Abstract
The recA gene from Thermus thermophilus HB27 was cloned and engineered to obtain insertion (recA::kat) and deletion (ΔrecA) derivatives. Transcription of recA in this extreme thermophile was induced by mitomycin C, leading to the synthesis of a monocistronic mRNA. This DNA damage-mediated induction was dependent on the integrity of recA. In addition to UV sensitivity, the recA mutants of T. thermophilus showed severe pleiotropic defects, ranging from irregular nucleoid condensation and segregation to a dramatic reduction in viability during culture. An increase in the frequency of both carotenoidless and auxotrophic mutants within surviving cells of the ΔrecA strain indicated a high mutation rate. As RecA is not required for plasmid transformation, we have used the α-lacZ gene fragment and the ampicillin resistance gene from Escherichia coli as passenger reporters to confirm such high mutation rates. Our data support the idea that the absence of RecA results in a hypermutational phenotype in T. thermophilus. Furthermore, a direct relationship is deduced between the growth temperature and mutation rate, which finally has a deleterious effect on cell survival in the absence of RecA.
RecA is a multifunctional protein that plays key roles in several cellular processes, such as recombination and DNA repair in Bacteria (10, 23). Accordingly, the sequences of RecA proteins from different origins are well conserved, allowing the construction of robust phylogenetic trees (8). Homologues to the RecA protein have also been described in Eucarya and Archaea (17, 32).
The functions of RecA have been analyzed in great detail in Escherichia coli, where different mutants are available (42). RecA controls the expression of the SOS regulon through its inducible coprotease activity. This activity is acquired when RecA binds to single-stranded DNA regions originated by the arrest of DNA replication or as a consequence of DNA damage. After a conformational change, activated RecA promotes autocleavage of the SOS repressor LexA, which in the E. coli LexA protein occurs at the Ala84-Gly85 peptide bond. This allows access by the RNA polymerase to at least 30 gene promoters, including those of the recA and lexA genes (42). The final goal of the overexpression of the SOS genes is to permit the cell to grow by repairing most of the damage and also by translesion DNA synthesis using the specialized DNA polymerase V. This DNA polymerase requires the help of RecA and single-stranded DNA-binding proteins to act (28, 37). The translesion synthesis mechanism is responsible for the hypermutational effect that the induction of the SOS system has in E. coli and other bacteria. In fact, recA mutants of E. coli are defective in DNA damage-mediated mutagenesis and show a low spontaneous mutation rate (42).
RecA is also required for homologous recombination in normal cells (30). Therefore, and despite their extreme UV sensitivity, recA mutants of E. coli are commonly used as hosts for genetic engineering, since the stability of plasmid constructs is relatively great. In the gram-positive Bacillus subtilis, the RecA protein is also required for chromosomal transformation by natural competence (3), and its induction is under the control of DinR, a functional homologue of the LexA repressor (44, 45).
Deinococcus and Thermus spp. form a coherent phylogenetic cluster close to the root of most bacterial phylogenetic trees (12), far away, from an evolutionary point of view, from Proteobacteria and gram-positive bacteria. Therefore, Thermus spp. show distinctive properties, like coding of 23S and 16S RNAs in separate transcription units (11), a class V H+ ATPase (48), and the presence of an external membrane covering a gram-positive-like peptidoglycan (2). Thus, it should not be unexpected to find peculiarities and differences in the specific functions of central components of the SOS system in isolates of the Thermus-Deinococcus group and those of Proteobacteria or gram-positive bacteria. As a matter of fact, the RecA protein from Deinococcus binds much better to double-stranded DNA than its E. coli counterpart (19) and promotes DNA strand exchange via a pathway that is the inverse of that catalyzed by the RecA protein of E. coli (18). Moreover, a LexA homologue exists in Deinoccoccus, but it is not required for recA induction (27), which seems to be mediated by the IrrE protein (7). Also, the RecA protein of T. thermophilus shows differences from that of E. coli, e.g., in binding to single-stranded DNA (16), despite being able to complement an E. coli ΔrecA mutation (15).
In order to analyze putative differences between the in vivo roles of the RecA protein of T. thermophilus and those of other organisms, the isolation and analysis of insertion and deletion recA mutants from T. thermophilus HB27 are described in this report. Essentially, it is demonstrated that the absence of RecA in Thermus results in severe pleiotropic defects that lead to an extremely low cell survival ratio and to a hypermutational phenotype. Such a phenotype is probably related to a requirement for a specific functional adaptation of RecA to repair a naturally high mutation rate produced by the high temperatures at which these bacteria grow.
MATERIALS AND METHODS
Strains and growth conditions.
T. thermophilus HB27 was a generous gift from Y. Koyama. The E. coli strains DH5αF′ [F′ (lacIq lacZ+ proAB+) supE4 Δ(lacZYA-argF)U169 (φ80lacZΔM15)hsdR17 recA1 endA1 gyrA96 thi1 relA1] (Bethesda Research Laboratories, Gaithersburg, Md.) and JM109 [F′ (traD36 proAB lacIq lacZΔM15) endA1 hsdR17 (rK− mK+) supE44 thimcrA recA1 gyrA96 (Nalr) relA1 Δ(lac-proAB)] (46) were used as hosts for plasmid construction and β-galactosidase complementation assays, respectively.
