Abstract
Interactions between the MinD and MinE proteins are required for proper placement of the Escherichia coli division septum. The site within MinE that is required for interaction with MinD was mapped by studying the effects of site-directed minE mutations on MinD-MinE interactions in yeast two-hybrid and three-hybrid experiments. This confirmed that the MinE N-terminal domain is responsible for the interaction of MinE with MinD. Mutations that interfered with the interaction defined an extended surface on one face of the α-helical region of the MinE N-terminal domain, consistent with the idea that the MinE-MinD interaction involves formation of a coiled-coil structure by interaction with a complementary helical surface within MinD.
Proper placement of the Escherichia coli division site requires the coordinated action of the three Min proteins—MinC, MinD, and MinE. The proteins ensure that division occurs at the midpoint of the long axis of the rod-shaped cell by preventing septation at aberrant sites elsewhere along the cylinder (reviewed in reference 17).
MinC is an inhibitor of septation that is given specificity for sites away from midcell by the combined action of MinD and MinE. Expression of the Min proteins is associated with a cycle of Min protein redistribution in which a membrane-associated polar zone containing MinC, MinD, and MinE is formed at one end of the cell. MinE then forms an annular, or ring-like, structure, the E ring, that is located adjacent to the medial edge of the polar zone. This is followed by disassembly of the polar zone and E ring and by formation of a new polar zone at the opposite end of the cell (17). As a result of this repetitive cycle, the proteins oscillate from pole to pole many times during each division cycle. It has been suggested that this process maintains the MinC division inhibitor away from midcell and near the ends of the cell for sufficient time during the division cycle to prevent the aberrant polar-division events that cause the formation of anucleate minicells in the absence of the Min proteins (14).
In vivo studies have suggested that MinD and MinE interact during these events. Thus, MinD is required for the membrane association of MinE, and MinE is required for membrane-associated MinD to redistribute itself into the polar zones, which undergo the pole-to-pole oscillation cycle. In vitro studies have shown that MinE activates the low-level ATPase activity of MinD when phospholipid is present, also implying direct interaction between the two proteins (5). This leads to release of MinD and MinE from phospholipid vesicles (4, 11) and to alterations in MinD organization in vitro (20) and presumably explains the cyclic binding and release of the Min proteins during the oscillation cycle.
Genetic and biochemical studies have shown that MinE consists of two structurally and functionally distinct domains. The C-terminal domain of MinE (MinE32-88) is responsible for its topological specificity function. Thus, this domain is required for the ability of MinE to prevent minicell formation, to form MinE rings, and to induce formation of MinCDE polar zones, but it does not counteract the action of the MinC division inhibitor in MinCD+ cells (13, 22). In contrast, the N-terminal segment of MinE (MinE1-22 or MinE1-31) is unable to prevent minicell formation but is similar to full-length MinE in its ability to counteract the action of the MinC division inhibitor in the presence of MinD (13, 22). The N-terminal MinE fragment suppresses the filamentation that occurs when MinC and MinD are expressed at normal levels but does not suppress the filamentation that occurs when MinC is expressed at high levels in the absence of MinD. This led to the suggestion that the suppression of the filamentation phenotype requires interaction between MinD and a site within the N-terminal MinE domain (22).
MinE6-35 is predicted to exist as an α-helix, with a tendency to form a coiled-coil structure in which two α-helices coil around each other (9). Consistent with this hypothesis, nuclear magnetic resonance studies have shown that MinE1-22 exists in solution as a nascent α-helix (9) and that a portion of the predicted MinE6-35 helix is visible at the N terminus of the MinE31-88 three-dimensional structure, as determined by multidimensional nuclear magnetic resonance analysis (10). These observations led to the suggestion that the MinE-MinD interaction during the normal division cycle involves the association of one face of the MinE6-35 α-helix with a complementary helical site within MinD (9, 10). In the present work, site-directed mutagenesis within the MinE N-terminal domain was used to test certain predictions of this model.
MATERIALS AND METHODS
E. coli strains and growth conditions.
The E. coli host strain used in this study was RC1 (ΔminCDE) (18). Unless otherwise noted, plasmid-containing E. coli strains were grown at 37°C in Luria-Bertani (LB) medium containing the required antibiotics. For determination of the cell division phenotype, an overnight culture in LB containing 0.25% glucose was diluted 200-fold in LB containing 10 μM IPTG (isopropyl-β-d-thiogalactopyranoside) and grown to an optical density at 600 nm (OD600) of ∼0.9.
Plasmids.
Subscripts in the names of genes indicate the protein products (e.g., minE1-31 indicates a minE fragment coding for amino acids 1 to 31; minEL22R indicates a mutant allele coding for a mutant protein in which amino acid 22 has been changed from leucine to arginine). Tables 1 and 2 list the plasmid genotypes.
TABLE 1.
Plasmids used for determination of cell division phenotype and fluorescence microscopya
| Plasmid | Relevant genotype | Source or reference |
|---|---|---|
| pSY1083 | Plac-minC minD minE | 2 |
| pEM4 | Plac-minC minD minEL4R | This study |
| pEM8 | Plac-minC minD minEL8R | This study |
| pEM15 | Plac-minC minD minEA15R | This study |
| pEM18 | Plac-minC minD minEA18T | This study |
| pEM19 | Plac-minC minD minEK19A | This study |
| pEM20 | Plac-minC minD minEE20A | This study |
| pEM22 | Plac-minC minD minEL22R | This study |
| pEM22S | Plac-minC minD minEL22S | This study |
| pEM23 | Plac-minC minD minEQ23R | This study |
| pEM24 | Plac-minC minD minEI24R | This study |
| pEM25 | Plac-minC minD minEI25R | This study |
| pEM26 | Plac-minC minD minEV26R | This study |
| pEM27 | Plac-minC minD minEA27R | This study |
| pEM28 | Plac-minC minD minEE28A | This study |
| pEM30 | Plac-minC minD minER30A | This study |
| pYEC15 | Plac-yfp::minD minEA15R::cfp | This study |
| pYEC18 | Plac-yfp::minD minEA18T::cfp | This study |
| pYEC19 | Plac-yfp::minD minEK19A::cfp | This study |
| pYEC22 | Plac-yfp::minD minEL22R::cfp | This study |
| pYEC22S | Plac-yfp::minD minEL22S::cfp | This study |
| pYEC25 | Plac-yfp::minD minEI25R::cfp | This study |
| pYEC30 | Plac-yfp::minD minER30A::cfp | This study |
| pMEC1 | Plac-minE::cfp | This study |
| pMEC22 | Plac-minEL22R::cfp | This study |
| pMEC22S | Plac-minEL22S::cfp | This study |
| pMEC25 | Plac-minEI25R::cfp | This study |
| pYLS41G | Plac-minE::gfp | 19 |
| pMEG25 | Plac-minEI25R::gfp | This study |
| pMA101 | Plac-minE1-31::gfp | This study |
| pMA102 | Plac-minE32-88::gfp | This study |
See Materials and Methods for further details.
