Abstract
The ability of gram-negative bacterial cells to transport cobalamin and iron-siderophore complexes and their susceptibility to killing by some bacteriophages and colicins are characteristics routinely used to assay mutations of proteins in the TonB-dependent energy transduction system. These assays vary greatly in sensitivity and are subject to perturbation by overexpression of TonB and, perhaps, other proteins that contribute to the process. Thus, the choice of assay and the means by which a potential mutant is expressed can greatly influence the interpretation and recognition of a given mutant. In the present study, we expressed TonB at several different quantified levels in cells that were then subjected to a panel of assays. Our results suggest that it is reasonable to regard the assays as having windows of sensitivity. Thus, while no single assay satisfactorily spans the potential range of TonB activity, it is evident that certain assays are better suited for resolving small deviations from wild-type levels of activity, with others most useful when activity levels are very low. It is apparent from the results that the application of all possible assays to the characterization of new mutants will yield the most meaningful results.
The TonB-dependent transport system of gram-negative bacteria provides for the harvest of cobalamin and iron-sequestering molecules from the external environment. Central to this system is the TonB protein, which transduces the potential energy of the cytoplasmic membrane proton gradient to TonB-gated transporters in the essentially unenergized outer membrane, to drive the pumping of specific ligands across the outer membrane (for a review, see reference 31). The mechanism of energy transduction appears to be a dynamic process in which TonB shuttles between the cytoplasmic and outer membrane to collect and then deliver conformationally stored potential energy (23, 26). These associations at the cytoplasmic membrane involve specific interactions with at least two proteins, ExbB and ExbD (9, 12, 24). At the outer membrane, TonB associates not only with the TonB-gated transporters (5, 14, 37), but also with several other proteins that currently lack obvious roles in energy transduction (16).
Much of what we currently understand regarding how the interplay of TonB and these components contributes to energy transduction comes from specific mutations and their effects on function (3, 4, 22, 24, 39). While this approach has proven fruitful, several features of the system complicate the interpretation of such studies. First, energy transduction itself is not yet a quantifiable event, with the common assays actually scoring events that occur following energy transduction, such as ligand transport, ligand-dependent growth, or susceptibility to agents such as colicins and bacteriophage that parasitize the transport system. Thus, the assays currently in use are indirect and have a wide range of sensitivities. This has long been recognized. For example, in one early study, tonB mutants unable to demonstrably transport cobalamin were nonetheless capable of cobalamin-dependent growth, reflecting the wide difference in sensitivity between two assays (2). A second complication to such studies arises from the use of plasmid-encoded proteins as mutagenesis targets. While the use of plasmids facilitates mutant generation and mobilization, it also results in consideration of proteins expressed at higher levels than they would be under wild-type conditions. Beyond the likelihood that the severity of a given mutation is downplayed by its overexpression, it has been established that stoichiometric imbalance of TonB system components results in their accelerated degradation (10, 38).
In the present study, we evaluated the sensitivity and discriminating power of assays commonly used to detect TonB activity. To determine if the accuracy of a given assay extended beyond simply distinguishing between the presence and absence of TonB, cells in which TonB was expressed at two intermediate levels were included. Also included were cells in which TonB was expressed at levels that were severalfold higher than chromosomally encoded TonB. This allowed us to determine the degree to which perturbations of stoichiometry influenced the outcomes of individual assays. The data indicate that the different assays can be assigned windows of sensitivity, with only the nutritional disk assays failing (in most cases) to give a response proportional to intermediate TonB levels.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The plasmids and bacterial strains used are summarized in Table 1. All bacteria are derivatives of Escherichia coli K-12. The bacterial strains generated in the present study were constructed by standard transduction techniques with phage P1 vir (30).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics | Source |
|---|---|---|
| E. coli | ||
| RK4349 | metE163::Tn10 | R. Kadner |
| RP7947 | pcnB::kan | 27 |
| W3110 | F− IN(rrnD-rrnE)1 | 17 |
| KP1033 | W3110 metE::Tn10 | Present study |
| KP1270 | W3110 aroB zhf-5::Tn10 | 23 |
| KP1344 | W3110 tonB::blaM | 23 |
| KP1402 | W3110 tonB::blaM metE::Tn10 | Present study |
| KP1406 | W3110 tonB::blaM aroB zhf-5::Tn10 | Present study |
| KP1407 | W3110 tonB::blaM metE::Tn10 pcnB::kan | Present study |
| KP1408 | W3110 tonB::blaM aroB zhf-5::Tn10 pcnB::kan | Present study |
| KP1413 | W3110 metE::Tn10 pcnB::kan | Present study |
| KP1414 | W3110 aroB zhf-5::Tn10 pcnB::kan | Present study |
| Plasmids | ||
| pACYC184 | Cmr Tcr p15A ori | 6 |
| pKP299 | pACYC184 tonB+ | 25 |
| pKP325 | pACYC184 PBAD-regulated tonB araC | 23 |
Media.
Bacterial strains and plasmids were maintained on Luria-Bertani (LB) agar plates (25), supplemented with chloramphenicol at 34 μg ml−1 where appropriate. Individual assays were performed with cells grown in or on tryptone (T) or M9 minimal medium (30), supplemented as described for each assay. Chromazurol S (CAS) plates were made as described (35).
Chemicals and reagents.
The anti-TonB monoclonal antibodies were prepared as described previously (21), while the anti-FhuA monoclonal antibody was the kind gift of James Coulton. Group B colicins were produced as previously described (25), with cells disrupted by passage through a French pressure cell. Horseradish peroxidase-conjugated goat-anti-mouse immunoglobulin G was purchased from Caltag Laboratories and used at 1:5,000 dilution. 55Fe was purchased from Amersham. Medium components were purchased from Difco Laboratories.
Establishment of TonB expression levels.