T. thermophilus HB27 was grown in rich (TB) (29) or minimal (M162) medium (6) at the required temperature (55 to 70°C). Plates containing agar (1.5% [wt/vol]) were usually incubated in a wet chamber for 24 to 72 h. E. coli was grown in Luria broth (22) at 37°C. Ampicillin (100 mg/liter) and/or kanamycin (30 mg/liter) was added to plates or liquid media when needed. For the induction of the response to DNA damage, exponential cultures of T. thermophilus HB27 were treated with mitomycin C (2 mg/liter; Kyowa Hakko Kogyo Co., Tokyo, Japan), and samples were taken at appropriate times for processing (see below). To determine the stability of a specific mRNA, cells grown at the required temperatures were treated with rifampin (200 mg/liter), and samples were taken at the appropriate times.
Plasmids.
Plasmid pKT1 was used as a source of the kat gene, encoding thermostable resistance to kanamycin (21). Plasmids pUC119 and pUC118 (40) were used in several constructs. Plasmid pGEM-T (Promega, Madison, Wis.) was used to clone the amplified recA gene. The E. coli-Thermus bifunctional plasmids EM2S (4) and pMK18 (5) were used for analysis of the mutation frequency in vivo of the ampicillin resistance gene and the α-lacZ gene fragment, respectively.
Northern blotting.
Total RNA was purified by using a protocol derived from that described in the FastRNA Kit-Blue from Bio 101 (La Jolla, Calif.). Samples were analyzed in agarose-formaldehyde gels (31) and transferred by capillarity to Hybond-N+ filters (Amersham-Pharmacia Biotech, Little Chalfont, United Kingdom). Detection of recA and slpA was carried out with the dUTP-fluorescein-labeled oligonucleotides OrecR1 (5′GTCCAGAAGGGACAT3′), complementary to positions +1 to +15 from the recA stop codon, and OslpA1 (5′CAGGGCCTCCACGGC3′), respectively. The ECL system (Amersham-Pharmacia) was used to reveal the labeled mRNA.
Isolation of recA mutants.
The recA gene of T. thermophilus HB27 was amplified with the oligonucleotides TrecAup (5′GATATCGGATCCCTGGCTGGGACAAGC3′) and TrecAdwn (5′GATATCCCAAGCCCCAGGACCAGGAGC3′), starting at positions −192 and +45 with respect to the translational start and stop codons of the T. thermophilus HB8 recA gene, respectively. The amplified 1.14-kb DNA fragment was cloned into the pGEM3 vector to give pGEMTRecA and was then sequenced in both DNA strands by the dideoxy method on an ALF sequencer (Pharmacia Biotech).
The kat gene cassette was inserted into recA by partial digestion of the pGEMTRecA plasmid with BglII and ligation with the BamHI-digested kat cassette, producing the pGEMTRECK plasmid. Thus, the kat gene was located in the same transcriptional direction as recA to allow any putative downstream gene to be expressed. Plasmid pGEMTRECK was linearized by SphI digestion and used to transform the T. thermophilus HB27 strain by natural competence as described previously (5). Kanamycin-resistant colonies grown after 48 h at 70°C were further checked for UV sensitivity and the presence of kat.
For the isolation of deletion mutants, the recA gene cloned in pGEMTRecA was digested with BglII and religated. Transformation with lineal forms of the plasmid (PstI digestion) carrying the ΔrecA gene was performed as described above, and selection for the mutants was carried out by negative selection with dideoxynucleotides. In brief, transformed cells were diluted up to 10 ml in M162 medium on a sterile petri dish, subjected to a short (30-s) sublethal UV treatment (G8T5 [Sylvania]; 20-cm distance), and allowed to recover in the dark for 8 h at 70°C in the same medium containing a mixture of dideoxynucleotides (100 mM). The cells were then washed by centrifugation (5,000 × g; 5 min; room temperature) and extended on TB plates. Colonies that grew after 72 h at 70°C were checked for UV sensitivity. Total DNAs from UV-sensitive putative ΔrecA mutants were purified (24) and checked for the presence of the expected mutations by Southern blotting (31) and PCR with the oligonucleotides OrecR1 and OrecD1 (5′GGATCCCTGGCTGGGACA3′; positions −185 to −167 with respect to the coding region of recA).
Nucleotide sequence accession number.
The EMBL accession number of the nucleotide sequence of the T. thermophilus HB27 recA gene is AF331800.
RESULTS
Isolation of recA mutants.
The sequence of the 340-amino-acid RecA protein from T. thermophilus HB27 was very similar to that of the HB8 strain. Seven amino acid differences were found with respect to the sequence P48297 (15) (A75→G, G151→R, G239→A, A321→T, and tripeptide SDQ329→AGE). The G151→R change was previously identified (43) on the sequence of the recA gene from the HB8 strain, suggesting that it could represent an error in the former sequence.