TABLE 2.
Plasmids used in yeast two- and three-hybrid assaysa
| Plasmid | Relevant genotype | Source |
|---|---|---|
| pBridge | PADH1-BD; PMet25b | Clontech |
| pGBKT7 | PADH1-BD | Clontech |
| pGAD424 | PADH1-AD | Clontech |
| pGADT7 | PADH1-AD | Clontech |
| pYW9 | PADH1-AD::minD in pGAD424 | This study |
| pMDB1 | PADH1-BD::minD in pGBKT7 | This study |
| pMCA1 | PADH1-AD::minC in pGADT7 | This study |
| pYW10 | PADH1-BD::minC; PMet25 in pBridge | This study |
| pBE1 | PADH1-BD::minE; PMet25 in pBridge | This study |
| pBE2 | PADH1-BD::minE1-31; PMet25 in pBridge | This study |
| pBE3 | PADH1-BD::minE32-88; PMet25 in pBridge | This study |
| pBEC1 | PADH1-BD::minE1-31; PMet25-minC in pBridge | This study |
| pBCE1 | PADH1-BD::minC; PMet25-minE in pBridge | This study |
| pBCE2 | PADH1-BD::minC; PMet25-minE1-31 in pBridge | This study |
| pBCE3 | PADH1-BD::minC; PMet25-minE32-88 in pBridge | This study |
| pBCE11 | PADH1-BD::minC; PMet25-minEL22R in pBridge | This study |
| pBCE12 | PADH1-BD::minC; PMet25-minE1-31,L22R in pBridge | This study |
| pBCE13 | PADH1-BD::minC; PMet25-minE1-31,125R in pBridge | This study |
| pBCE15 | PADH1-BD::minC; PMet25-minEK19A in pBridge | This study |
| pBEM16 | PADH1-BD::minC; PMet25-minEL22S in pBridge | This study |
| pBEM4 | PADH1-BD::minE1-31,L4R; PMet25 in pBridge | This study |
| pBEM8 | PADH1-BD::minE1-31,L8Rl PMet25 in pBridge | This study |
| pBEM15 | PADH1-BD::minE1-31,A15R; PMet25 in pBridge | This study |
| pBEM18 | PADH1-BD::minE1-31,A18T; PMet25 in pBridge | This study |
| pBEM19 | PADH1-BD::minE1-31,K19A; PMet25 in pBridge | This study |
| pBEM20 | PADH1-BD::minE1-31,E20A; PMet25 in pBridge | This study |
| pBEM22 | PADH1-BD::minE1-31,L22R; PMet25 in pBridge | This study |
| pBEM22S | PADH1-BD::minE1-31,L22S; PMet25 in pBridge | This study |
| pBEM23 | PADH1-BD::minE1-31,Q23R; PMet25 in pBridge | This study |
| pBEM24 | PADH1-BD::minE1-31,I24R; PMet25 in pBridge | This study |
| pBEM25 | PADH1-BD::minE1-31,I25R; PMet25 in pBridge | This study |
| pBEM27 | PADH1-BD::minE1-31,A27R; PMet25 in pBridge | This study |
| pBEM28 | PADH1-BD::minE1-31,E28A; PMet25 in pBridge | This study |
| pBEM30 | PADH1-BD::minE1-31,R30A; PMet25 in pBridge | This study |
See Materials and Methods for constructions and further details.
PMet25, methionine promoter, repressible by growth in methionine.
Point mutations within minE were generated by mutagenic PCR of plasmid pSY1083 (Plac-minCDE) (18), as previously described (19), to generate the pEM series of plasmids (Table 1). For plasmids to be used in yeast two- and three-hybrid studies (Table 2), protein fusions to the Gal4 activation domain (AD) and Gal4 binding domain (BD) were obtained by insertion of DNA fragments into pBridge or pGBKT7 (for BD fusions) or into pGAD424 or pGADKT7 (for AD fusions); the plasmid vectors were obtained from Clontech. The DNA fragments were obtained by PCR, using pSY1083 as a template for wild-type minC, minD, and minE and the appropriate pEM plasmids (Table 1) as templates to generate mutant minE fragments. The pBridge vector, in addition to coding for the desired BD protein fusion, contains a second cloning site downstream of PMet. DNA coding for a second protein was inserted at this site, generating in-frame fusions in which the protein was downstream of PMet and a nuclear localization signal and hemagglutinin epitope (Clontech manual).
pYEC plasmids (Plac-yfp::minD minE::cfp), coding for proteins in which Yfp (yellow fluorescent protein) was fused to the N-terminal end of MinD and Cfp (cyan fluorescent protein) was fused to the C-terminal end of mutant MinE, were constructed by three fragment ligations between (i) the XbaI/XmnI fragment from pYLS68 (Plac-yfp::minD minE::cfp) (19), containing a minD fragment; (ii) the XbaI/BamHI fragment from pYLS68 that contains yfp, cfp, and the pMLB1113 vector; and (iii) XmnI/BamHI fragments from pEM plasmids (see above) containing mutant minE alleles. pMEC plasmids (Plac-minE::cfp) were constructed by ligating the XmnI/HindIII fragment containing minE::cfp from pYLS68 (19), or from pYEC plasmids containing mutant minE::cfp alleles (see above), into the SmaI/HindIII site of the pMLB1113 vector (1).
