically identical strains can rapidly acquire unknown secondary mutations (32, 76), especially if the strain carries a mutation that impairs growth or creates a condition which favors a compensatory mutation. The E. coli K-12 genome (4) contains a large number of transposable insertion elements which are known to give rise to frequent mutations (6, 37) and which may be especially problematic for genes near their insertion sites (34). We therefore examined at least two independent mutants for each system and considered only those phenotypes that the mutants had in common. In two cases (the phoBR and cbrBC mutants) we found agreement between two independent mutants but not with a third similar mutant. The basis of these anomalies is not understood. Nevertheless, the finding that two independent mutants had phenotypes in common provides confidence that the majority of the PM assay results are correct.
We also detected common phenotypes for mutants with mutations in individual genes belonging to the same regulatory system. While the arcA and arcB mutants each displayed about 50 phenotypic changes (Table 3), the majority of these changes were in common (Table 5). The barA and uvrY mutants had a new phenotype in common, enhanced utilization of melibiose (Table 5). The change in melibiose metabolism could be a secondary manifestation of a change in sodium metabolism, since this sugar is uniquely metabolized by a sodium cotransport system (75a). We also detected a dominant phenotype that the rcsB, yojN, and yojN-rcsBC mutants had in common, sodium chloride sensitivity (Table 4).
Besides the arcA and arcB mutants, four other mutants showed more than 10 phenotypic differences (Table 5). The cpxRA mutants displayed 23 changes with varied features that are coherent with the many gene regulatory targets of the cpxRA system (14). Mutants with mutations in the EnvZ/OmpR and NtrB/NtrC systems exhibited both expected and unexpected phenotypes. Curiously, the dcuSR and rssB mutants had many phenotypes in common (Table 4). The rssB mutants also had a large number of defects in the use of amino acids and peptides as N sources. Whether these phenotypes are related to a role in protein turnover (24) remains to be determined.
The mutants examined in this study included most of the mutants that were studied for global effects on gene expression (54). Mutations in the ArcA, AtoS/AtoC, DpiB/DpiA (CitA/CitB), EnvZ/OmpR, RcsB, UvrY, and YpdA/YpdB systems were shown to affect flagellar synthesis. Since PM assays do not measure motility, effects on flagellar synthesis would go unnoticed. It was also found that arcB, cpxRA, fimZ, ompR-envZ, rstAB, and yfhA mutations led to up-regulation of the ent operon (which encodes enzymes for enterochelin biosynthesis). Such phenotypes would also probably not be detected in PM assays. The maltose transport system was up-regulated in the arcB, dpiBA (citAB), rcsB, and uvrY mutants. Although we did see increased use of other C sources, we found no effects on maltose use in these or other mutants.
Several studies have now shown that there are regulatory connections among two-component systems and with other global regulators (25, 72). For example, the HK CreC can activate PhoB under certain conditions both in the absence of PhoR (82) and in the presence of PhoR (J. L. Masella and B. L. Wanner, unpublished results). The HK ArcB has also been shown to be involved in porin gene regulation in a manner requiring OmpR (44). In addition, results of DNA microarray studies provided evidence of regulatory interactions that are indicative of cross-regulation or overlapping regulons between the EnvZ/OmpR and AtoS/AtoC systems and among ArcB, RssB, UvrY, and the RpoS regulon (54).
We also found evidence which supports the hypothesis that there are regulatory networks, as many mutants had phenotypes in common with each other (Table 6). Here we found osmotic sensitivity of the ompR-envZ and atoSC mutants, except that they are sensitive to different agents. The cpxAR, ntrBC, and luxS mutants showed susceptibility to alanine at an elevated pH, which is indicative of a deamination defect. The arcA, arcB, luxS, and phnC-P mutants showed alkaline pH sensitivity. The arcB and rssB mutants were both sensitive to sodium nitrite. Is ArcB therefore the partner HK for RssB? The arcB, baeSR, and rssB mutants had sodium tungstate sensitivity in common. Clearly, many potentially interesting connections are suggested by the results summarized in Table 6. An understanding of the biochemical and physiological basis of these phenotypes requires further investigation.
TABLE 6.