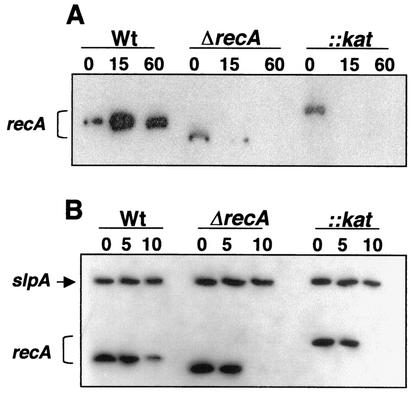
Two constructs were used to obtain recA mutants, one bearing the kat gene (recA::kat), which resulted in a truncated protein of 62 amino acids, and a deletion derivative (ΔrecA) lacking 200 residues between amino acid positions 62 and 262. The deleted region included the ATP- and DNA-binding domains. The Northern blots in Fig. 1 show the presence of the expected insertion and deletion in the mRNA from the recA::kat and ΔrecA mutants, respectively. The two mutants were phenotypically indistinguishable.
FIG. 1.
Genetic analysis of recA mutants from T. thermophilus HB27. (A) Northern blot to detect the recA mRNA of the wild type (Wt) and the deletion (ΔrecA) and insertion (::kat) recA mutants of T. thermophilus HB27, isolated 0, 15, and 60 min after the addition of mitomycin C (2 mg/liter) to exponential cultures of the three strains growing at 70°C. (B) Stability of recA mRNA. Exponential cultures of the indicated strains growing at 70°C were treated with rifampin (200 mg/liter). RNA was purified at the indicated times (in minutes) and subjected to Northern blotting. Detection of recA and slpA mRNAs was done with oligonucleotides OrecR1 and OslpA1, respectively.
The recA gene controls its own expression.
After 15 min of treatment with mitomycin C, cells growing at 70°C induced the transcription of the recA gene in the wild-type strain, leading to the overexpression of a monocistronic mRNA of ∼1 kb (Fig. 1A). After 60 min, the transcription level decreased to a nearly basal expression level. A similar induction profile was observed when cells were subjected to UV treatment (data not shown). By contrast, induction was observed in neither the ΔrecA nor the recA::kat mutant under these conditions (Fig. 1A). In fact, a decrease was observed after 15 min of mitomycin C treatment in both mutants. As shown in Fig. 1B, this decrease was not the consequence of a dramatic difference between the stabilities of the recA mRNAs in the wild type and the recA mutants. In fact, the recA mRNA was detected in the three strains 5 min after the addition of rifampin, an inhibitor of bacterial transcription, supporting similar stabilities in the three strains. The mRNA of the slpA gene (2.9 kb), which has a half-life of 15 min, was used as an internal control in the same experiment.
Segregation defects in recA mutants.
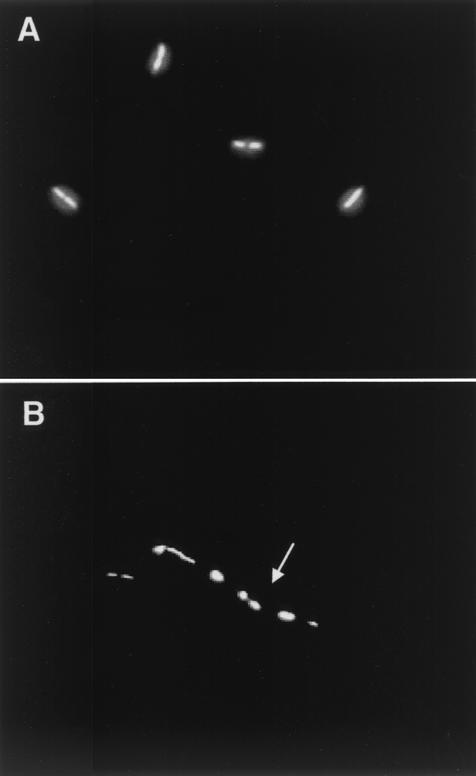
In order to discover if the absence of the RecA protein affected nucleoid segregation, culture samples of the ΔrecA1 mutant were stained with DAPI (4′,6-diamidino-2-phenylindole) as described previously (39) and analyzed by fluorescence microscopy. The results showed that defects in nucleoid condensation and distribution actually occur in the recA mutant (Fig. 2). However, these defects were detected in ∼10% of the cells, a value which is similar to that found in recA mutants from other bacteria (39, 49).
FIG. 2.
Nucleoid distribution in recA mutants of T. thermophilus. Shown is fluorescence microscopy of exponential cultures of the wild type (A) and the ΔrecA mutant (B) of T. thermophilus HB27. The cells were stained with DAPI. Note the irregular distribution of the nucleoids (arrow) in a filament of the mutant strain.
The ΔrecA mutant of T. thermophilus shows low viability.
After the preliminary observations, insertion and deletion recA mutants showed impaired growth and a smaller cell yield in liquid cultures in relation to the parental strain. Defective growth was increasingly obvious upon successive reinoculations and seemed to be dependent on the growth temperature.
