Abstract
Procrustes analysis involves finding the optimal superposition of two or more “forms” via rotations, translations, and scalings. Procrustes problems arise in a wide range of scientific disciplines, especially when the geometrical shapes of objects are compared, contrasted, and analyzed. Classically, the optimal transformations are found by minimizing the sum of the squared distances between corresponding points in the forms. Despite its widespread use, the ordinary unweighted least-squares (LS) criterion can give erroneous solutions when the errors have heterogeneous variances (heteroscedasticity) or the errors are correlated, both common occurrences with real data. In contrast, maximum likelihood (ML) estimation can provide accurate and consistent statistical estimates in the presence of both heteroscedasticity and correlation. Here we provide a complete solution to the nonisotropic ML Procrustes problem assuming a matrix Gaussian distribution with factored covariances. Our analysis generalizes, simplifies, and extends results from previous discussions of the ML Procrustes problem. An iterative algorithm is presented for the simultaneous, numerical determination of the ML solutions.
Keywords: heteroscedasticity, morphometrics, Procrustes analysis, superpositions, least-squares
The goal of Procrustes analysis is to superpose non-identical shapes in an optimal manner via scalings, translations, and rotations (1–3). Classical Procrustes analysis uses the unweighted least-squares (LS) criterion for finding the optimal transformations. LS implicitly assumes that the landmarks describing the forms' shapes are uncorrelated and that they have identical variances (i.e., that they are homoscedastic). However, in many practical applications these assumptions are known to be violated. For instance, when superpositioning macromolecular proteins, individual atoms (the landmarks) are connected via covalent chemical bonds, and thus the variance of a given atom can be correlated with the variance of atoms to which it is connected. Furthermore, certain atoms may have larger mobilities than others, or they may have spatial positions with relatively greater uncertainty due to experimental error. Similarly, when comparing the skulls from different members of a species, some homologous features may be highly variable relative to others. Hence, different landmarks can have widely different variances. Under these conditions, ordinary LS can give misleading results, even with large samples of data (4).
The method of maximum likelihood (ML) is a common alternative to LS and is widely considered to be fundamental for statistical modeling and parameter estimation (5). Given the proper model, ML can provide accurate and robust estimates of parameters in the presence of both heteroscedasticity and correlation. Incorporating heterogeneous variances and non-zero correlations into the ML Procrustes problem involves weighting by two covariance matrices (a landmark and a dimensional covariance matrix). However, estimation of these covariance matrices has been a major stumbling block prohibiting a viable ML Procrustes analysis (1, 3, 6–10). Without special assumptions, simultaneous estimation of the landmark covariance matrix and the translations is generally impossible. By parametrically constraining the landmark covariance matrix using a hierarchical, empirical Bayes likelihood model, we provide a relatively simple solution that permits valid covariance estimation.
In addition to allowing simultaneous estimation of the landmark covariance matrix and the translations, we address several additional challenges that have thus far impeded successful ML inference in the Procrustes problem. (i) As usually described, Procrustes analysis gives degenerate estimates of the covariance matrices even when the translations are known (7). We suggest a method for uniquely identifying the covariance matrices, using the fact that the absolute reference frame in Procrustes analysis is arbitrary. (ii) The dimensional covariance matrix introduces further difficulties for optimal rotation estimation, and current methods involve a circuitous and computationally intensive numerical procedure. We provide a simplified, analytic solution to estimating the optimal rotations when using an arbitrary dimensional covariance matrix. (iii) We give a simplified ML estimator for the scaling factors and also present a hierarchical model for estimating the scalings that can improve estimation in common biological cases. (iv) The ML estimator of the mean form is known to be inconsistent (i.e., it converges to the wrong value as the number of forms increases). Based on the approximate χ2 distribution of the Procrustes sum of squared distances, we derive an easily calculable bias-corrected ML estimator for the mean. Given that the errors are not large, this estimator is consistent for landmarks with uncorrelated, nonisotropic ellipsoidal variability. (v) Finally, we present a relatively simple and stable iterative algorithm for the simultaneous solution of these unknown parameters.
Formulation of the ML Procrustes Problem
In statistical shape theory, the shape of a geometrical configuration is commonly defined as the geometrical information that is unchanged after translation, rotation, and scaling of the object. Particular instances of an object, which are described by a set of points summarizing the object's shape, we refer to as a “form.” We also define “size-and-shape” as those geometrical aspects of a form that are unchanged after translation and rotation. In its most general formulation, Procrustes analysis involves the optimal matching of two or more form matrices. Here we consider only sets of forms where each form matrix has the same dimensions.
Consider N different forms (Xi, i = 1 … N), each with K corresponding points or “landmarks” that depict the form's shape. Each form is defined as a K × D matrix holding K rows of landmarks, where each landmark is a D-vector in ℜD. For example, when superposing protein structures, the forms are independent protein models and the landmarks are the three-dimensional atomic coordinates of each atom in the protein structures. Furthermore, each form is observed in a different, arbitrary, and unknown coordinate system (1, 10, 11). The absolute reference frame for the forms is arbitrary; only relative rotational and translational transformations are of interest. We wish to estimate the optimal orthogonal rotations, translations, and scaling factors that, when applied to this set of form matrices, predict the observed data with the highest likelihood.