Phenotypes that two-component and other mutants have in commona
| Mode of action | Phenotype | Genes deleted |
|---|---|---|
| Alanine deamination | Increased sensitivity to l-alanine at pH 9.5 | cpxRA, ntrBC, luxS |
| Alkaline pH sensitivity | Increased sensitivity to pH 10 | arcA, arcB, luxS, phnC-P |
| Amino acid analog | Increased resistance to β-chloro-l-alanine | arcA, arcB, ntrBC |
| Anion transport | Increased sensitivity to sodium nitrite | arcB, rssB |
| Anion transport | Increased sensitivity to sodium tungstate | arcB, baeSR, rssB |
| Antimicrobial | Increased sensitivity to myricetin | arcA, arcB, baeSR |
| C source | Decreased utilization of C4 acids | dcuRS, rssB |
| C source | Increased utilization of d-fructose and d-mannitol | ompR-envZ, phoPQ |
| C source | Increased utilization of d-melibiose | barA, uvrY |
| C source | Increased utilization of β-methyl-d-glucuronic acid | arcA, rpoS |
| Cation transport | Increased sensitivity to cobalt chloride | arcA, arcB, ompR-envZ, qseCB |
| Cation transport | Increased sensitivity to nickel chloride | baeSR, qseCB |
| Cell wall inhibitor | Increased sensitivity to cefoxitin | phnC-P, phoH |
| Cholinergic antagonist | Increased sensitivity to pridinol | phoH, rstAB |
| DNA damage, antifolate | Increased resistance to hydroxylamine | cbrA, uvrY |
| DNA synthesis inhibitor | Increased sensitivity to nitrofurazone | cbrBC, ntrBC, rcsB, uvrY |
| Folate antagonist | Increased sensitivity to trimethoprim | rcsB, yojN-rcsB |
| Lipophilic chelator | Increased sensitivity to 1,10-phenanthroline | arcA, arcB, cusRS, luxS |
| Lipophilic chelator | Increased sensitivity to 5,7-dichloro-8-hydroxyquinaldine | cbrA, luxS, phn |
| Lipophilic chelator | Increased sensitivity to 5-chloro-7-iodo-8-hydroxyquinoline | arcB, narP |
| Lipophilic chelator | Increased sensitivity to 8-hydroxyquinoline | arcB, psiE |
| Membrane agent, outer | Increased sensitivity to polymyxin B | atoSC, uvrY |
| Membrane perturbant | Increased sensitivity to membrane-active agents | arcA, arcB |
| Membrane, cationic detergent | Increased sensitivity to methyltrioctylammonium chloride | arcA, arcB, atoSC, uvrY |
| N source | Decreased utilization of Leu-Trp | cpxRA, rssB |
| N source | Decreased utilization of δ-amino-N-valeric acid | ntrBC, rssB |
| Osmotic sensitivity | Increased sensitivity to ethylene glycol | cpxRA, ompR-envZ, yojN |
| Osmotic sensitivity | Increased sensitivity to NaCl | atoSC, rcsB, yojN, yojN-rcsB |
| P sources | Decreased utilization of organophosphates as P sources | phoA, phoBR |
| Protein synthesis inhibitor | Increased resistance to troleandomycin | phn, phoH |
| Protein synthesis inhibitor | Increased sensitivity to amikacin | cpxRA, ntrBC |
| Protein synthesis inhibitor | Increased sensitivity to dihydrostreptomycin | atoSC, cpxRA, uvrY |
| Protein synthesis inhibitor | Increased sensitivity to Geneticin (G418) | arcA, cpxRA, ntrBC |
| Protein synthesis inhibitor | Increased sensitivity to hygromycin B | cpxRA, kdpFABCDE, rpoS |
| Protein synthesis inhibitor | Increased sensitivity to paromomycin | arcA, ntrBC, phoBR |
| Protein synthesis inhibitor | Increased sensitivity to tobramycin | cpxRA, ntrBC, phoBR |
| Protein synthesis inhibitor | Increased sensitivity to troleandomycin | arcB, rstAB |
| Respiration inhibitor | Increased sensitivity to iodonitrotetrazolium violet | atoSC, rcsB, uvrY |
| Respiration inhibitor | Increased sensitivity to thioridazine | arcA, arcB, ntrBC, ompR-envZ |
| S source | Decreased utilization of methane sulfonic acid | cpxRA, rssB |
| Transport, toxic cation | Increased sensitivity to lithium chloride | cpxRA, ompR-envZ |
See text.