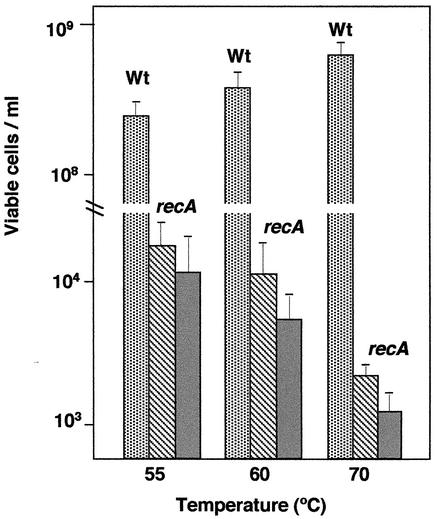
To check this, identical inocula (optical density at 550 nm, 0.1) from the wild type and from two consecutive growth cycles of the ΔrecA mutant were incubated for 6 h in TB medium at 55, 60, and 70°C, and the numbers of viable cells were determined after incubation at the same temperature. As shown in Fig. 3, while cell numbers of ∼108 to 109 per ml were detected for the wild type at the three temperatures, only 103 to 104 viable mutant cells per ml were recovered from the mutant cultures. Moreover, whereas in the wild type the increase in temperature resulted in the expected increase in cell viability, in the mutant, a decrease in viability of ∼1 order of magnitude was observed as the consequence of such a temperature increase.
FIG. 3.
Growth temperature and cell viability in a ΔrecA background. Identical inocula (optical density at 550 nm, 0.1) of the wild type (Wt) and the first (hatched) and second (shaded) growth cycles of the ΔrecA mutant (at 70°C) were incubated at 55, 60, and 70°C. After 6 h, serial dilutions were plated on TB and incubated at the same temperatures for 24 (70°C) or 48 (55 and 60°C) h before the number of viable cells was counted. The error bars indicate standard deviations.
In order to check whether the extremely low plate yield found for the mutant was the consequence of the absence of the RecA protein, a complementation experiment was carried out by transforming exponential cultures of the ΔrecA mutant with the plasmid pGEMTRecA. As this plasmid does not replicate in Thermus, an internal fragment of recA was used to detect the complemented strains by colony blotting. It was expected that once inside the cell the wild-type recA gene from the plasmid could be expressed transiently, allowing the replacement of the ΔrecA gene in the chromosome through homologous recombination. All nine of the colonies that hybridized with the probe were shown to have plate yields, morphologies, and UV resistances similar to that of the wild type (not shown). Therefore, the absence of RecA was responsible for these phenotypes.
Interestingly, one of these nine complemented clones presented a white phenotype (see below), and two more showed amino acid requirements (they were unable to grow in M162 containing alanine, valine, leucine, and glutamic acid). Consequently, these phenotypes are not complemented by a wild-type RecA protein.
A high number of carotenoid biosynthesis mutants are detected among T. thermophilus recA mutants.
A high percentage of the colonies of the recA mutants appeared colorless (“white”) on TB plates, in contrast to the bright-yellow color of the parent strain. In T. thermophilus HB27, the yellow color corresponds to the synthesis of at least seven carotenoid pigments, which are apparently connected, from a biosynthetic point of view (13, 47). Keeping in mind that many of these biosynthetic genes are carried within a megaplasmid (38) and that the recA mutants show defects in nucleoid distribution (Fig. 2), we wondered if the white phenotype could be the consequence of the loss of such a plasmid during division. To check this, DNA was purified directly from a few white colonies, and Southern blot assays were developed with a probe against crtB, a gene carried within this megaplasmid which is responsible for a limiting step in carotenoid biosynthesis (13, 38). In all of the colonies checked, the crtB gene was present (data not shown), leading to the conclusion that the white phenotype was most likely related to mutations in any of the yet-unknown genes coding for the biosynthetic pathway. Consequently, our data suggested that the recA mutants were suffering from a dramatic increase in the mutation rate, at least that of the carotenoid biosynthesis pathway.
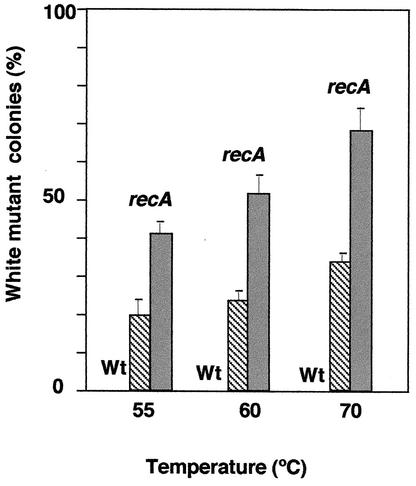
Interestingly, the percentage of white mutants increased dramatically with the incubation temperature, and also with the number of generations (Fig. 4). No white colonies were detected among the viable cells (∼104 colonies) analyzed from the wild-type strain independently of the incubation temperature. In contrast, percentages of white colonies ranging from ∼20 up to 33% were observed in the mutant at 55 and 70°C, respectively, for cells inoculated from a first growth cycle and ∼40 to ∼70% for cells inoculated from a second growth cycle. Therefore, if the white colony phenotype was a consequence of any mutation in the genes for carotenoid biosynthesis, it follows that there is a direct relationship between the temperature and mutation rate.
FIG. 4.