Gaussian Perturbation Model with Factored Covariances
To analyze the Procrustes problem from a likelihood perspective, one must choose a statistical model that is assumed to generate the observed data. We assume a perturbation model in which each form Xi is distributed according to a Gaussian (normal) probability density (7, 11). In this Gaussian perturbation model, each Xi can be considered to be an arbitrarily scaled, rotated, and translated Gaussian perturbation of a mean form M
where Ri is a D × D rotation matrix, βi is a scaling parameter, ti is a 1 × D row vector for the translational offset, and 1K denotes the K × 1 column vector of ones. The matrix Ei is a zero-mean matrix of Gaussian random errors Ei ≈ NK,D (0, Σ, Ξ) with the same dimensions as the mean form M, where Σ and Ξ are covariance matrices described in detail below. In other words, to generate each of the observed forms in D-dimensional Euclidean space, each landmark in an idealized mean form M is randomly perturbed according to a Gaussian distribution. The perturbations for each landmark may be unequal on average and may be correlated with each other, as specified by the covariance matrices. Each perturbed form is then arbitrarily scaled, rotated, and translated, the results of which constitute the observed data.
Covariance matrices describe the variability and correlations among the landmarks in the forms. In the above model, we use factored covariances by assuming that the overall KD × KD covariance matrix Ω can be decomposed adequately as the Kronecker product of Σ and Ξ (i.e., Ω = Σ ⊗ Ξ). Here, Σ is a K × K covariance matrix for the rows of the forms (the landmarks), and Ξ is a constrained D × D covariance matrix for the columns of the forms (the dimensions). In the protein structure example, the landmark covariance matrix Σ describes the variances and correlations among the atoms. Additionally, the dimensional covariance matrix Ξ describes the variances and correlations between the three coordinate axes, for example, if atomic positions are on average more variable in one dimension than another. We note that, in many cases, the factored covariance model is physically unrealistic, because it assumes that each of the landmarks have the same ellipsoidal principal axes of variability. However, we accept this assumption as a simplifying approximation, and note that it is more general than the assumptions underlying classical LS Procrustes analysis.
Although the product Σ ⊗ Ξ is uniquely identified, Σ and Ξ independently are not, because for all c > 0, Ω = Σ ⊗ Ξ = cΣ ⊗ 1/c Ξ. Thus, to jointly identify both Σ and Ξ, we impose the constraint that trΞ = D. Furthermore, as a generality in Procrustes analysis, the absolute rotational frame of reference is arbitrary, and only the relative rotations among the forms are of interest. As a result, Ξ is only identifiable up to an arbitrary orthogonal similarity transformation (7). However, any given Ξ can be spectrally decomposed such that Ξ = VΔV′. Then, the diagonal matrix of eigenvalues Δ is equivalent to the Ξ that would result if the mean form M were rotated with the orthogonal matrix V′ of left eigenvectors. Thus, for unique identifiability, we require that the axes of the mean form's reference frame be aligned with the principle axes of the dimensional covariance matrix Ξ. This requirement also guarantees that Ξ is diagonal.
Procrustes Matrix Gaussian Likelihood Equation
A likelihood analysis requires an explicit statement of the likelihood function derived from the probability density function that describes the assumed statistical model. The full joint likelihood equation for the Gaussian Procrustes model given in Eq. 1 is obtained from a multivariate matrix normal distribution (12, 13). To simplify the appearance of the likelihood equation, we define the squared Frobenius Mahalanobis matrix norm as ‖U‖V,W2 = tr{VU′WU}, which can be regarded as the sum of the squared elements of U with columns weighted by V and rows weighted by W. Letting |U| be the determinant of matrix U, the full Procrustes log-likelihood ℓ(β, R, t, M, Ξ, Σ; X) = ℓP is given by
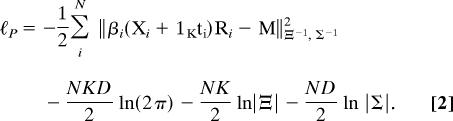 |
The Procrustes log-likelihood equation above (Eq. 2) is the key conceptual contribution underlying the ML analysis that follows. In order to solve the ML Procrustes problem, ML estimators must be derived for each of the unknown parameters in the Procrustes log-likelihood equation. We provide the individual ML estimates for each of these unknowns: the Ξ, Σ, M, and the N βi, ti, and Ri (for derivations see Appendices).
Regularization of the Covariance Matrices Allows Simultaneous Estimation
Estimation of the factored covariance matrices has been the primary impediment for a successful, practical implementation of a joint non-isotropic ML treatment of the Procrustes problem (1, 3, 6–10). We first present the unrestricted ML estimators of the covariance matrices and then discuss the difficulties that our approach remedies.
The unrestricted ML estimators of the covariance matrices, Ξ̂U and Σ̂U, are given by straightforward variants of the usual covariance estimators for the matrix normal distribution (Appendix I) (4, 13).