Acknowledgments
We thank Peter Gadzinski and Eric Olender for help with computer software and data analysis and Mary Berlyn for guidance on nomenclature. We are also grateful to Sydney Kustu and Valley Stewart for advice on the Ntr and Nar mutants.
This research was supported by NIH award GM62107 to B.R.B and by NSF award MCB0110656 and NIH award GM62662 to B.L.W.
REFERENCES
- 1.Aiba, H., and T. Mizuno. 1990. Phosphorylation of a bacterial activator protein, OmpR, by a protein kinase, EnvZ, stimulates the transcription of the ompF and ompC genes in Escherichia coli. FEBS Lett. 261:19-22. [DOI] [PubMed] [Google Scholar]
- 2.Baranova, N., and H. Nikaido. 2002. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 184:4168-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bearson, S. M., J. A. Albrecht, and R. P. Gunsalus. 2002. Oxygen and nitrate-dependent regulation of dmsABC operon expression in Escherichia coli: sites for Fnr and NarL protein interactions. BMC Microbiol. 2:13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 4a.Bochner, B. R. 2003. New technologies to assess genotype-phenotype relationships. Nat. Rev. Genet. 4:309-314. [DOI] [PubMed] [Google Scholar]
- 5.Bochner, B. R., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadesus, J., and J. R. Roth. 1989. Absence of insertions among spontaneous mutants of Salmonella typhimurium. Mol. Gen. Genet. 216:210-216. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 8.Conter, A., R. Sturny, C. Gutierrez, and K. Cam. 2002. The RcsCB His-Asp phosphorelay system is essential to overcome chlorpromazine-induced stress in Escherichia coli. J. Bacteriol. 184:2850-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosloy, S. D., and E. McFall. 1973. Metabolism of d-serine in Escherichia coli K-12: mechanism of growth inhibition. J. Bacteriol. 114:685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davalos-Garcia, M., A. Conter, I. Toesca, C. Gutierrez, and K. Cam. 2001. Regulation of osmC gene expression by the two-component system rcsB-rcsC in Escherichia coli. J. Bacteriol. 183:5870-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Wulf, P., and E. C. C. Lin. 2000. Cpx two-component signal transduction in Escherichia coli: excessive CpxR-P levels underlie CpxA* phenotypes. J. Bacteriol. 182:1423-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Wulf, P., A. M. McGuire, X. Liu, and E. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277:26652-26661. [DOI] [PubMed] [Google Scholar]
- 15.Dwyer, F. P., I. K. Reid, A. Shulman, G. M. Laycock, and S. Dixson. 1969. The biological actions of 1,10-phenanthroline and 2,2′-bipyridine hydrochlorides, quaternary salts and metal chelates and related compounds. 1. Bacteriostatic action on selected gram-positive, gram-negative and acid-fast bacteria. Aust. J. Exp. Biol. Med. Sci. 47:203-218. [DOI] [PubMed] [Google Scholar]
- 16.Epstein, W., E. Buurman, D. McLaggan, and J. Naprstek. 1993. Multiple mechanisms, roles and controls of K+ transport in Escherichia coli. Biochem. Soc. Trans. 21:1006-1010. [DOI] [PubMed] [Google Scholar]
- 17.Galán, J. E., and R. Curtiss III. 1989. Virulence and vaccine potential of phoP mutants of Salmonella typhimurium. Microb. Pathog. 6:433-443. [DOI] [PubMed] [Google Scholar]
- 18.Gassel, M., T. Mollenkamp, W. Puppe, and K. Altendorf. 1999. The KdpF subunit is part of the K(+)-translocating Kdp complex of Escherichia coli and is responsible for stabilization of the complex in vitro. J. Biol. Chem. 274:37901-37907. [DOI] [PubMed] [Google Scholar]
- 19.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the Arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 20.Golby, P., S. Davies, D. J. Kelly, J. R. Guest, and S. C. Andrews. 1999. Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli. J. Bacteriol. 181:1238-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groisman, E. A., E. Chiao, C. J. Lipps, and F. Heffron. 1989. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc. Natl. Acad. Sci. USA 86:7077-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guina, T., E. C. Yi, H. Wang, M. Hackett, and S. I. Miller. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 182:4077-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hengge-Aronis, R. 2002. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:341-346. [PubMed] [Google Scholar]
- 25.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoch, J. A., and T. J. Silhavy. 1995. Two-component regulatory systems. ASM Press, Washington, D.C.