pYLS41G (Plac-minE::gfp) was generated by ligation of the XmnI/HindIII fragment from pSY1083G (19) that contains minE::gfp into the SmaI/HindIII site of the pMLB1113 vector. pMEG25 (Plac-minEI25R::gfp) was constructed similarly, using the XmnI/HindIII fragment containing minEI25R::gfp from pEG25, which was obtained by religation of BamHI-treated pEM25 to generate an in-frame minEI25R::gfp fusion. pMA101 (Plac-minE1-31::gfp), pMA102 (Plac- minE32-88::gfp), and pMA22 (Plac-minE1-31,L22S::gfp) were constructed by ligation of the large EcoRI/BamHI fragment from pSY1083 that contains gfp and the pMLB1113 vector to fragments containing minE1-31, minE32-88, or minE1-31,L22S, which were obtained by PCR using pSY1083 and pEM22S as templates.
Yeast two- and three-hybrid systems.
Plasmids coding for the appropriate BD and AD fusion proteins (Table 2) were transformed into Saccharomyces cerevisiae SFY526 containing the Gal4-inducible reporter gene lacZ (Clontech). Plasmid-containing cells were obtained and characterized, and further analysis was performed as described in the Clontech manual. Interaction between the BD and AD probes was monitored after 3 days of growth at 30°C by β-galactosidase activity, as estimated from colony color on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates and by liquid assay (Clontech manual, protocol PT3024-1); specific activity is given in Miller units (12) and represents the average of three independent measurements. Three-hybrid assays utilized the pBridge vector; the third protein inserted downstream of PMet was induced by growth in the absence of methionine.
Fluorescence microscopy.
Cells were grown in the presence of 10 μM IPTG, and Yfp-, Cfp-, and Gfp-labeled proteins were detected by fluorescence microscopy as previously described (19). Oscillation rates were determined as previously described (19) and are given as the average for 12 to 15 cells. The oscillation rate of the MinD polar zone in pYLS68 (Plac-yfp::minD minE::cfp) was 0.15 ± 0.07 cycles/min, similar to values previously described for cells containing doubly labeled MinD and MinE (3, 19).
Cell fractionation.
Cells were suspended in SHD buffer (20% sucrose, 10 mM HEPES, 5 mM dithiothreitol), broken using a French pressure cell, and centrifuged at 3,000 rpm at 4°C for 10 min in a Beckman Microfuge 18 centrifuge to remove unbroken cells (8). The supernatant fraction was centrifuged for 90 min at 100,000 rpm at 4°C in a Beckman TL100 centrifuge to obtain the crude cytoplasmic (supernatant) and membrane (pellet) fractions for Western blot analysis. Coomassie blue-stained sodium dodecyl sulfate (SDS) gels confirmed that the pellet contained all of the detectable major outer membrane protein bands with negligible amounts of the protein bands that characterized the cytoplasmic fraction.
Western blot analysis.
Western blot analysis was performed as previously described on SDS extracts of intact cells (21) or of membrane and cytoplasmic fractions. Protein concentrations were determined by bicinchoninic acid assay (Pierce) of the SDS extracts. For quantitation of individual proteins, 10-, 20-, or 30-μg samples were subjected to Western blot analysis as previously described, using polyclonal anti-MinD or anti-MinE2-19 antibody (21). The relative intensities of the stained bands were determined on digitized images using the ImageQuant program (Molecular Dynamics). For assays of hybrid proteins in yeast, cultures were grown in synthetic dropout (minus Leu, minus Trp, minus Met) medium (Clontech), and proteins were extracted by the trichloroacetic acid method (Clontech manual, protocol PT3024-1). Protein extracted from 0.5 OD600 unit of cells was analyzed using polyclonal antibody (Clontech) directed against the hemagglutinin tag that is part of the fusion protein.
RESULTS
Min protein interactions.
The yeast two-hybrid system was used to study interactions between MinD and MinE, using β-galactosidase expression as an indicator of interaction. In these experiments, MinD was fused to the Gal4 AD (AD-MinD) and MinE was fused to the Gal4 BD (BD-MinE). Consistent with previous reports (7), full-length MinE (MinE1-88) interacted very weakly with MinD, as indicated by color development on X-Gal plates and direct enzyme assay (Table 3). We therefore separately examined the abilities of the two functional domains of MinE (MinE1-31 and MinE32-88) to interact with MinD in the two-hybrid system (Table 3). This revealed a strong interaction of MinD with MinE1-31, the MinE domain that was predicted from genetic experiments to functionally interact with MinD (9, 13, 22). The interaction, as judged by β-galactosidase activity, was >100-fold greater than the interaction of full-length MinE with MinD. As expected, the MinE32-88 domain, which appears not to be involved in MinE-MinD interactions in vivo, showed no interaction. The demonstration that MinE1-31 interacts strongly with MinD in the two-hybrid system provided a convenient assay for studying determinants involved in the MinD-MinE interaction.
TABLE 3.
Min protein interactions in yeast two-hybrid system
| Fusion to BDa | Fusion to ADa,b | Color on X-Gal platec | β-Galactosidase activityd |
|---|---|---|---|
| −b | MinD | 0 | <0.2 |
| MinE1-88 | − | 0 | ND |
| MinE1-88 | MinD | ± | 0.9 ± 0.09 |
| MinE1-31 | − | 0 | ND |
| MinE1-31 | MinD | +++ | 112 ± 5 |
| MinE32-88 | − | 0 | ND |
| MinE32-88 | MinD | 0 | <0.2 |
| MinE1-31 | MinE32-88 | 0 | <0.2 |
| MinE1-31 | MinE1-31 | 0 | <0.2 |
| MinE32-88 | MinE32-88 | 0 | <0.2 |
| MinD | MinD | ++ | 42 ± 1 |
| MinC | MinD | ++++ | 715 ± 5 |
| MinC | MinC | + | 6 ± 1 |
See Materials and Methods and Table 1 for names of plasmids.