To determine relative levels of TonB expression, cells were grown to mid-log phase (A550 of 0.5 as determined in a Spectronic 20 spectrophotometer with a 1.5-cm path length) in liquid medium corresponding to the medium used for specific assays, harvested, and prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (24). Relative amounts of sample (based on A550 at harvest) were loaded on 11% gels, with serial twofold dilutions of samples from cells with chromosomally encoded TonB used as a standard for comparison. Resolved proteins were electrotransferred to polyvinylidene difluoride membranes and probed with TonB-specific monoclonal antibody, with subsequent visualization by enhanced chemiluminescence as previously described (25). Individual exposures were evaluated by scanning densitometry, and relative TonB levels were estimated from samples with densities within the linear range of the chromosomal standards.
Cobalamin-dependent growth assays.
Cells were grown to mid-log phase in M9 medium supplemented with 0.2% (wt/vol) d-glucose, 0.4% (wt/vol) vitamin-free Casamino Acids, 40 μg tryptophan ml−1, 0.4 μg thiamine ml−1, 10 mM MgSO4, 0.5 mM CaCl2, 88 μM iron (provided as FeCl3 · 6H2O), and 34 μg chloramphenicol ml−1, harvested, and then washed twice in unsupplemented M9. For colony growth assays, cells were diluted in M9, and about 1,000 CFU were spread in triplicate on M9 plates supplemented as above, except that Casamino Acids were replaced by a defined mix of amino acids lacking methionine, containing either 5 × 10−7, 5 × 10−9, 5 × 10−10, 5 × 10−11, or 5 × 10−12 M freshly prepared cobalamin or 0.67 mM methionine as a positive control. Plates were scored for growth (as indicated by colony presence) at 18 and 48 h postinoculation.
For disk assays, washed cells were suspended in an original volume of unsupplemented M9, with 200 μl then added to 5 ml of melted methionine-free supplemented M9 top agar containing 0.01% tetrazolium, overlaid on methionine-free supplemented M9 plates, and allowed to solidify. The use of tetrazolium allowed enhanced detection of growth zones but did not alter the apparent degree or extent of growth. Sterile disks containing either 5 μl of 100 or 500 nM cobalamin or 27 mM methionine were then added in triplicate, with a single, blank disk included as a sterility control. Resultant growth zones were measured at 18 h postinoculation.
Enterochelin secretion.
Cells were grown to mid-log phase in LB supplemented with 0.2% (wt/vol) glucose and harvested, and a uniform number (≈8 × 105) of cells was applied in triplicate to sterile cellulose disks on CAS plates. Plates were incubated 18 h at 37°C, and the zones of clearing were then measured.
Siderophore-dependent growth.
Cells were grown to mid-log phase in M9 medium supplemented as for the cobalamin-dependent growth assays except that iron was present at 1.85 μM, harvested, washed twice in unsupplemented M9 containing 0.1 mM nitrilotriacetate, and suspended to their original volume. Washed cells (100 μl) were added to 2.5 ml of iron-depleted M9 top agar supplemented as above except for the exclusion of iron and the presence of 0.1 mM diethylenetriaminepentaacetic acid (DTPA) as an iron chelator, and poured onto identically supplemented iron-depleted M9 plates. To measure ferrichrome-dependent growth, sterile cellulose disks containing 3 μl of iron-charged ferrichrome (0.5 mM, made by mixing a 1.0 mM solution of deferrated ferrichrome with an equal volume of 10 mM FeSO4 in 10 mM HCl) or a 5 mM FeSO4 control were added to the plates in triplicate, with growth zones measured after 18 h of incubation at 37°C. Ferric dicitrate-dependent growth was measured similarly, with sterile cellulose disks containing 5 μl of either 5 mM ferric dicitrate generated as described (40) or 5 mM FeSO4 or 490 mM sodium citrate as controls.
Transport of radiolabeled ligands.
Ferrichrome transport assays were performed with 55Fe-loaded ferrichrome as previously described (22), with cells grown as for siderophore-dependent growth assays. Immediately prior to labeling, samples were taken for SDS-PAGE and immunoblot analysis with an FhuA-specific monoclonal antibody to determine the relative levels of the cognate TonB-gated transporter.
Phage and colicin assays.
Colicin and phage spot titer assays were performed essentially as described previously (23) with serial fivefold dilutions of group B colicins and 10-fold serial dilutions of bacteriophage φ80. Cells were grown in T broth and plated onto T plates in T top agar supplemented with 0.2% (wt/vol) glucose. For relative φ80 titers, cells were mixed with aliquots of diluted phage, allowed to adsorb for 10 min at 37°C, and then plated in glucose-supplemented T top agar with 88 μM iron. In all cases the plates were inoculated in triplicate, incubated for 18 h at 37°C, and then scored for clearing or plaque number. Irreversible φ80 adsorption assays were performed as previously described (25), with cells grown as for siderophore-dependent growth assays.
RESULTS
To determine the relative efficacies of various assays for TonB activity, cells in which TonB was expressed at several different levels were examined. In addition to cells expressing chromosomally encoded TonB at native levels, cells in which plasmid-encoded TonB was either over- or underexpressed were used. Overexpression of TonB was achieved with plasmid pKP299, a pACYC184 derivative carrying the tonB gene expressed from its native promoter and under iron-dependent Fur regulation (33). Underexpression of TonB was achieved with plasmid pKP325, a pACYC184 derivative carrying the portion of the tonB gene from its transcriptional initiation site through its Rho-independent terminator (23), under stringent regulation of the PBAD promoter (11). This plasmid also carries a copy of the araC gene under its normal regulation to provide the regulatory protein for the PBAD promoter. Cells were grown under repressing conditions, i.e., in the absence of arabinose and presence of 0.2% glucose. TonB expressed under these conditions resulted from the “leakiness” of the PBAD promoter. To further limit TonB production, pKP325 was also placed in isogenic pcnB strains, in which the plasmid copy number per cell was greatly reduced.
Establishment of TonB expression levels.
Because the TonB assays tested in this study used more than one type of culture medium, it was necessary to establish expression levels for both a defined (M9) and a complex (T-broth) medium. To exclude potential contributions by or competition with any endogenous TonB-dependent substrate, many assays were performed with aroB strains, which cannot synthesize the siderophore enterochelin or its siderophore precursors. TonB expression was quantified by resolving cell lysates by SDS-PAGE, immunoblotting with TonB-specific monoclonal antibody, and subsequent enhanced chemiluminescence visualization and scanning densitometry.