Temperature-dependent appearance of mutants in carotenoid biosynthesis. The percentage of mutant colonies defective in carotenoid biosynthesis (white), from the experiments described in Fig. 3, was represented against the incubation temperature. The error bars indicate standard deviations.
The mutation rate of the ΔrecA mutant depends on temperature.
In order to confirm the above-mentioned interpretation of a temperature-dependent increase in the mutation rates of the recA mutants, we decided to analyze three additional genetic markers on cells grown at 55 and 70°C (Table 1). Two of these were heterologous genes carried within E. coli-Thermus bifunctional plasmids.
TABLE 1.
Effects of growth temperature on appearance of different mutant phenotypes in ΔrecA mutants of T. thermophilus
| Phenotype | % With phenotype
|
|||
|---|---|---|---|---|
| 55°C
|
70°C
|
|||
| wte | recA | wt | recA | |
| Carotenoid-free | <0.1 | 22 ± 3 | <0.1 | 33 ± 2 |
| Mcaasa,b | 6.5 | 25 | 7 | 36 |
| β-Galactosidase-freeb,c | 0.1 | 5 ± 1 | 0.1 | 13 ± 2 |
| Ampicillin sensitived | <0.1 | 14 | <0.1 | 47 |
Mcaas, percentage of cells that did not grow in minimal M162 medium with a complete set of amino acids.
Data from single experiment.
Results of alpha complementation in E. coli JM109 transformed with plasmid pMK18 isolated from wild-type or ΔrecA mutants grown at the indicated temperatures.
Results obtained after replica plating of E. coli JM109 transformed with plasmid EMS2 isolated from wild-type or ΔrecA mutants grown at the indicated temperatures.
wt, wild type.
One of the markers used was the presence of auxotrophic mutants. As shown in Table 1, at 56°C ∼25% of the colonies of the ΔrecA mutant were unable to grow in M162 medium containing a complete set of amino acids. At 70°C, this percentage increased to 36%. In the wild type, 6 to 7% of the colonies did not grow in the medium. Furthermore, of those ΔrecA colonies that grew on the amino acid mixture, most were auxotrophic for one or more amino acids (not shown).
As a second approach, the α-fragment of the E. coli β-galactosidase gene was used as a “neutral” reporter gene. This gene was included in the bifunctional plasmid pMK18 (5), which can be selected by kanamycin resistance in T. thermophilus at 70°C. The ΔrecA mutant was transformed by natural competence during the first growth cycle at 60°C. Transformation efficiencies (selection on kanamycin) of ∼103 transformants per μg of plasmid DNA were obtained for the ΔrecA1 mutant, whereas values of 3 × 106 to 5 × 106 were obtained for the wild-type strain. After transformation, plasmids were obtained from 2-ml cultures of a number of kanamycin-resistant recA colonies grown at 55 or 70°C for 48 and 24 h, respectively. The plasmids were mixed and used to transform E. coli JM109 in parallel with a similar mixture and concentration of plasmids isolated from the wild-type strain. The results (Table 1) supported the existence of a dramatic increase in the number of β-galactosidase-free colonies of E. coli when plasmid preparations isolated from the ΔrecA mutant (5%) were used for transformation, in comparison with the low percentage detected when the source of the plasmid was the wild-type strain (0.1%). At 70°C, the number of β-galactosidase-free colonies observed in the mutant, but not in the wild type, was even higher, again supporting a relationship between the growth temperature and mutation rate.
Finally, our data on the ampicillin-resistant marker encoded by the plasmid EM2S (4) also support the existence of a dramatic increase in the mutation frequency in recA mutants. Essentially, the experiment was similar to that developed with pMK18, but this time we checked how many of the kanamycin-resistant E. coli transformant colonies still carried an active ampicillin resistance gene. As shown in Table 1, ∼14% of the plasmids were ampicillin sensitive compared to <0.1% in the wild type. As with β-galactosidase, the number of ampicillin-sensitive clones increased dramatically (47%) with temperature.
DISCUSSION
Despite its primary sequence conservation and ubiquity in bacteria, the roles that the RecA protein plays may be subtly different among members belonging to distant branches of the bacterial phylogenetic tree. For instance, RecA is required for adherence and colonization in Vibrio cholerae (20), phenotypic switching in Pseudomonas tolaasii (34), nucleoid segregation in B. subtilis (33), plasmid maintenance in Streptococcus pneumoniae (25), and nucleoid condensation in Leptospira biflexa (39). Moreover, there are mechanistic differences among RecA proteins from different origins, even in their well-known role in recombination and repair. As an example, it has been shown that RecA from Deinococcus radiodurans, a microorganism closely related to Thermus spp., promotes DNA strand exchange via a pathway that is the inverse of that catalyzed by E. coli RecA (18).
As could be expected from the phylogenetic relationship of the organisms, the RecA protein of T. thermophilus HB27 is more similar in sequence to that of D. radiodurans (68.4% identity) than to that of E. coli (62.3% identity). However, despite the phylogenetic relationship, there are also clear differences between the radiation-resistant and the thermophilic microorganisms in the ways in which they respond to DNA damage. Whereas in D. radiodurans recA is expressed as part of a three-gene operon (26), our results (Fig. 1) support the idea that recA is transcribed as a monocistronic mRNA in T. thermophilus HB27. The inactivation of recA described here constitutes a first step toward the in vivo analysis of the peculiarities of the response to DNA damage in this extreme thermophile.