−, no gene fusion.
0, white colonies; ± to ++++, intensity of blue color of colonies.
Miller units. ND, not done.
As previously shown (7), MinD interacted strongly with MinC in the two-hybrid system (Tables 3 and 4). MinE strongly interfered with the MinC-MinD interaction, as shown by a 99% decrease in β-galactosidase expression when MinE was expressed together with AD-MinD and BD-MinC (Table 4); a similar result was obtained when MinE was replaced by MinE1-31, as shown by a 94% reduction in β-galactosidase expression (Table 4). The ability of MinE and MinE1-31 to block the MinC-MinD interaction is likely to reflect interaction between MinD and MinE, but interaction between MinE and MinC cannot be excluded, despite the fact that MinE-MinC (7) and MinE1-31-MinC (unpublished data) interactions have not been observed in two-hybrid experiments. MinE32-88 partially interfered with the MinC-MinD interaction, as indicated by a decrease of ∼50% in β-galactosidase activity (Table 4). This may reflect the presence in MinE32-88 of the C-terminal portion of the MinE6-35 α-helix that is believed to be responsible for the MinE-MinD interaction (9) (see below). The ability to perturb the MinC-MinD interaction provided a second assay for mapping the interacting site.
TABLE 4.
Min protein interactions in yeast three-hybrid system
| Fusion to BDa | Fusion to ADa | Downstream of PMetb | Color on X-Gal platec | β-Galactosidase activityd |
|---|---|---|---|---|
| MinC | MinD | − | ++++ | 987 ± 40 |
| MinC | MinD | MinE1-88 | ± | 2 ± 0.1 |
| MinC | MinD | MinE1-31 | +++ | 64 ± 2 |
| MinC | MinD | MinE32-88 | ++++ | 470 ± 29 |
| MinC | MinD | MinE1-88,L22R | ++++ | 912 ± 108 |
| MinC | MinD | MinE1-31,L22R | ++++ | 1,096 ± 144 |
| MinC | MinD | MinE1-88,L22S | ++++ | 400 ± 6 |
| MinC | MinD | MinE1-88,I25R | ++++ | 987 ± 117 |
| MinC | MinD | MinE1-31,I25R | ++++ | 882 ± 37 |
| MinC | MinD | MinE1-88,K19A K19A | +++ | 61 ± 2 |
See Table 2 for names of plasmids.
Cells grown in absence of methionine. −, no gene fusion.
± to ++++, intensity of blue color of colonies.
Miller units.
The ability of MinE to interfere with the MinC-MinD interaction presumably explains in vitro observations that MinE can promote the release of MinC from a MinC-MinD-phospholipid complex without the release of MinD (6, 11).
The full-length MinE protein (MinE1-88) was highly effective in interfering with the MinC-MinD interaction, although it showed very little activity when assayed for the ability to interact directly with MinD (Table 3). The cause of the difference in behavior of full-length MinE in the two assays is not known. Further work will be needed to determine whether it reflects an inhibitory effect of the Gal4 BD, when fused to full-length MinE, on the accessibility of the MinD BD or whether the difference in behavior has implications for the in vivo functions of the Min proteins in E. coli.
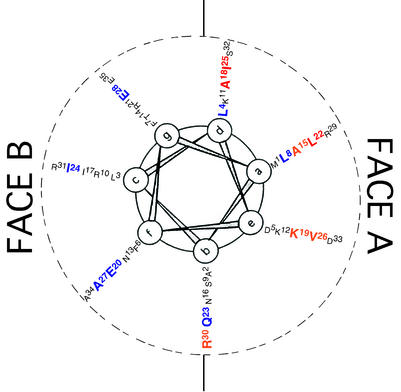
If the region of MinE1-31 that interacts with MinD exists in an α-helical conformation, residues within MinE1-31 that functionally interact with MinD would be expected to cluster on one face of the helix. To examine this prediction, we mutagenized conserved residues (10) that are distributed around the predicted MinE1-35 helix. In each case, the mutation changed a nonpolar to a polar amino acid, or vice versa. The mutant proteins were then assayed in the two-hybrid and three-hybrid assay systems for the ability to interact with MinD or to disrupt the MinC-MinD interaction. For convenience, we divided the predicted helical surface into two faces, called the A and B faces (Fig. 1).
FIG. 1.
Helical-wheel projection of MinE1-35 amino acid residues. The positions of MinE amino acid residues 1 to 35 are indicated on the surface of the predicted N-terminal α-helix. Mutated amino acids are indicated in large type. The colors indicate the effects of mutations on MinE-MinD interaction in yeast two-hybrid and three-hybrid assays (Tables 3, 4, and 5). Red, complete loss of activity in both assays; orange, partial loss of activity; blue, no effect. a through g, positions of amino acids in the helical wheel.
Mutations in 7 (Ala15, Ala18, Lys19, Leu22, Ile25, Val26, and Arg30) of the 14 mutagenized residues led to a complete or partial loss of MinE function in the two-hybrid and three-hybrid assay systems. This was shown by the failure of the mutated proteins to induce β-galactosidase expression in the MinE1-31-MinD system (Table 5) and/or by inability to interfere with the increase in β-galactosidase expression that results from interaction between MinC and MinD (Table 4). The other mutated proteins did not differ from unmutagenized MinE in these assays. In the case of Leu22, three substitutions that led to conversion of the leucine residue to glutamate (L22E), arginine (L22R), and serine (L22S) were individually studied. The L22E and L22R mutations led to complete loss of interaction in the two-hybrid and three-hybrid experiments (Tables 4 and 5). The conversion to serine, in MinEL22S, was associated with loss of detectable interaction with MinD (Table 5) but only partial loss of the ability to interfere with the MinC-MinD interaction (Table 4). The results were not due to differences in cellular concentrations of the proteins, as shown by quantitative immunoblots (data not shown).