- 27.Imamura, A., N. Hanaki, A. Nakamura, T. Suzuki, M. Taniguchi, T. Kiba, C. Ueguchi, T. Sugiyama, and T. Mizuno. 1999. Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 40:733-742. [DOI] [PubMed] [Google Scholar]
- 28.Iuchi, S., and E. C. C. Lin. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. USA 85:1888-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iuchi, S., and E. C. C. Lin. 1991. Adaptation of Escherichia coli to respiratory conditions: regulation of gene expression. Cell 66:5-7. [DOI] [PubMed] [Google Scholar]
- 30.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins, L. S., and W. D. Nunn. 1987. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J. Bacteriol. 169:42-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jishage, M., and A. Ishihama. 1997. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J. Bacteriol. 179:959-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung, K., M. Krabusch, and K. Altendorf. 2001. Cs+ induces the kdp operon of Escherichia coli by lowering the intracellular K+ concentration. J. Bacteriol. 183:3800-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamath, A. V., K. Gish, and C. Yanofsky. 1994. A copy of insertion element IS5 is present within tnaB in the Kohara library of Escherichia coli W3110. J. Bacteriol. 176:1546-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato, A., H. Tanabe, and R. Utsumi. 1999. Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J. Bacteriol. 181:5516-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kier, L. D., R. M. Weppelman, and B. N. Ames. 1979. Regulation of nonspecific acid phosphatase in Salmonella: phoN and phoP genes. J. Bacteriol. 138:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitamura, K., Y. Torii, C. Matsuoka, and K. Yamamoto. 1995. DNA sequence changes in mutations in the tonB gene on the chromosome of Escherichia coli K12: insertion elements dominate the spontaneous spectra. Jpn. J. Genet. 70:35-46. [DOI] [PubMed] [Google Scholar]
- 38.LaRossa, R. A. 1996. Mutant selections linking physiology, inhibitors, and genotypes, p. 2527-2587. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 39.Lee, A. I., A. Delgado, and R. P. Gunsalus. 1999. Signal-dependent phosphorylation of the membrane-bound NarX two-component sensor-transmitter protein of Escherichia coli: nitrate elicits a superior anion ligand response compared to nitrite. J. Bacteriol. 181:5309-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, Y. S., J. S. Han, Y. Jeon, and D. S. Hwang. 2001. The arc two-component signal transduction system inhibits in vitro Escherichia coli chromosomal initiation. J. Biol. Chem. 276:9917-9923. [DOI] [PubMed] [Google Scholar]
- 41.Liu, X., and T. Ferenci. 2001. An analysis of multifactorial influences on the transcriptional control of ompF and ompC porin expression under nutrient limitation. Microbiology 147:2981-2989. [DOI] [PubMed] [Google Scholar]
- 42.Maeda, T., S. M. Wurgler-Murphy, and H. Saito. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242-245. [DOI] [PubMed] [Google Scholar]
- 43.Majdalani, N., D. Hernandez, and S. Gottesman. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46:813-826. [DOI] [PubMed] [Google Scholar]
- 44.Matsubara, M., S. I. Kitaoka, S. I. Takeda, and T. Mizuno. 2000. Tuning of the porin expression under anaerobic growth conditions by His-to-Asp cross-phosphorelay through both the EnvZ-osmosensor and ArcB-anaerosensor in Escherichia coli. Genes Cells 5:555-569. [DOI] [PubMed] [Google Scholar]
- 45.Metcalf, W. W., W. Jiang, L. L. Daniels, S.-K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 46.Metcalf, W. W., P. M. Steed, and B. L. Wanner. 1990. Identification of phosphate-starvation-inducible genes in Escherichia coli K-12 by DNA sequence analysis of psi::lacZ(Mu d1) transcriptional fusions. J. Bacteriol. 172:3191-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer, M., P. Dimroth, and M. Bott. 1997. In vitro binding of the response regulator CitB and of its carboxy-terminal domain to A+T-rich DNA target sequences in the control region of the divergent citC and citS operons of Klebsiella pneumoniae. J. Mol. Biol. 269:719-731. [DOI] [PubMed] [Google Scholar]