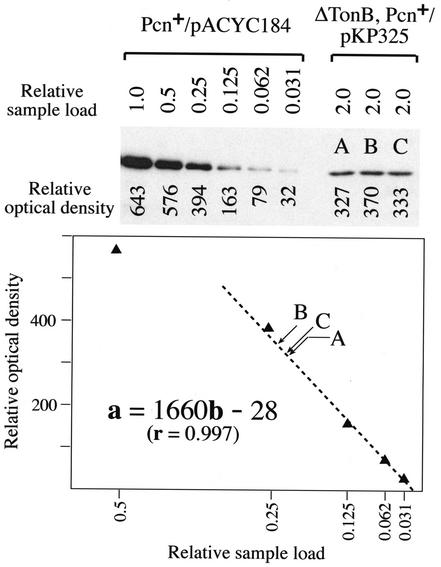
Figure 1 shows a sample analysis. Here, a triplicate load of sample in which a plasmid-encoded, PBAD-regulated TonB was expressed in an AroB− PcnB+ strain grown in T-broth (Fig. 1, lanes A to C, right panel) is compared to a standard comprised of a serial twofold dilution of sample from an identically grown isogenic strain in which TonB was expressed from the chromosome (left panel). In this example, the four greatest dilutions of the chromosomally encoded TonB fell within the linear range of detection. Interpolation of the triplicate samples (labeled A to C, with an A550-based relative sample loading of 2.0) provides a mean value corresponding to a relative sample load of 0.22, resulting in a value of 0.11 for the amount of TonB expressed from the plasmid relative to the level expressed from the chromosome (Fig. 1).
FIG. 1.
Determination of TonB expression level relative to a chromosomal standard. An immunoblot of SDS-PAGE-resolved samples probed with a TonB-specific monoclonal antibody and visualized by enhanced chemiluminescence is shown at the top. Here, triplicate samples from KP1406 (ΔtonB aroB pcnB+) carrying PBAD-regulated tonB (on plasmid pKP325) and grown in glucose-supplemented T-broth (lanes A, B, and C, right panel) were compared to a standard consisting of a twofold serial dilution of sample from KP1270 (aroB pcnB+) carrying a control plasmid (pACYC184) and grown under identical conditions (left panel). The film presented was evaluated by scanning densitometry, with the relative optical density of each band indicated. Values for the chromosomal standard corresponding to relative loads from 0.031 to 0.5 are plotted at the bottom. In this example, the four lowest dilutions gave values (0.031 to 0.25) proportional to the amount of sample loaded (with a range that bracketed the values of the samples to be compared), whereas the undiluted (not shown) and 0.5 dilution samples gave values less than predicted, indicating that the signal detected exceeded the linear range of the detection system (film saturation). Linear regression of data for the four lowest dilutions predicted a line (as shown) with a slope and intercepts as indicated in the equation. The optical density of each test sample could then be superimposed on the line (as indicated), and the relative amount of TonB could be determined by interpolation; likewise, the optical density can be entered into the equation as a, with the relative TonB level predicted by solving for b. In the example presented, test samples were loaded at a level twofold that of the undiluted chromosomal standard, and thus the predicted levels for these samples are then divided by 2 to determine the actual relative level of TonB for the test samples.
The results obtained by this approach for the three strain and culture medium combinations used in this study are summarized in Table 2. For each data set, the values are presented relative to the level of chromosomally encoded TonB. Among the individual sets, the relative levels of TonB from plasmid-borne, PBAD-regulated tonB were not significantly different, with the levels in PcnB+ strains being about eightfold less than chromosomally encoded levels, while the levels in PcnB− strains were about 200-fold less. This uniformity did not hold for plasmid-borne tonB under native Fur-mediated regulation, with the levels of full-length TonB ranging from 2.5- to ninefold that of chromosomally encoded TonB. The absolute levels of chromosomally encoded TonB among sets were 2.5-fold greater for the AroB− strains under either culture regimen (data not shown), consistent with previous observations of increased TonB expression under conditions of iron limitation (15, 33).
TABLE 2.
Relative levels of TonB expressiona
| TonB source | Relative TonB level
|
||
|---|---|---|---|
| AroB+ | AroB−
|
||
| M9 | M9 | T | |
| Chromosomal | 1 | 1 | 1 |
| pKP299 | 8 | 2.5 | 9 |
| pKP325, PcnB+ | 0.13 | 0.12 | 0.11 |
| pKP325, PcnB− | 0.004 | 0.004 | 0.005 |
Values are expressed as ratios relative to the level of chromosomally encoded TonB under the given genetic and culture circumstances. Note that for the AroB− strains, the absolute values for chromosomally encoded TonB were about 2.5-fold that of the AroB+ strains. M9 medium was supplemented with 0.2% d-glucose, 0.4% vitamin-free Casamino Acids, and 40 μg of tryptophan ml−1, 0.4 μg of thiamine ml−1, 10 mM MgSO4, 0.5 mM CaCl2, 88 μM iron (provided as FeCl3 · 6H2O), and 34 μg chloramphenicol ml−1. T medium was supplemented with 0.2% d-glucose and 34 μg of chloramphenicol ml−1.
Cobalamin-based assays.
In the absence of the methionine synthase encoded by metE, the ability of E. coli to methylate homocysteine to produce methionine (and thus grow in the absence of exogenously provided methionine) depends on the activity of the cobalamin-dependent methionine synthase isozyme encoded by metH (2). Because outer membrane active transport of cobalamin requires a TonB-gated transporter (BtuB), this assay provides a measure of TonB activity. Originally, this assay was performed on a minimal medium lacking methionine but supplemented with various concentrations of cobalamin, with results scored as colony size. In that study, very low concentrations of cobalamin (50 pM) were sufficient to support full growth of a TonB+ metE strain, whereas tonB strains could be divided into two groups, those requiring about 10- and those requiring about 1,000-fold more cobalamin to form colonies (2).