TABLE 5.
Effects of MinE mutations on MinD-MinE interaction in two-hybrid systema
| Fused to BD | Fused to AD | Color on X-Gal plateb | β-Galactosidase activityc |
|---|---|---|---|
| MinE1-31 | 0 | ND | |
| MinE1-31 | MinD | +++ | 104 |
| MinE1-31,L4R | MinD | +++ | ND |
| MinE1-31,L8R | MinD | +++ | ND |
| MinE1-31,A15R | MinD | ++ | 41 |
| MinE1-31,A18T | MinD | 0 | <1 |
| MinE1-31,K19A | MinD | ± | <1 |
| MinE1-31,E20A | MinD | +++ | 162 |
| MinE1-31,L22R | MinD | 0 | <1 |
| MinE1-31,L22E | MinD | 0 | <1 |
| MinE1-31,L22S | MinD | 0 | <1 |
| MinE1-31,Q23R | MinD | +++ | ND |
| MinE1-31,I24R | MinD | +++ | ND |
| MinE1-31,I25R | MinD | 0 | <1 |
| MinE1-31,V26E | MinD | + | ND |
| MinE1-31,V26R | MinD | +++ | ND |
| MinE1-31,A27R | MinD | +++ | ND |
| MinE1-31,E28A | MinD | +++ | ND |
| MinE1-31,R30A | MinD | ++ | 8 |
See Materials and Methods and Table 2 for plasmid names and experimental details.
0, white colonies; ± to ++++, intensity of blue color of colonies.
Miller units. ND, not done.
Strikingly, the seven residues that were implicated in MinE function in these assays were all located on the A face of the helix, clustered in the middle portion of the predicted MinE6-35 helix (Fig. 1). The results are consistent with the idea that the interaction between MinD and the N-terminal domain of MinE involves an interaction between one face (the A face) of the MinE6-35 α-helix and a complementary site within MinD (9).
It has been shown that mutation of Lys19 (K19Q) or double mutations that included Lys19 or Ala18 were associated with a significant decrease in the ability of MinE to stimulate MinD ATPase activity (5). These observations are consistent with the yeast two- and three-hybrid results.
Effects of MinE mutations on Min protein localization.
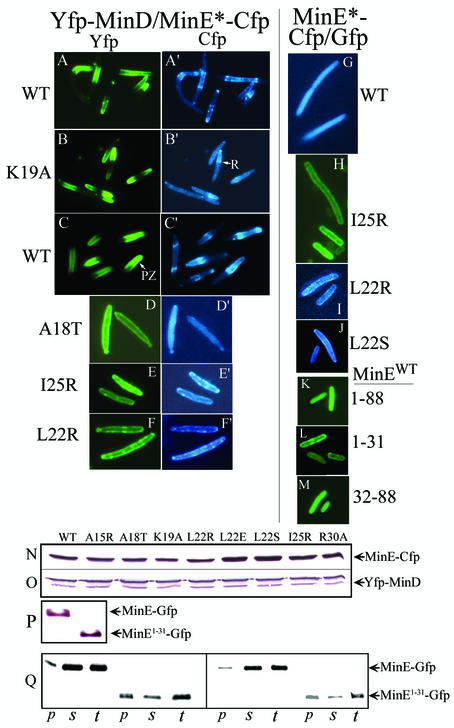
Proteins labeled with Gfp, Yfp, and Cfp (19) were used to study the effects of the MinE mutations on the cellular localization of fluorescently labeled MinD and MinE when minE and minD were coexpressed in RC1 (ΔminCDE) cells. These studies showed that mutations that led to complete loss of interaction in the two-hybrid assay systems (minEA18T, minEI25R, and minEL22R) were associated with loss of the normal distribution patterns of the proteins, as shown by the absence of MinD polar zones and MinE rings and polar zones (Fig. 2D and D ′, E and E′, and F and F′).
FIG. 2.
Fluorescence microscopy of labeled MinD and MinE. Cells were prepared from strain RC1 (ΔminCDE) containing the following plasmids: pYLS68 (Plac-yfp::minD minE::cfp) (A and A′); pYEC19 (Plac-yfp::minD minEK19A::cfp) (B and B′); pYLS68 (C and C′); pYEC18 (Plac-yfp::minD minEA18T::cfp) (D and D′); pYEC25 (Plac-yfp::minD minEI25R::cfp) (E and E′); pYEC22 (Plac-yfp::minD minEL22R::cfp) (F and F′); pWEC1 (Plac-minE::cfp) (G); pMEG25 (Plac-minEI25R::gfp) (H); pMEC22 (Plac-minEL22R::cfp) (I); pMEC22S (Plac-minEL22S::cfp) (J); pYLS41G (Plac-minE::gfp) (K); pMA101 (Plac-minE1-31::gfp) (L); and pMA102 (Plac-minE32-88::gfp) (M). (A, A′, B, and B′) Fixed cells. (C to M) Unfixed cells. Yfp and Gfp are green, and Cfp is blue. MinE-Cfp images were acquired 10 s after the corresponding Yfp-MinD images. The labels to the left and right of the fluorescence images indicate the minE alleles of the corresponding images. WT, wild type. (N and O) Western blot analyses of cells from the experiments described in panels A to F and the corresponding experiments described in the text. (P) Western blot analysis of cells from the experiments described in panels K and L. (Q) Western blot analysis of cell fractions from cells expressing minE::gfp and minE1-31::gfp from plasmids pYLS41G (Plac-minE::gfp) and pMA101 (Plac-minE1-31::gfp), respectively. p, membrane (pellet fraction); s, cytoplasm (supernatant fraction); t, total extract. The aliquots applied to each lane were derived from 0.04 OD600 unit of cells for lanes 1 to 6 and 0.02 OD600 unit of cells for lanes 7 to 12. Anti-MinE2-19 antibody was used for panels N, P, and Q; anti-MinD antibody was used for panel O.