The first evidence of a consequence of recA inactivation in Thermus was the extremely low viability of the mutants, as only 1 in 103 to 105 cells, depending on the growth temperature, was able to form a colony. In recA mutants of organisms like E. coli, B. subtilis, and L. biflexa, a moderate decrease in viability, ranging between 50 and 90%, has been described previously (1, 33, 39). In E. coli, the primary cause of death appears to be chromosomal degradation, presumably at sites of stalled DNA replication forks. In fact, ∼10% of E. coli recA mutants are anucleate, and an additional fraction show signs of chromosomal degradation (35, 49). By contrast, Streptomyces lividans recA mutants are not viable at all unless additional compensatory mutations are selected that suppress the lethal effect of the absence of RecA (41). In T. thermophilus recA mutants, there are also defects in chromosome condensation and asymmetric partitioning, but this could represent only a minor fraction (∼10 to 15%) of the cells and cannot by itself explain the dramatic decrease in viability observed (Fig. 2).
The direct relationship between the decrease in viability and the absence of RecA is clearly demonstrated by two facts. First, the complementation experiments support the idea that the phenotype is ligated to the gene contained in the DNA fragment used for transformation (the recA gene). Second, any hypothetical polar effect on the expression of downstream genes should be compensated for in the recA::kat mutant by the transcription from the kat gene promoter, which does not have any transcription terminator.
Our hypothesis to explain this recA-associated decrease in viability is that massive accumulation of mutations during growth makes most cells nonviable. This is supported by the increase in the frequency of appearance of two “internal” (carotenoid-free and auxotrophic mutants) and two “external” genetic markers. With these four markers, an increase of 2 or 3 orders of magnitude in the appearance of the mutant phenotypes was observed between the wild type and the ΔrecA strain (Table 1). Therefore, and in contrast to what was expected from the extrapolation of data on the recA mutants of E. coli (42), the absence of RecA in T. thermophilus apparently results in a dramatic and generalized increase in the mutation rate.
The reason for this “hypermutator” phenotype could be related to a requirement for RecA in the repair of a high frequency of replicative errors concomitant to the extreme temperatures at which this bacterium grows. This is experimentally suggested by the fact that an increase in the temperature of incubation of the recA mutants results in higher mutation frequencies, independently of the marker used (Fig. 4 and Table 1).
Two putative mechanisms could explain the relationship between high growth temperatures and mutation rates. On one hand, the base pairing during replication is necessarily less efficient, and consequently more error prone, at high temperatures. On the other hand, spontaneous hydrolysis, such as deamination of DNA cytosine and 5-methylcytosine residues, which is a frequent source of C/G-to-T/A transitions, must accelerate with increasing temperatures (36). Consequently, it could be expected that thermophilic microorganisms have to counter the correspondingly more pronounced challenge to their genome integrity through specific adaptations of their repair systems. This can be done with more or less success, as in the hyperthermophilic archaeon Pyrobaculum aerophilum, in which the frequencies of A/T-to-G/C and G/C-to-A/T mutations are between 50- and 200-fold higher than in E. coli (9; http://cgb.utmem.edu/meeting_reports/redwards_11_06_01.htm, 2001).
Therefore, it is likely that the hypermutational phenotype in recA mutants of T. thermophilus is not the consequence of the activation of a mutational pathway but the result of the impairment of a RecA-dependent repair system devoted to countering a naturally high mutation rate. We are aware that this hypermutational effect contrasts with the low mutability of recA mutants of E. coli (42), in which it is the induction of the SOS responses by a functional RecA protein that promotes hypermutability through its role in translesion synthesis (37). However, even in E. coli, RecA enhances the fidelity of in vitro DNA synthesis by the Klenow fragment of DNA polymerase I (14), supporting the idea that RecA could also be relevant for fidelity during DNA synthesis in other, nonthermophilic bacterial systems. Thus, the hypermutational effect of recA mutations in Thermus might be the consequence of a specific adaptation of this thermophilic RecA protein to repair, to the detriment of a putative role in translesion synthesis after DNA damage.
Acknowledgments
This work was supported by grants BIO2001-1267 to J. Berenguer and BMC2001-2065 and 2001SGR-206 to J. Barbé and by an institutional grant from Fundación Ramón Areces to CBMSO. Pablo Castán held a fellowship from the Ministerio de Educación y Cultura.