In the present study, with a minimal medium similar to the original but supplemented with 88 μM Fe, a TonB+ metE strain (KP1033) formed visible colonies on plates containing as little as 5 pM cobalamin (with 50 pM cobalamin producing colonies similar to those produced on methionine-replete medium), whereas a ΔtonB metE strain (KP1402) required about 10,000-fold more cobalamin to form visible colonies (data not shown). Strains in which TonB was expressed at levels 8- or 200-fold less than the chromosomal level also formed visible colonies on plates with 5 pM cobalamin and were thus indistinguishable from cells with wild-type levels of expression when assayed under these conditions (data not shown).
Whereas the above assay allowed detection of low levels of TonB activity, it did not discriminate between the TonB levels tested here. We tried the assay in a disk format. Here, cells were suspended in a minimal top agar, to which was applied sterile cellulose disks containing 5 μl of either 100 or 500 μM cobalamin or 27 mM methionine as a positive control. TonB+ metE strains (KP1033 and KP1413) formed a growth zone around cobalamin-laden disks, whereas a ΔtonB metE strain (KP1402) did not (Table 3). Surprisingly, strains with lower relative levels of TonB gave larger but more diffuse growth zones, as did the strain in which TonB was overexpressed (Table 3).
TABLE 3.
Cobalamin-dependent growth of metE strains with various levels of TonB expression
| Strain/plasmid | Relative TonB level | Growth (mean diam [mm] ± SD) with:
|
||
|---|---|---|---|---|
| 27 mM methionine | 100 nM cobalamin | 500 nM cobalamin | ||
| KP1402/pACYC184 | 0.0 | 27 ± 0.5 | No growth | No growth |
| KP1033/pACYC184 | 1.0 | 26 ± 0.5 | 11 ± 0.5 | 19 ± 0.5 |
| KP1413/pACYC184 | 1.0 | 27 ± 0.5 | 11 ± 0.5 | 20 ± 0.8 |
| KP1402/pKP325 | 0.13 | 26 ± 0.5 | 13 ± 0.5 | 24 ± 0.5 |
| KP1407/pKP325 | 0.004 | 29 ± 1.0 | 20 ± 0.5 | 27 ± 0.0 |
| KP1402/pKP299 | 8.0 | 27 ± 0.5 | 14 ± 0.5 | 21 ± 0.5 |
Siderophore-based assays.
Disruption of TonB function decreases cell iron levels, which in turn triggers the upregulation of virtually all of the genes involved in siderophore-mediated iron uptake. The enhanced secretion of the E. coli siderophore enterochelin could provide an indirect measure of TonB activity. Siderophore secretion can be visualized with CAS indicator plates (35). In the present study, expression of TonB at levels either 8- or 200-fold less than the chromosomal level provided for zones of clearing on CAS plates similar in extent to those produced by the ΔtonB control, whereas little or no clearing was evident for strains with TonB at chromosomally encoded levels (Table 4). Cells in which TonB was overproduced also showed clearing, but these zones were not as large as those resulting from underexpression of TonB (Table 4).
TABLE 4.
Siderophore secretion by strains with various levels of TonB expression
| Strain/plasmid | Relative TonB level | Mean cleared zone diam (mm) ± SD |
|---|---|---|
| KP1402/pACYC184 | 0.0 | 13 ± 0.5 |
| KP1033/pACYC184 | 1.0 | No zone |
| KP1413/pACYC184 | 1.0 | No zone |
| KP1402/pKP325 | 0.13 | 12 ± 0.0 |
| KP1407/pKP325 | 0.004 | 13 ± 0.0 |
| KP1402/pKP299 | 8.0 | 10 ± 0.5 |
A second assay of TonB-dependent iron uptake is siderophore-dependent growth. These studies were performed similarly to the cobalamin disk assays, here examining the ability of iron chelates to support the growth of aroB strains (unable to make their own siderophore) on iron-limited medium. Iron-charged ferrichrome (ligand of the TonB-gated transporter FhuA) provided for robust growth of TonB+ strains (KP1270 and KP1414), while the ΔtonB strain (KP1406) showed only faint growth, similar to that achieved with unchelated iron alone (Table 5). As noted above for cobalamin-dependent growth, cells expressing TonB at an eightfold reduction had larger growth zones than cells expressing TonB from the chromosome, but in contrast to the cobalamin results, the phenomenon did not extend to cells with 200-fold-reduced TonB levels, for which growth was indistinguishable from that of the ΔtonB strain (Table 5).
TABLE 5.
Ferrichrome and ferric dicitrate-dependent growth of aroB strains with various levels of TonB expression
| Strain/plasmid | Relative TonB level | Growth (mean diam [mm] ± SD) with:
|
|||
|---|---|---|---|---|---|
| 0.5 mM ferrichrome | 5.0 mM FeSO4 | 5.0 mM ferric dicitrate | 490 mM sodium citrate | ||
| KP1406/pACYC184 | 0.0 | Faint growtha | Faint growth | 11 ± 1.0 | 10 ± 0.0 |
| KP1270/pACYC184 | 1.0 | 18 ± 1.0 | 7 ± 0.0 | 15 ± 0.0 | 11 ± 1.0 |
| KP1414/pACYC184 | 1.0 | 20 ± 1.0 | 8 ± 0.5 | 16 ± 0.5 | 12 ± 0.0 |
| KP1406/pKP325 | 0.12 | 25 ± 0.0 | 7 ± 0.5 | 13 ± 0.5 | 9 ± 0.0 |
| KP1408/pKP325 | 0.004 | Faint growth | Faint growth | 12 ± 1.0 | 9 ± 0.0 |
| KP1406/pKP299 | 2.5 | 22 ± 1.0 | 7 ± 0.0 | 15 ± 0.5 | 13 ± 0.5 |
Faint growth indicates a diffuse zone of growth lacking a clear boundary and thus not readily measured in this assay.
As in the cobalamin disk assays, cells in which TonB was overproduced produced larger, slightly more diffuse growth zones than cells expressing TonB from the chromosome (Table 5). For ferric dicitrate (ligand of the TonB-gated transporter FecA), the results were less distinct, with sodium citrate alone supporting significant growth. This result suggests that the citrate levels used here were sufficient to compete effectively with DTPA for residual iron in the medium (and thus, rather than a control, actually represented either a lower but undefined concentration of citrate-chelated iron or the potential ability of citrate to render some iron available for low-affinity TonB-independent transport). It is also likely that the levels of citrate used here allowed some diffusion of ferric dicitrate through porin channels. Together, these complications make interpretation of ferric dicitrate-dependent growth difficult.