The minEL22S mutation, which led to partial loss of interaction in the three-hybrid system (Table 4), was associated with a significant decrease in the number of cells containing visible MinD polar zones and E rings compared with cells that expressed wild-type MinE (Table 6). Only 20% of minEL22S cells contained visible MinD polar zones, and 5% contained visible MinE rings, whereas nearly all cells expressing wild-type MinE contained E rings and MinD polar zones.
TABLE 6.
Phenotypes of MinE mutants
| MinEa | MinD-MinE interactionb | MinD polar zones
|
E ringse | Phenotype | ||
|---|---|---|---|---|---|---|
| 1c | 2d | |||||
| WT | + | 92 | 3 | 95 | WT | |
| A15R | ± | 96 | 1 | 97 | WT | |
| K19A | ± | 27 | 70 | 74 | Sep− Min− | |
| L22S | ± | 20 | <1 | 5 | Sep− | |
| R30A | ± | 95 | <1 | 94 | WT | |
| A18T | 0 | <1 | <1 | <1 | Sep− | |
| L22R | 0 | <1 | <1 | <1 | Sep− | |
| I25R | 0 | <1 | <1 | <1 | Sep− | |
The mutated amino acids in mutant MinE proteins are shown; 100 cells of each type were analyzed to determine the percentage of cells with polar zones or MinE rings. WT, wild type.
MinD-MinE interaction as indicated by induction of β-galactosidase in two-hybrid assays (Table 5) and/or by ability to inhibit MinC-MinD interaction in three-hybrid assays (Table 4). 0, no interaction in either assay; ±, partial interaction compared with wild-type MinE in one or both assay systems; +, normal interaction compared with wild-type MinE.
Percentage of cells containing one MinD polar zone.
Percentage of cells containing polar zones at both poles.
Percentage of cells with E rings.
Immunoblots confirmed that the failure to form E rings or MinD polar zones was not due to a decrease in cellular concentrations of MinE or MinD in the mutant strains (Fig. 2N and O).
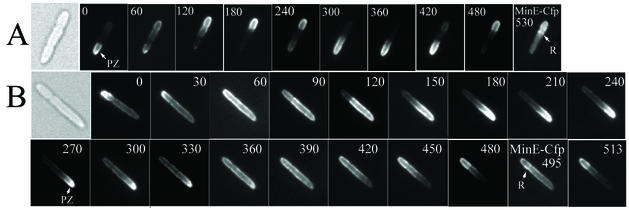
Many of the minEL22S cells that contained polar zones showed marked abnormalities in their patterns of assembly and disassembly. In cells expressing wild-type MinE together with MinD (Fig. 3A), the assembly of a polar zone at one end of the cell is followed by its shrinkage from midcell toward the pole, terminating in its disappearance from the original cell pole (2), apparently by release into the cytoplasm (16). A new polar zone is then assembled at the opposite end of the cell. In contrast, in the minEL22S cells (Fig. 3B), the disappearance of MinD from the polar zone was often not followed by formation of a new polar zone at the opposite cell pole. Instead, after the disappearance of the polar zone, MinD was present around the entire cell periphery, presumably membrane associated, suggesting that the disappearance of the polar zone might be accomplished by the random redistribution of MinD within the cytoplasmic membrane. This was followed by retraction of the membrane-associated MinD from the original pole, leading to accumulation of MinD within a “new” polar zone at the opposite cell pole (Fig. 3B). Thus, in contrast to the normal situation, the new MinD polar zones in many minEL22S cells appeared not to be formed de novo but often reflected membrane-associated MinD that was left at one cell pole as a result of the disappearance or redistribution of membrane-associated MinD from the other end of the cell. The rate of disassembly of the membrane-associated MinD was much lower than that of the disassembly of MinD polar zones in cells expressing wild-type MinE (Fig. 3A) (19). Interestingly, this phenotype resembled in part the phenotype resulting from mutation at the MinED45/V49 site within the C-terminal topological-specificity domain of MinE that is required for E-ring formation, where a general peripheral distribution of MinE was often followed by retraction from one end of the cell (19). The possible implications of this similarity remain to be explored.
FIG. 3.
Oscillation of Yfp-MinD in living cells. Time lapse fluorescence micrographs of Yfp-MinD in strain RC1 containing pYLS68 (Plac- yfp::minD minE::cfp) (A) and pYEC22S (Plac-yfp::minD minEL22S::cfp) (B). The time at which the image was acquired (in seconds) is indicated in each micrograph. The cells were observed at 15-s intervals between micrographs to confirm that no dramatic change occurred during the intervals. In one image in each panel, as indicated, fluorescence from MinE-Cfp was captured. The arrows indicate the MinE ring (R) and the MinD polar zone (PZ).
The minEK19A mutation, which was also associated with a partial loss of interaction with MinD in the two-hybrid and three-hybrid assay systems (Tables 4 and 5), showed an unusually high percentage of cells that contained MinD polar zones at both poles (70% of the cells) (Fig. 2B and Table 6). In comparison, in cells expressing wild-type MinE in parallel cultures, only 3% of the cells contained visible polar zones at both poles (Table 6). The rate of pole-to-pole oscillation in the minEK19A cells was lowered to ∼40% of the rate in wild-type cells. A similar observation was made by Hu and Lutkenhaus (5), who observed a decrease in the oscillation rates of ∼50% in the presence of two other mutations affecting the Lys19 residue (minEK19Q and minEK19A,E20A). In the remaining mutants, essentially all cells contained polar zones and E rings and the oscillation rates of the polar zones were not significantly perturbed.
Effect of MinE mutations on cell division phenotype.