- 48.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munson, G. P., D. L. Lam, F. W. Outten, and T. V. O'Halloran. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182:5864-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muse, W. B., and R. A. Bender. 1998. The nac (nitrogen assimilation control) gene from Escherichia coli. J. Bacteriol. 180:1166-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagakubo, S., K. Nishino, T. Hirata, and A. Yamaguchi. 2002. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 184:4161-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagasawa, S., K. Ishige, and T. Mizuno. 1993. Novel members of the two-component signal transduction genes in Escherichia coli. J. Biochem. (Tokyo) 114:350-357. [DOI] [PubMed] [Google Scholar]
- 53.Nixon, B. T., C. W. Ronson, and F. M. Ausubel. 1986. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc. Natl. Acad. Sci. USA 83:7850-7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oshima, T., H. Aiba, Y. Masuda, S. Kanaya, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptome analysis of all two-component regulatory systems of Escherichia coli. Mol. Microbiol. 46:281-291. [DOI] [PubMed] [Google Scholar]
- 55.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parkinson, J. S. 1988. Protein phosphorylation in bacterial chemotaxis. Cell 53:1-2. [DOI] [PubMed] [Google Scholar]
- 57.Pernestig, A. K., D. Georgellis, T. Romeo, K. Suzuki, H. Tomenius, S. Normark, and O. Melefors. 2003. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J. Bacteriol. 185:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pernestig, A. K., O. Melefors, and D. Georgellis. 2001. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem. 276:225-231. [DOI] [PubMed] [Google Scholar]
- 59.Pratt, L. A., W. H. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 60.Prohinar, P., S. A. Forst, D. Reed, I. Mandic-Mulec, and J. Weiss. 2002. OmpR-dependent and OmpR-independent responses of Escherichia coli to sublethal attack by the neutrophil bactericidal/permeability increasing protein. Mol. Microbiol. 43:1493-1504. [DOI] [PubMed] [Google Scholar]
- 61.Pruss, B. M., J. W. Campbell, T. K. Van Dyk, C. Zhu, Y. Kogan, and P. Matsumura. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raffa, R. G., and T. L. Raivio. 2002. A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 45:1599-1611. [DOI] [PubMed] [Google Scholar]
- 63.Rainwater, S., and P. M. Silverman. 1990. The cpx proteins of Escherichia coli K-12: evidence that cpxA, ecfB, ssd, and eup mutations all identify the same gene. J. Bacteriol. 172:2456-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rieger, K. J., M. el Alama, G. Stein, C. Bradshaw, P. P. Slonimski, and K. Maundrell. 1999. Chemotyping of yeast mutants using robotics. Yeast 15:973-986. [DOI] [PubMed] [Google Scholar]
- 66.Rieger, K. J., A. Kaniak, J. Y. Coppee, G. Aljinovic, A. Baudin-Baillieu, G. Orlowska, R. Gromadka, O. Groudinsky, J. P. di Rago, and P. P. Slonimski. 1997. Large-scale phenotypic analysis—the pilot project on yeast chromosome III. Yeast 13:1547-1562. [DOI] [PubMed] [Google Scholar]
- 67.Ross-Macdonald, P., P. S. R. Coelho, T. Roemer, S. Agarwal, A. Kumar, R. Jansen, K. H. Cheung, A. Sheehan, D. Symoniatis, L. Umansky, M. Heldtman, F. K. Nelson, H. Iwasaki, K. Hager, M. Gerstein, P. Miller, G. S. Roeder, and M. Snyder. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402:413-418. [DOI] [PubMed] [Google Scholar]
- 68.Schneider, B. L., S.-P. Shiau, and L. J. Reitzer. 1991. Role of multiple environmental stimuli in the control of transcription from a nitrogen-regulated promoter in Escherichia coli with weak or no activator binding sites. J. Bacteriol. 173:6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sigman, D. S., M. D. Kuwabara, C. H. Chen, and T. W. Bruice. 1991. Nuclease activity of 1,10-phenanthroline-copper in study of protein-DNA interactions. Methods Enzymol. 208:414-433. [DOI] [PubMed] [Google Scholar]
- 70.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809-821. [DOI] [PubMed] [Google Scholar]