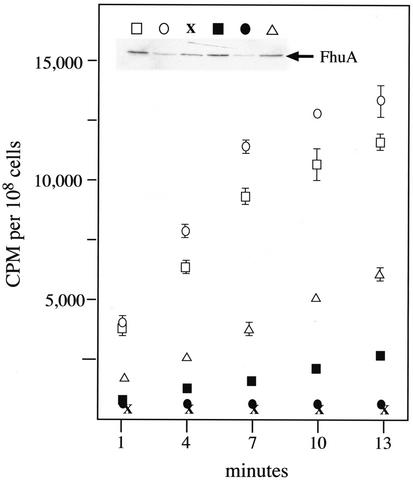
The most direct measure of TonB-dependent iron uptake is to follow the transport of labeled substrates. Ferrichrome transport was diminished in cells in which TonB expression was reduced by eightfold and not detectible in cells with 200-fold-reduced TonB levels (Fig. 2). Preliminary studies with cobalamin gave similar results (data not shown). Overexpression of TonB by only 2.5-fold also resulted in a significant reduction of ferrichrome transport. While the level of the TonB-gated ferrichrome transporter FhuA varied between the strains used in this experiment (Fig. 2. inset), this variance did not correlate with the differences observed in ferrichrome transport.
FIG. 2.
Transport of [55Fe]ferrichrome. aroB strains expressing TonB at various levels were grown and assayed for uptake of [55Fe]ferrichrome as described in Materials and Methods, with data represented as cpm per 108 cells. A portion of each sample was removed just prior to assay and subjected to SDS-PAGE and immunoblot analysis with an FhuA-specific monoclonal antibody to establish levels of the TonB-gated ferrichrome transporter (inset). Strain-plasmid combinations (and relative TonB levels) are keyed as follows: open square, KP1270/pACYC184 (TonB = 1.0); open circle, KP1414/pACYC184 (TonB = 1.0); X, KP1406/pACYC184 (TonB = 0.0); solid square, KP1406/pKP325 (TonB = 0.12); solid circle, KP1408/pKP325 (TonB = 0.004); open triangle, KP1406/pKP299 (TonB = 2.5). Relative transport rates, calculated with the data collected at 1 and 7 min and expressed as cpm per 108 cells per minute, are as follows: KP1270/pACYC184 (TonB = 1), 811 ± 128; KP1414/pACYC184 (TonB =1), 1,220 ± 16; KP1406/pACYC184 (TonB = 0), 14 ± 8; KP1406/pKP325 (TonB = 0.12), 141 ± 12; KP1408/pKP325 (TonB = 0.004) 5 ± 27; and KP1406/pKP299 (TonB = 2.5), 385 ± 38.
Colicin- and bacteriophage-based assays.
The exploitation of the TonB-dependent transport system by group B colicins (7) and bacteriophages such as φ80 (29) has provided a means for selecting tonB strains and for assaying TonB activity. In the present study, a ninefold reduction in TonB level resulted in a roughly fivefold drop in resistance to all group B colicins tested (colicins B, D, Ia, and M), as measured by a spot titer assay (Table 6). When TonB was expressed at a level 200-fold less than normal levels of chromosomally encoded TonB, a 25- to 125-fold drop in resistance was evident, depending on the colicin tested. Overexpression resulted in a slight (less than fivefold) decrease in apparent TonB activity for colicins B, D, and Ia but not for colicin M.
TABLE 6.
Sensitivity to group B colicins of aroB strains with various levels of TonB expression
| Strain/plasmid | Relative TonB level | Sensitivitya
|
|||
|---|---|---|---|---|---|
| Colicin B | Colicin D | Colicin Ia | Colicin M | ||
| KP1406/pACYC184 | 0.0 | R R R | R R R | R R R | R R R |
| KP1270/pACYC184 | 1.0 | 999 | 877 | 888 | 776 |
| KP1414/pACYC184 | 1.0 | 999 | 888 | 889 | 676 |
| KP1406/pKP325 | 0.11 | 888 | 776 | 788 | 666 |
| KP1408/pKP325 | 0.005 | 777 | 555 | 667 | 454 |
| KP1406/pKP299 | 9.0 | 889 | 777 | 777 | 777 |
Scored as the highest fivefold dilution of a standard colicin preparation that provided an evident zone of clearing on a cell lawn. Thus, 9 represents a dilution of 5 to the 9th power (59 = 1/1,953,125). R indicates resistance (i.e., no clearing) with undiluted colicin. The values of three platings are presented for each strain/plasmid and colicin pairing.
Spot titers with bacteriophage φ80 showed surprisingly little differences, with a ninefold reduction in relative TonB levels giving values near those of normal TonB levels. Only with the 200-fold reduction was a clear effect on TonB activity seen. In this assay, ninefold overexpression of TonB also had no apparent affect on TonB activity (Table 7). Similar results were obtained when relative titers were determined, with only the 200-fold reduction in TonB level having a significant effect on activity (Table 7). Initial attempts to titer phage on the strain with a 200-fold reduction in TonB resulted in no distinct plaques; only when T top agar was supplemented with 88 μM iron was plaque formation evident.
TABLE 7.
Sensitivity to bacteriophage φ80 of aroB strains with various levels of TonB expression
| Strain/plasmid | Relative TonB level | Sensitivity
|
|
|---|---|---|---|
| Spot titera | Relative titer (PFU) | ||
| KP1406/pACYC184 | 0.0 | R R R | 0 |
| KP1270/pACYC184 | 1.0 | 888 | 2.8 × 108 ± 0.1 × 108 |
| KP1414/pACYC184 | 1.0 | 778 | 2.2 × 108 ± 0.1 × 108 |
| KP1406/pKP325 | 0.11 | 887 | 2.1 × 108 ± 0.1 × 108 |
| KP1408/pKP325 | 0.005 | 776 | 4.8 × 106 ± 0.4 × 106 |
| KP1406/pKP299 | 9.0 | 877 | 2.7 × 108 ± 0.1 × 108 |
Scored as the highest 10-fold dilution of a phage preparation that provided an evident zone of clearing on a cell lawn. R indicates resistance (i.e., no clearing) with undiluted phage. For spot titers, the values of three platings are presented for each strain/plasmid and phage pairing.