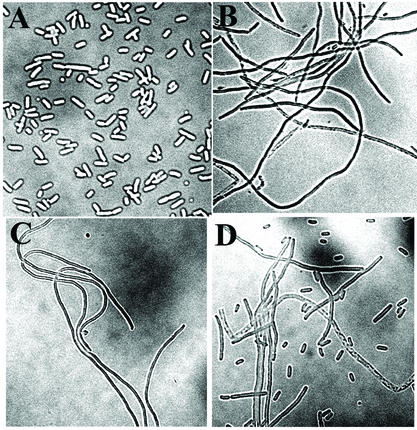
Expression of normal levels of MinCD in the absence of MinE leads to a general inhibition of septation, leading to formation of long nonseptate filaments (1). The filamentation can be prevented by expression of either full-length MinE or N-terminal MinE fragments (1, 13, 22). To determine the effect of the minE mutations on the cell division phenotype, the mutant alleles were substituted for wild-type minE in plasmid pSY1083 (Plac-minCDE), and the cell division phenotype was determined in host strain RC1 (ΔminCDE) after IPTG induction. The ability to counteract the division inhibition is thought to require interaction between MinD and a site within the N-terminal MinE domain (22). Consistent with this view, MinE mutations that led to complete loss of MinD-MinE interaction in the two-hybrid system (mutations A18T, I25R, L22R, and L22S) were associated with a filamentation phenotype (Fig. 4B to D and Table 6). Mutation K19A, which led to significant but not total loss of detectable interaction in the two-hybrid assay (Table 5), showed a mixture of filaments, minicells, and cells of approximately normal length. We assume that this mixed phenotype represented the effects of different levels of expression of the partially active MinEK19A protein in different cells within the culture.
FIG. 4.
Effects of minE mutations on cell division patterns. Micrographs of Nomarski images of RC1 containing pSY1083 (Plac-minC minD minE) (A), pEM22S (Plac-minC minD minEL22S) (B), pEM25 (Plac-minC minD minEI25R) (C), and pEM19 (Plac-minC minD minEK19A) (D) are shown. The cells were prepared and analyzed as previously described (1).
The possibility that the loss of ability to counteract the MinCD division inhibition was due to a decrease in the cellular concentrations of the mutant proteins was excluded by quantitative immunoblot analysis (data not shown).
Membrane localization of MinE in the absence of MinD.
It has been shown that full-length MinE (MinE1-88) fails to localize to the peripheral portion of the cell unless MinD is coexpressed, indicating that MinD is required for the membrane association of MinE (2, 15). These observations were confirmed in the present study, where MinE-Cfp and MinE-Gfp, when expressed in the absence of MinD, were diffusely distributed within the cell (Fig. 2G and K), indicating a cytoplasmic localization. However, in contrast to full-length MinE, MinE1-31-Gfp showed a peripheral distribution even when expressed in the absence of MinD (Fig. 2L). This unexpected result was obtained in several independent experiments and was seen in living cells as well as fixed cells. This suggested that the MinE1-31 protein was able to associate with the cell membrane in the absence of MinD. The possibility that the difference between the behaviors of full-length MinE and MinE1-31 was due to differences in cellular concentration was excluded by immunoblot analysis, which showed that the cellular concentrations of MinE1-88-Gfp and MinE1-31-Gfp were similar (Fig. 2P).
The distributions of MinE1-88-Gfp and MinE1-31-Gfp were also examined in crude cytoplasmic and membrane fractions of cells that expressed minE1-88::gfp or minE1-31::gfp in the absence of minD (Fig. 2Q). The results were consistent with the fluorescence localization studies in intact cells (Fig. 2L). Nearly all of the MinE1-88-Gfp was recovered in the cytoplasmic fraction, whereas >70% of the MinE1-31-Gfp was recovered in the crude membrane fraction.
In the case of several of the mutant alleles (minEI25R, minEL22R, and minEL22S), the full-length MinE-Cfp protein also showed a peripheral distribution when expressed in the absence of MinD (Fig. 2H to J). The observations were reproducible. These unexpected results, which are surprising in view of the fact that MinD is required to bring wild-type MinE1-88 to the membrane, are discussed further below.
DISCUSSION
It is likely that interactions between MinE and MinD play a role in several aspects of Min protein function. These include the MinD-dependent membrane association of MinE (15), the MinE-dependent formation and oscillation of MinD polar zones (2, 16), the ability of MinE to counteract the MinCD division inhibitor (1), and the ability of MinE to stimulate MinD ATPase activity (5) and thereby promote release of membrane-associated MinD (4, 11, 20). The present study provides direct evidence that the MinE-MinD interaction involves the N-terminal MinE1-31 domain and maps the site within the N-terminal MinE domain that is responsible for the interaction.
Although the mutations that were originally introduced into the N-terminal MinE domain were designed to be distributed on both faces of the MinE1-31 helical region, it was striking that the mutations that affected the MinD-MinE interaction were located exclusively on one face of the helix, the A face, where they defined an extended contiguous surface domain. Because the MinE6-35 helical region is predicted to have a propensity to form coiled-coil structures (9), it is reasonable to speculate that the site on the A face interacts with a corresponding surface on an α-helix within MinD to form a supercoiled structure. The relatively large area of the putative MinD-binding site on the A face, involving two or three turns of the helix, is consistent with the idea that it represents a surface that interacts in a side-by-side configuration with a surface-exposed α-helix in MinD. We presume that this interaction plays an essential role in the ability of MinD to bring MinE to the membrane and possibly in other MinE/MinD-dependent functions of the Min system. Mutations that interfered with MinD-MinE interactions in the two-hybrid system were associated with the complete or partial absence of MinD polar zones and E rings. This is consistent with the idea that interaction between MinD and the putative binding site on the A face of MinE1-31 plays an important role in the ability of MinE to promote the membrane redistribution events that lead to formation of the MinD polar zone. The mutations also interfered with the ability of MinE to counteract the MinCD division inhibitor, consistent with the idea that this function requires that MinE be brought to the membrane in a reaction that needs interaction with MinD (15).