REFERENCES
- 1.Capaldo, F. N., G. Ramsey, and S. D. Barbour. 1974. Analysis of the growth of recombinant-deficient strains of Escherichia coli K-12. J. Bacteriol. 118:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castán, P., O. Zafra, R. Moreno, M. A. De Pedro, C. Valles, F. Cava, E. Caro, H. Schwarz, and J. Berenguer. 2002. The periplasmic space in Thermus thermophilus: evidence from a regulation-defective S-layer mutant overexpressing an alkaline phosphatase. Extremophiles 6:225-232. [DOI] [PubMed] [Google Scholar]
- 3.Cheo, D. L., K. W. Bayles, and R. E. Yasbin. 1992. Molecular characterization of regulatory elements controlling expression of the Bacillus subtilis recA+ gene. Biochimie 74:755-762. [DOI] [PubMed] [Google Scholar]
- 4.de Grado, M., I. Lasa, and J. Berenguer. 1998. Characterization of a plasmid replicative origin from an extreme thermophile. FEMS Microbiol. Lett. 165:51-57. [DOI] [PubMed] [Google Scholar]
- 5.de Grado, M., P. Castán, and J. Berenguer. 1999. A high-transformation-efficiency cloning vector for Thermus thermophilus. Plasmid 42:241-245. [DOI] [PubMed] [Google Scholar]
- 6.Degryse, E., N. Glansdorff, and A. Piérard. 1978. A comparative analysis of extreme thermophilic bacteria belonging to the genus Thermus. Arch. Microbiol. 117:189-196. [DOI] [PubMed] [Google Scholar]
- 7.Earl, A. M., M. M. Mohundro, I. S. Mian, and J. Battista. 2002. The IrrE protein of Deinococcus radiodurans R1 is a novel regulator of recA expression. J. Bacteriol. 184:6216-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisen, J. A. 1995. The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16S rRNAs from the same species. J. Mol. Evol. 41:1105-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitz-Gibbon, S. T., H. Ladner, U.-J. Kim, K. O. Stetter, M. V. Simon, and J. H. Miller. 2002. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. USA 99:984-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedberg, E. C. 1995. Out of the shadows and into the light: the emergence of DNA repair. Trends Biochem. Sci. 20:381-384. [DOI] [PubMed] [Google Scholar]
- 11.Hartmann, R. K., and V. A. Erdmann. 1989. Thermus thermophilus 16S rRNA is transcribed from an isolated transcription unit. J. Bacteriol. 171:2933-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartmann, R. K., B. Wolters, B. Kroger, S. Shultze, T. Spetch, and V. A. Erdmann. 1989. Does Thermus represent another deep eubacterial branching? Syst. Appl. Microbiol. 11:243-249. [Google Scholar]
- 13.Hoshino, T., R. Fujii, and T. Nakahara. 1993. Molecular cloning and sequence analysis of the crtB gene of Thermus thermophilus HB27, an extreme thermophile producing carotenoid pigments. Appl. Environ. Microbiol. 59:3150-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karthikeyan, G., G. S. Lakshmikant, M. D. Wagle, G. Krishnamoorthy, and B. J. Rao. 1999. RecA interacts with Klenow and enhances fidelity of DNA synthesis in vitro. J. Mol. Microbiol. Biotechnol. 1:149-156. [PubMed] [Google Scholar]
- 15.Kato, R., and S. Kuramitsu. 1999. Characterization of thermostable RecA protein and analysis of its interaction with single-stranded DNA. Eur. J. Biochem. 259:592-601. [DOI] [PubMed] [Google Scholar]
- 16.Kato, R., and S. Kuramitsu. 1993. RecA protein from an extremely thermophilic bacterium, Thermus thermophilus HB8. J. Biochem. (Tokyo) 114:926-929. [DOI] [PubMed] [Google Scholar]
- 17.Kawabata, M., and K. Saeki. 1998. Sequence analysis and expression of a novel mouse homolog of Escherichia coli recA gene. Biochim. Biophys. Acta 1398:353-358. [DOI] [PubMed] [Google Scholar]
- 18.Kim, J. I., and M. M. Cox. 2002. The RecA proteins of Deinococcus radiodurans and Escherichia coli promote DNA strand exchange via inverse pathways. Proc. Natl. Acad. Sci. USA 99:7917-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, J. I., A. K. Sharma, S. N. Abbott, E. A. Wood, D. W. Dwyer, A. Jambura, K. W. Minton, R. B. Inman, M. J. Daly, and M. M. Cox. 2002. RecA protein from the extremely radioresistant bacterium Deinococcus radiodurans: expression, purification, and characterization. J. Bacteriol. 184:1649-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar, K. K., R. Srivastava, V. B. Sinha, J. Michalski, J. B. Kaper, and B. S. Srivastava. 1994. recA mutations reduce adherence and colonization by classical and El Tor strains of Vibrio cholerae. Microbiology 140:1217-1222. [DOI] [PubMed] [Google Scholar]
- 21.Lasa, I., J. R. Castón, L. A. Fernandez-Herrero, M. A. Pedro, and J. Berenguer. 1992. Insertional mutagenesis in the extreme thermophilic eubacteria Thermus thermophilus. Mol. Microbiol. 11:1555-1564. [DOI] [PubMed] [Google Scholar]
- 22.Lennox, E. X. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190-206. [DOI] [PubMed] [Google Scholar]
- 23.Lusetti, S. L., and M. M. Cox. 2002. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 71:71-100. [DOI] [PubMed] [Google Scholar]
- 24.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 25.Martin, B., P. Garcia, M. P. Castanie, and J. P. Claverys. 1995. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol. Microbiol. 15:367-379. [DOI] [PubMed] [Google Scholar]