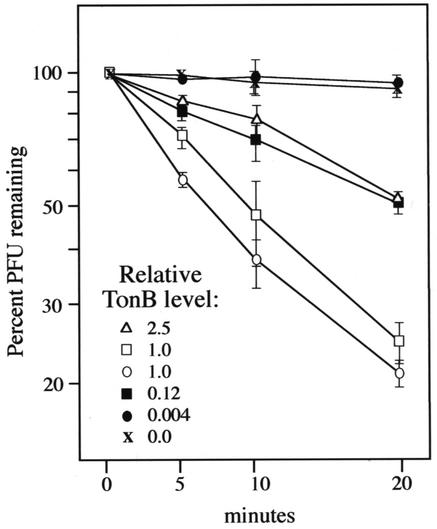
Whereas changes in TonB level had relatively minor effects in phage titer experiments, profound differences in activity were evident in irreversible phage adsorption assays (Fig. 3). Here, an eightfold reduction in TonB level greatly diminished phage adsorption, and a 200-fold reduction in TonB level rendered cells indistinguishable from the ΔtonB control. By this assay, moderate overexpression of TonB was again seen to impact TonB activity, to a degree similar to that of the eightfold reduction (Fig. 3).
FIG. 3.
Irreversible φ80 adsorption. aroB strains expressing TonB at various levels were grown as indicated in Materials and Methods and assayed for the ability to irreversibly adsorb bacteriophage φ80 as previously described (27), with 3 × 108 cells and φ80 at a multiplicity of infection of 1.0. Values are presented as the percentage of phage remaining following the indicated incubation; thus, the lower the value, the higher the relative TonB activity. Relative transport rates, calculated with the data collected at 0 and 10 min and expressed as 107 PFU adsorbed per minute, are as follows: KP1270/pACYC184 (TonB = 1), 1.6 ± 0.3; KP1414/pACYC184 (TonB =1), 1.9 ± 0.2; KP1406/pACYC184 (TonB = 0), 0.2 ± 0.2; KP1406/pKP325 (TonB = 0.12), 0.9 ± 0.4; KP1408/pKP325 (TonB = 0.004), −0.1 ± 0.2; and KP1406/pKP299 (TonB = 2.5), 0.7 ± 0.2.
DISCUSSION
The ability to transport cobalamin and various iron-siderophore complexes and sensitivity to group B colicins and certain bacteriophages provide the basis for assays routinely used to characterize mutations of proteins in the TonB system. Because each assay involves multiple independent components, it is generally recognized that the degree of sensitivity varies between assays and that individual assays give relative rather than absolute measures of TonB activity. For example, we have stated that in the absence of ExbB and ExbD, TonB activity is reduced 10-fold, based on irreversible φ80 adsorption (38) and cobalamin transport (37); yet when we measure by sensitivity to colicin B, the reduction in activity appears much greater (1). In this particular comparison, the assays are not only indirect, involving distinct components, but also occur on different time scales and consider different numbers of events, with transport assays measuring thousands of events over the course of minutes, while colicin sensitivity measures many fewer events over the course of hours.
TonB contacts the outer membrane transporters directly (for a recent review, see reference 34) and has been shown to be the limiting factor in transport processes (20). Certainly it is not an abundant protein. The number of energy transduction complexes under iron-replete conditions would be calculated at ≈175 per cell (15), slightly less than the least abundant transporter, BtuB, estimated at ≈200 per cell (41). In the present study, in which wild-type TonB was expressed at different levels to determine the correspondence between TonB activity and TonB protein level, the results from decreased TonB levels thus reflected the unique characteristics of each assay. We found that the various standard assays fell into several distinct groups based on their ability to discriminate among the various levels of TonB tested. These groups can be characterized as having different functional ranges or “windows” of sensitivity. A clear understanding of the limitations of the assays will prevent future misattribution to specific protein structural features (39).
The most sensitive assay is cobalamin-dependent growth, with the ability to discriminate between no TonB and 1/200-fold TonB but in which the 1/200-fold level confers a phenotype indistinguishable from that of cells in which TonB is encoded from the chromosome. Clearly this assay would not detect minor dysfunctions in the TonB system but does provide an exquisitely sensitive method for detecting very low levels of TonB activity, with the 1/200 level corresponding to ≈2 copies of TonB per cell under the conditions assayed. It has also historically proven useful in identifying cells with very low levels of TonB activity (14). This assay may in fact provide resolution when levels of TonB function are even lower. In its original application (2), this approach identified two classes of tonB strains, those indistinguishable from a known deletion, and a second class with a level of activity intermediate between that of the deletion strain and that of the wild-type strain. We examined one of these intermediate strains (RK4129) and found that it encoded a truncated TonB, suggesting the possibility of a nonsense mutation, with the minimal amount of TonB function evident potentially reflecting the rare translational readthrough of the premature stop (R. A. Larsen and K. Postle, unpublished observations).
We attempted to modify the cobalamin-dependent growth assay to a disk format to potentially provide more resolution at the intermediate levels of TonB activity. We were surprised that growth zone size was not proportional to the amount of TonB present. Rather, cells expressing TonB at levels only ≈1/8 and ≈1/200 of the wild-type level formed growth zones that were larger than those formed by cells with wild-type levels of TonB (Table 3). Similar results were obtained when we used a standard disk assay of ferrichrome-dependent growth, although in this case cells expressing TonB at ≈1/200 of the chromosomal level were unable to sustain growth (Table 5). This phenomenon of larger zones for strains with less activity has been seen previously in other disk-based iron-siderophore-dependent growth assays (32, 36). Presumably, less TonB results in larger growth zones because slowed transport and cell growth allow the nutrient to diffuse further in the plate before being consumed. The effect was greater for cobalamin, because so little cobalamin is needed to sustain growth (8), whereas for ferrichrome, TonB levels of ≈1/200 are unable to sustain growth at the concentration of ferrichrome that occurs adjacent to the disk, let alone at a distance. For these assays, it would thus appear that the interplay between diffusion rate, growth rate, and absolute minimal concentrations able to sustain growth complicates interpretation of the results. As indicated in Table 5, disk assays with ferric dicitrate did not display this phenomenon. However, because the entire ferric citrate transport system is upregulated by extracellular citrate (41), the individual contribution of TonB in this assay is difficult to interpret.