- 71.Stewart, V., Y. Lu, and A. J. Darwin. 2002. Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J. Bacteriol. 184:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suzuki, K., X. Wang, T. Weilbacher, A. K. Pernestig, O. Melefors, D. Georgellis, P. Babitzke, and T. Romeo. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 184:5130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Swanson, R. V., L. A. Alex, and M. I. Simon. 1994. Histidine and aspartate phosphorylation: two-component systems and the limits of homology. Trends Biochem. Sci. 19:485-490. [DOI] [PubMed] [Google Scholar]
- 74.Takeda, S. I., Y. Fujisawa, M. Matsubara, H. Aiba, and T. Mizuno. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC→YojN→RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40:440-450. [DOI] [PubMed] [Google Scholar]
- 75.Tracy, B. S., K. K. Edwards, and A. Eisenstark. 2002. Carbon and nitrogen substrate utilization by archival Salmonella typhimurium LT2 cells. BMC E vol. Biol. 2:14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75a.Tsuchiya, T., and T. H. Wilson. 1978. Cation-sugar cotransport in the melibiose transport system of Escherichia coli. Membr. Biochem. 2:63-79. [DOI] [PubMed] [Google Scholar]
- 76.Verma, M., and J. B. Egan. 1985. Phenotypic variations in strain AB1157 cultivars of Escherichia coli from different sources. J. Bacteriol. 164:1381-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walderhaug, M. O., J. W. Polarek, P. Voelkner, J. M. Daniel, J. E. Hesse, K. Altendorf, and W. Epstein. 1992. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J. Bacteriol. 174:2152-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang, N., G. Shaulsky, R. Escalante, and W. F. Loomis. 1996. A two-component histidine kinase gene that functions in Dictyostelium development. EMBO J. 15:3890-3898. [PMC free article] [PubMed] [Google Scholar]
- 79.Wanner, B. L. 1992. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J. Bacteriol. 174:2053-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wanner, B. L. 1994. Gene expression in bacteria using TnphoA and TnphoA′ elements to make and switch phoA gene, lacZ (op), and lacZ (pr) fusions, p. 291-310. In K. W. Adolph (ed.), Methods in molecular genetics, vol. 3. Academic Press, Orlando, Fla.
- 81.Wanner, B. L. 1996. Phosphorus assimilation and control of the phosphate regulon, p. 1357-1381. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. ASM Press, Washington, D.C.
- 82.Wanner, B. L., W. Jiang, S.-K. Kim, S. Yamagata, A. Haldimann, and L. L. Daniels. 1996. Are the multiple signal transduction pathways of the Pho regulon due to cross talk or cross regulation?, p. 297-315. In E. C. C. Lin and A. S. Lynch (ed.), Regulation of gene expression in Escherichia coli. R. G. Landes Co., Austin, Tex.
- 83.Wanner, B. L., and P. Latterell. 1980. Mutants affected in alkaline phosphatase expression: evidence for multiple positive regulators of the phosphate regulon in Escherichia coli. Genetics 96:242-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Webber, C. A., and R. J. Kadner. 1997. Involvement of the amino-terminal phosphorylation module of UhpA in activation of uhpT transcription in Escherichia coli. Mol. Microbiol. 24:1039-1048. [DOI] [PubMed] [Google Scholar]
- 85.Wurgler-Murphy, S. M., and H. Saito. 1997. Two-component signal transducers and MAPK cascades. Trends Biochem. Sci. 22:172-176. [DOI] [PubMed] [Google Scholar]
- 86.Zhou, L., S.-K. Kim, L. Avramova, K. A. Datsenko, and B. L. Wanner. 2003. Use of conditional-replication, integration, and modular CRIM plasmids to make single-copy lacZ fusions, p. 65-89. In M. Blot (ed.), Methods and tools in biosciences and medicine. Prokaryotic genomics. Birkhäuser Verlag, Basel, Switzerland.
- 87.Zhou, Y., S. Gottesman, J. R. Hoskins, M. R. Maurizi, and S. Wickner. 2001. The RssB response regulator directly targets σS for degradation by ClpXP. Genes Dev. 15:627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zientz, E., J. Bongaerts, and G. Unden. 1998. Fumarate regulation of gene expression in Escherichia coli by the DcuSR (dcuSR genes) two-component regulatory system. J. Bacteriol. 180:5421-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]