It is not known whether binding to MinD is the only function of the N-terminal domain of MinE. Full-length MinE does not localize to the cytoplasmic membrane of E. coli in the absence of MinD, as shown by the diffuse distribution of MinE-Gfp within the cell. This indicates that MinE requires MinD to move from cytoplasm to membrane (references 2 and 15 and this study). In contrast to the behavior of full-length MinE, it was observed in the present study that MinE1-31-Gfp expressed in the absence of MinD was located around the periphery of the cell, in a pattern that is usually interpreted to indicate association with the membrane or cell envelope. This interpretation was supported by the finding in extracts of these cells that MinE1-31 was recovered primarily in the membrane fragment whereas full-length MinE was found almost exclusively in the cytoplasmic fraction. Several of the MinE mutant proteins showed similar peripheral cellular localization patterns when expressed in the absence of MinD, even when present as full-length proteins. The apparent membrane localization of MinE1-31 and of the mutant proteins may represent experimental artifacts, in which the mutations or the removal of the MinE C-terminal domain uncovers nonspecific and biologically irrelevant membrane-binding sites within the N-terminal MinE1-31 domain. This would be consistent with current models, in which MinD is the sole membrane anchor for membrane-associated MinE. However, the present observations may instead provide a clue that MinE1-31 contains membrane-binding sites that function after MinE is brought to the membrane by MinD. According to this view, the membrane-binding sites within MinE1-31 are normally occluded by the C-terminal domain and can be exposed as a result of conformational changes during the MinD-dependent membrane assembly process or experimentally by removal of the C-terminal domain or by certain mutations. The putative MinE-membrane interaction could act to stabilize the membrane association of the E ring or could play another role in MinE function. Further work will be needed to test these ideas.
Acknowledgments
This work was supported by National Institutes of Health grants GM60632 to L.R. and GM45583 to G.K.
REFERENCES
- 1.de Boer, P. A. D., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641-649. [DOI] [PubMed] [Google Scholar]
- 2.Fu, X., Y.-L. Shih, Y. Zhang, and L. I. Rothfield. 2001. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 98:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hale, C., H. Meinhardt, and P. de Boer. 2001. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 20:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu, Z., E. Gogol, and J. Lutkenhaus. 2002. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc. Natl. Acad. Sci. USA 99:6761-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu, Z., and J. Lutkenhaus. 2001. Topological regulation of cell division in E. coli: spatiotemporal oscillation of MinD occurs through stimulation of its ATPase activity by MinE and phospholipid. Mol. Cell 7:1337-1343. [DOI] [PubMed] [Google Scholar]
- 6.Hu, Z., C. Saez, and J. Lutkenhaus. 2003. Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. J. Bacteriol. 185:196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang, J., C. Cao, and J. Lutkenhaus. 1996. Interaction between FtsZ and inhibitors of cell division. J. Bacteriol. 178:5080-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishidate, K., E. S. Creeger, J. Zrike, S. Deb, B. Glauner, T. J. MacAlister, and L. I. Rothfield. 1986. Isolation of differentiated membrane domains from Escherichia coli and Salmonella typhimurium, including a fraction containing attachment sites between the inner and outer membranes and the murein skeleton of the cell envelope. J. Biol. Chem. 261:428-443. [PubMed] [Google Scholar]
- 9.King, G. F., S. L. Rowland, B. Pan, J. Mackay, G. Mullen, and L. I. Rothfield. 1999. The dimerization and topological specificity functions of MinE reside in a structurally autonomous C-terminal domain. Mol. Microbiol. 31:1161-1169. [DOI] [PubMed] [Google Scholar]
- 10.King, G. F., Y.-L. Shih, M. W. Maciejewski, N. P. S. Bains, B. Pan, S. Rowland, G. P. Mullen, and L. I. Rothfield. 2000. Structural basis for the topological specificity function of MinE. Nat. Struct. Biol. 7:1013-1017. [DOI] [PubMed] [Google Scholar]
- 11.Lachner, L., D. Raskin, and P. de Boer. 2003. ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J. Bacteriol. 185:735-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 13.Pichoff, S., B. Vollrath, C. Touriol, and J.-P. Bouché. 1995. Deletion analysis of gene minE which encodes the topological specificity factor of cell division in Escherichia coli. Mol. Microbiol. 18:321-329. [DOI] [PubMed] [Google Scholar]
- 14.Raskin, D., and P. de Boer. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichioa coli. J. Bacteriol. 181:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raskin, D., and P. de Boer. 1997. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91:685-694. [DOI] [PubMed] [Google Scholar]
- 16.Raskin, D., and P. de Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothfield, L. I., Y.-L. Shih, and G. F. King. 2001. Polar explorers: membrane proteins that determine division site placement. Cell 106:13-16. [DOI] [PubMed] [Google Scholar]
- 18.Rowland, S. L., X. Fu, M. A. Sayed, Y. Zhang, W. R. Cook, and L. I. Rothfield. 2000. Membrane redistribution of the Escherichia coli MinD protein induced by MinE. J. Bacteriol. 182:613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih, Y.-L., X. Fu, G. F. King, T. Le, and L. I. Rothfield. 2002. Division site placement in E. coli: mutations that prevent formation of the MinE ring lead to loss of the normal midcell arrest of growth of polar MinD membrane domains. EMBO J. 21:3347-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suefuji, K., R. Valluzzi, and D. RayChaudhuri. 2002. Dynamic assembly of MinD into filament bundles modulated by ATP, phospholipids, and MinE. Proc. Natl. Acad. Sci. USA 99:16776-16781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, Y., S. Rowland, G. King, and L. Rothfield. 1998. Relation of the oligomeric structure of MinE to its topological specificity function. Mol. Microbiol. 30:265-273. [DOI] [PubMed] [Google Scholar]
- 22.Zhao, C.-R., P. de Boer, and L. Rothfield. 1995. Proper placement of the E. coli division site requires two functions that are associated with different domains of the MinE protein. Proc. Natl. Acad. Sci. USA 92:4313-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]