The second most sensitive assay tested was cell susceptibility to killing by bacteriophage φ80, with a window of sensitivity that overlapped that of cobalamin-dependent growth but was still useful with slightly higher (in this study, >1/200-fold but <1/eightfold) levels of TonB activity. Sensitivity to φ80 can detect vanishingly small amounts of TonB activity (4, 23), presumably because entry of a single phage is sufficient to result in cell death and, like the growth assays, occurs over a course of hours.
The colicin-dependent killing assays had a moderate degree of sensitivity to TonB activity and offer a mid-range window that overlaps both phage sensitivity and transport-based assays. For the four group B colicins tested here, colicins B, D, and Ia distinguished each level of TonB examined, although the apparent decrease in TonB function was minimal in this assay (Table 6). The resolving power of colicin M was similar, although the distinction between cells expressing wild-type levels of TonB and cells expressing TonB at ≈1/8 this level was less evident than that obtained with the other colicins (Table 6). Like the φ80 killing assay, colicin sensitivity is believed to measure a small number of events over a course of hours, as it is generally held that a few colicin molecules are sufficient for killing.
Transport assays (including irreversible φ80 adsorption) could distinguish among chromosomal, 1/8-, and 1/200-fold levels of TonB, but cells with a 1/200-fold level of TonB were indistinguishable from those lacking TonB (Fig. 2 and 3). These assays offer the advantage of producing more quantitative data than the growth-dependent or killing assays but are also more labor intensive.
The least sensitive assay examined was enterochelin secretion. In this assay, cells with TonB levels as high as ≈1/8 of the chromosomal level gave results identical to those of TonB null cells (Table 4). This narrow window of sensitivity limits the assay usefulness to simply determining if TonB is functioning to a degree sufficient to avoid upregulation of siderophore biosynthesis.
An important factor when assaying the activity of a TonB allele is the maintenance of system component stoichiometry. Obviously, if TonB or some other essential component is present at decreased levels, the amount of activity observed in a given assay should be decreased. Interestingly, this also appears to be the case when TonB is overexpressed. The dominant negative gene dosage effect was first noted in cobalamin transport studies (13), in which the inclusion of tonB on a multicopy plasmid resulted in reduced transport. A subsequent study by these researchers found that the presence of tonB on a multicopy plasmid also reduced transport of ferrichrome and cell sensitivity to group B colicins (28). In the present study, for an approximately eight- to ninefold overexpression of TonB, the dominant negative gene dosage effect was evident even with the relatively insensitive assay of siderophore secretion (Table 4). Similarly, 2.5-fold overexpression of TonB reduced ferrichrome transport (Fig. 2) and irreversible φ80 adsorption (Fig. 3) activities to less than 50% of wild-type levels, similar to the values corresponding to the expression of TonB at a level approximately one-eighth that of the chromosomally encoded wild-type level.
Because we overexpressed TonB from a multicopy plasmid with a wild-type tonB regulatory region, one possible explanation for the higher level of siderophore secretion is titration of Fur protein by multiple copies of the tonB Fur box. This would have the effect of inducing expression of the enterochelin biosynthetic genes. While the present study does not exclude this possibility, such a mechanism would not explain the overexpression-induced decreases in TonB activity observed in transport assays. An explanation consistent with the results of each of these assays is based on the instability of TonB when its stoichiometry with ExbB/D is perturbed, either by a relative increase in TonB (10) or by a relative decrease in ExbB/D (38). In either case, TonB is subject to proteolysis, with an accumulation of partially degraded, nonfunctional fragments. It is likely that at least some of these fragments compete with functional TonB for interaction sites, with the resultant decrease in overall TonB-dependent function—a negative gene dosage effect—reported previously (28). Evidence supporting such a mechanism includes the observation that fragments generated by the cleavage of TonB at an engineered leader peptidase site interfere with the ability of wild-type TonB to support either cobalamin transport or irreversible φ80 adsorption (19) and the finding that expression of the carboxyl-terminal 118 residues of TonB in the periplasmic space inhibits TonB-dependent transport of both ferrichrome and ferric dicitrate, infection by φ80, and killing by colicin M (18).
Acknowledgments
Strain KP1033 was constructed by J. Jaskula. We thank James Coulton for providing the FhuA-specific monoclonal antibody. We thank Joydeep Ghosh for helpful discussions and the suggestion that the rate of substrate depletion influenced growth zone size.
This work was supported by National Institute of General Medical Sciences research grant GM42146 (to K.P.).