- 26.Narumi, I., K. Satoh, M. Kikuchi, T. Funayama, S. Kitayama, T. Yanagisawa, H. Watanabe, and K. Yamamoto. 1999. Molecular analysis of the Deinococcus radiodurans recA locus and identification of a mutation site in a DNA repair-deficient mutant, rec30. Mutat. Res. 435:233-243. [DOI] [PubMed] [Google Scholar]
- 27.Narumi, I., K. Satoh, M. Kikuchi, T. Funayama, T. Yanagisawa, Y. Kobayashi, H. Watanabe, and K. Yamamoto. 2001. The LexA protein from Deinococcus radiodurans is not involved in RecA induction following gamma irradiation. J. Bacteriol. 183:6951-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pham, P., E. M. Seitz, S. Saveliev, X. Shen, R. Woodgate, M. M. Cox, and M. F. Goodman. 2002. Two distinct modes of RecA action are required for DNA polymerase V-catalyzed translesion synthesis. Proc. Natl. Acad. Sci. USA 99:11061-11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramírez-Arcos, S., L. A. Fernández-Herrero, and J. Berenguer. 1998. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochem. Biophys. Acta 1396:215-227. [DOI] [PubMed] [Google Scholar]
- 30.Roca, A. I., and M. M. Cox. 1997. RecA protein: structure, function, and role in recombinational DNA repair. Prog. Nucleic Acids Res. Mol. Biol. 56:129-223. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sandler, S. J., L. H. Satin, H. S. Samra, and A. J. Clark. 1996. recA-like genes from three archaean species with putative protein products similar to Rad51 and Dmc1 proteins of the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 24:2125-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sciochetti, S. A., G. W. Blakely, and P. J. Piggot. 2001. Growth phase variation in cell and nucleoid morphology in a Bacillus subtilis recA mutant. J. Bacteriol. 183:2963-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha, H., A. Pain, and K. Johnstone. 2000. Analysis of the role of recA in phenotypic switching of Pseudomonas tolaasii. J. Bacteriol. 182:6532-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skarstad, K., and E. Boye. 1993. Degradation of individual chromosomes in recA mutants of Escherichia coli. J. Bacteriol. 175:5505-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Starkuviene, V., and H.-J. Fritz. 2002. A novel type of uracyl-DNA glycosilase mediating repair of hydrolytic DNA damage in the extremely thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 30:2097-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34:479-497. [DOI] [PubMed] [Google Scholar]
- 38.Tabata, K., S. Ishida, T. Nakahara, and T. Hoshino. 1994. A carotenogenic gene cluster exists on a large plasmid in Thermus thermophilus. FEBS Lett. 341:251-255. [DOI] [PubMed] [Google Scholar]
- 39.Tchamedeu Kameni, A. P., E. Couture-Tosi, I. Saint-Girons, and M. Picardeau. 2002. Inactivation of the spirochete recA gene results in a mutant with low viability and irregular nucleoid morphology. J. Bacteriol. 184:452-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vieira, J., and J. Messing. 1987. Production of single stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 41.Vierling, S., T. Weber, W. Wohlleben, and G. Muth. 2001. Evidence that an additional mutation is required to tolerate insertional inactivation of the Streptomyces lividans recA gene. J. Bacteriol. 182:4374-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker, G. C. 1984. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol. Rev. 48:60-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wetmur, J. G., D. M. Wong, B. Ortiz, J. Tong, F. Reichert, and D. H. Gelfand. 1994. Cloning, sequencing, and expression of RecA proteins from three distantly related thermophilic eubacteria. J. Biol. Chem. 269:25928-25935. [PubMed] [Google Scholar]
- 44.Winterling, K. W., D. Chafin, J. J. Hayes, J. Sun, A. S. Levine, R. E. Yasbin, and R. Woodgate. 1998. The Bacillus subtilis DinR binding site: redefinition of the consensus sequence. J. Bacteriol. 180:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wojciechowski, M. F., K. R. Peterson, and P. E. Love. 1991. Regulation of the SOS response in Bacillus subtilis: evidence for a LexA repressor homolog. J. Bacteriol. 173:6489-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanish-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pU19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 47.Yokoyama, A., Y. Shizuri, T. Hoshino, and G. Sandmann. 1996. Thermocryptoxanthins: novel intermediates in the carotenoid biosynthetic pathway of Thermus thermophilus. Arch. Microbiol. 165:342-345. [DOI] [PubMed] [Google Scholar]
- 48.Yokoyama, K., S. Ohkuma, H. Taguchi, T. Yasunaga, T. Wakabayashi, and M. Yoshida. 2000. V-type H+-ATPase/synthase from a thermophilic eubacterium, Thermus thermophilus. Subunit structure and operon. J. Biol. Chem. 275:13955-13961. [DOI] [PubMed] [Google Scholar]
- 49.Zyskind, J. W., A. L. Svitil, W. B. Stine, M. C. Biery, and D. W. Smith. 1992. RecA protein of Escherichia coli and chromosome partitioning. Mol. Microbiol. 6:2525-2537. [DOI] [PubMed] [Google Scholar]