REFERENCES
- 1.Ahmer, B. M. M., M. G. Thomas, R. A. Larsen, and K. Postle. 1995. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J. Bacteriol. 177:4742-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassford, P. J. J., C. Bradbeer, R. J. Kadner, and C. A. Schnaitman. 1976. Transport of vitamin B12 in tonB mutants of Escherichia coli. J. Bacteriol. 128:242-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, V., S. Gaisser, C. Herrman, K. Kampfenkel, H. Killman, and I. Traub. 1996. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178:2836-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cadieux, N., C. Bradbeer, and R. J. Kadner. 2000. Sequence changes in the ton box region of BtuB affect its transport activities and interaction with TonB protein. J. Bacteriol. 182:5954-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadieux, N., and R. J. Kadner. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA 96:10673-10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies, J. K., and P. Reeves. 1975. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group B. J. Bacteriol. 123:96-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Girolamo, P. M., R. J. Kadner, and C. Bradbeer. 1971. Isolation of vitamin B12 transport mutants of Escherichia coli. J. Bacteriol. 106:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eick-Helmerich, K., and V. Braun. 1989. Import of biopolymers into Escherichia coli: nucleotide sequences of the exbB and exbD genes are homologous to those of the tolQ and tolR genes, respectively. J. Bacteriol. 171:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer, E., K. Günter, and V. Braun. 1989. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J. Bacteriol. 171:5127-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose P-BAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Held, K. G., and K. Postle. 2002. ExbB and ExbD do not function independently in TonB-dependent energy transduction. J. Bacteriol. 184:5170-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heller, K., B. J. Mann, and R. J. Kadner. 1985. Cloning and expression of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. J. Bacteriol. 161:896-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heller, K. J., R. J. Kadner, and K. Günter. 1988. Suppression of the btuB451 mutation by mutations in the tonB gene suggests a direct interaction between TonB and TonB-dependent receptor proteins in the outer membrane of Escherichia coli. Gene 64:147-153. [DOI] [PubMed] [Google Scholar]
- 15.Higgs, P. I., R. A. Larsen, and K. Postle. 2002. Quantitation of known components of the Escherichia coli TonB-dependent energy transduction system: TonB, ExbB, ExbD, and FepA. Mol. Microbiol. 44:271-281. [DOI] [PubMed] [Google Scholar]
- 16.Higgs, P. I., T. E. Letain, K. K. Merriam, N. S. Burke, H. Park, C. Kang, and K. Postle. 2002. TonB interacts with nonreceptor proteins in the outer membrane of Escherichia coli. J. Bacteriol. 184:1640-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill, C. W., and B. W. Harnish. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. USA 78:7069-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard, S. P., C. Herrmann, C. W. Stratilo, and V. Braun. 2001. In vivo synthesis of the periplasmic domain of TonB inhibits transport through the FecA and FhuA iron siderophore transporters of Escherichia coli. J. Bacteriol. 183:5885-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaskula, J. C., T. E. Letain, S. K. Roof, J. T. Skare, and K. Postle. 1994. Role of the TonB amino terminus in energy transduction between membranes. J. Bacteriol. 176:2326-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadner, R. J., and K. J. Heller. 1995. Mutual inhibition of cobalamin and siderophore uptake systems suggests their competition for TonB function. J. Bacteriol. 177:4829-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen, R. A., P. S. Myers, J. T. Skare, C. L. Seachord, R. P. Darveau, and K. Postle. 1996. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J. Bacteriol. 178:1363-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen, R. A., and K. Postle. 2001. Conserved residues Ser(16) and His(20) and their relative positioning are essential for TonB activity, cross-linking of TonB with ExbB, and the ability of TonB to respond to proton motive force. J. Biol. Chem. 276:8111-8117. [DOI] [PubMed] [Google Scholar]
- 23.Larsen, R. A., M. G. Thomas, and K. Postle. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31:1809-1824. [DOI] [PubMed] [Google Scholar]
- 24.Larsen, R. A., M. T. Thomas, G. E. Wood, and K. Postle. 1994. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (ΔV17) by a missense mutation in ExbB. Mol. Microbiol. 13:627-640. [DOI] [PubMed] [Google Scholar]
- 25.Larsen, R. A., G. E. Wood, and K. Postle. 1993. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol. Microbiol 10:943-953. [DOI] [PubMed] [Google Scholar]
- 26.Letain, T. E., and K. Postle. 1997. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in gram-negative bacteria. Mol. Microbiol. 24:271-283. [DOI] [PubMed] [Google Scholar]
- 27.Liu, J., and J. S. Parkinson. 1989. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J. Bacteriol. 171:1254-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mann, B. J., C. D. Holroyd, C. Bradbeer, and R. J. Kadner. 1986. Reduced activity of TonB-dependent functions in strains of Escherichia coli. FEMS Lett. 33:255-260. [Google Scholar]
- 29.Matsushiro, A. 1963. Specialized transduction of tryptophan markers in Escherichia coli K-12 by bacteriophage φ80. Virology 19:475-482. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Moeck, G. S., and J. W. Coulton. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol 28:675-681. [DOI] [PubMed] [Google Scholar]
- 32.Newton, S. M., J. D. Igo, D. C. Scott, and P. E. Klebba. 1999. Effect of loop deletions on the binding and transport of ferric enterobactin by FepA. Mol. Microbiol 32:1153-1165. [DOI] [PubMed] [Google Scholar]
- 33.Postle, K. 1990. Aerobic regulation of the Escherichia coli tonB gene by changes in iron availability and the fur locus. J. Bacteriol. 172:2287-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Postle, K., and R. J. Kadner. Touch and go: tying TonB to transport. Mol. Microbiol., in press. [DOI] [PubMed]
- 35.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 36.Scott, D. C., Z. Cao, Z. Qi, M. Bauler, J. D. Igo, S. M. Newton, and P. E. Klebba. 2001. Exchangeability of N termini in the ligand-gated porins of Escherichia coli. J. Biol. Chem. 276:13025-13033. [DOI] [PubMed] [Google Scholar]
- 37.Skare, J. T., B. M. M. Ahmer, C. L. Seachord, R. P. Darveau, and K. Postle. 1993. Energy transduction between membranes, - TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J. Biol. Chem. 268:16302-16308. [PubMed] [Google Scholar]
- 38.Skare, J. T., and K. Postle. 1991. Evidence for a TonB-dependent energy transduction complex in Escherichia coli. Mol. Microbiol. 5:2883-2890. [DOI] [PubMed] [Google Scholar]
- 39.Traub, I., S. Gaisser, and V. Braun. 1993. Activity domains of the TonB protein. Mol. Microbiol. 8:409-423. [DOI] [PubMed] [Google Scholar]
- 40.Velayudhan, J., N. J. Hughes, A. A. McColm, J. Bagshaw, C. L. Clayton, S. C. Andrews, and D. J. Kelly. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol 37:274-286. [DOI] [PubMed] [Google Scholar]
- 41.White, J. C., P. M. DiGirolamo, M. L. Fu, Y. A. Preston, and C. Bradbeer. 1973. Transport of vitamin B12 in Escherichia coli: isolation and properties of the initial B12-binding site. J. Biol. Chem. J. Biol. Chem. 248:3978-3986. [PubMed] [Google Scholar]