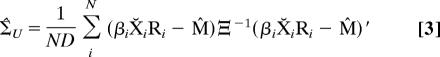 |
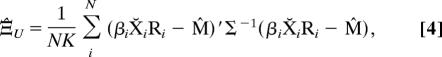 |
where X̆ is a row-weighted centered form.† Because one of the covariance matrices must be normalized for identifiability, we (arbitrarily) choose to normalize Ξ̂U by
and use Ξ∼−1 in place of Ξ−1 in calculating Σ̂U given in Eq. 3.
Both of these covariance matrix estimators involve an inverse of a covariance matrix, as do several of the other ML solutions, a fact that presents three important and distinct difficulties for ML estimation in the Procrustes problem. First, in general, the ML estimators of the matrix normal covariance matrices only exist when the estimators are invertible, a requirement that obtains when N ≥ (K/D) + 1 and N ≥ (D/K) + 1 (13). In practice, these conditions are usually unsatisfied due to a paucity of available data. Second, Procrustes analysis requires centering the form matrices (e.g., Eq. 6), which imparts a common linear constraint on the columns of the forms. Even with ample data, then, the sample landmark covariance matrix is collinear and rank deficient, with at least one zero eigenvalue (3, 6), and consequently it is noninvertible. Third, and most fundamentally, the displacements due to Σ and the translations ti are linearly entangled (see Eq. 1), and therefore unrestricted simultaneous estimation of both is impossible (6, 10). This latter problem is especially grave, because the unrestricted Procrustes estimators of the translations ti and of the covariance matrix Σ are strongly interdependent; translations that closely superpose a given landmark decrease the estimated variance, and a small variance in turn preferentially weights the translations to center on that landmark. For example, it is always possible to find translation vectors that will perfectly superpose any selected landmark, thereby rendering its sample variance exactly zero, Σ̂ singular, and the likelihood infinite.
To overcome the first two of these problems it is sufficient to guarantee the invertibility of the estimated landmark covariance matrix, i.e., all eigenvalues must be positive. In the third difficulty, the possibility of an infinite likelihood immediately points to a need for regularization, either of the translations (6) or of the covariance matrix, or of both.
In real data sets, the variances often are unable to take arbitrary values, but rather they describe coupled components of a larger, integrated system. For instance, the atoms in a macromolecule are linked by chemical bonds, and the bones in a skull are physically joined by ligaments and at sutures. In such cases, the variances for each landmark are similar in magnitude, and extremely small or large variances are improbable and physically unrealistic. Thus, the problem of simultaneous estimation of the covariance matrices and the translations can be circumvented by realistically restricting the variances of the landmarks.
An alternative, then, is to view each of the eigenvalues (λj) of the landmark covariance matrix as a draw from a positive distribution (where λj > 0). The eigenvalues of the covariance matrix can be considered as variances with all correlations eliminated, e.g., the eigenvalues of a diagonal covariance matrix are the variances themselves. The inverse gamma (Iγ) distribution is used conventionally in Bayesian statistics as a natural proper prior for the variances in a multivariate Gaussian model (14). The flexibility of the Iγ distribution is convenient as it can adopt a wide variety of shapes approximating other distributions. Therefore, we regularize our matrix Gaussian Procrustes model using a hierarchical empirical Bayes approach (an extended likelihood) (5, 15) by assuming that the eigenvalues of the landmark covariance matrix belong to a common Iγ distribution. The full, regularized Procrustes log-likelihood (ℓr) is then the sum of the Procrustes log-likelihood from Eq. 2 and the log-likelihood of an Iγ distribution of the eigenvalues (see Eq. 15 in Appendix II): ℓr = ℓP + ℓ(λ)Iγ.
Assuming an Iγ distribution for the eigenvalues of the landmark covariance matrix (with scale parameter α and shape parameter γ), the extended ML estimator Σ̂Iγ of Σ is a simple linear function of the unrestricted ML estimate Σ̂U from Eq. 3 (see Appendix II)
In this hierarchical model, the Iγ parameters α and γ are point estimates determined by the data, unlike when using a bona fide Bayesian prior. Eq. 6 can be interpreted as a shrinkage estimator that contracts the eigenvalues of the covariance matrix towards the mode of the inverse gamma distribution. Given positive α and γ parameters, this regularized model constrains all eigenvalues (and variances) of the landmark covariance matrix to be positive, thereby guaranteeing that Σ̂Iγ is invertible. Furthermore, this treatment enables simultaneous estimation of the landmark covariance matrix Σ and the translations by decoupling the strong dependence of their estimators. Thus, the inverse gamma regularization is sufficient to overcome all three of the difficulties listed above. In our experience, the Σ̂Iγ estimator is also well conditioned. We note that our regularized hierarchical estimator of the multivariate Gaussian covariance matrix also has potential for wide applicability beyond Procrustes analysis.
Simplified ML Estimates of the Rotations
Maximizing the Procrustes likelihood with respect to the rotations is equivalent to maximizing the first term of Eq. 2, which requires weighting by the inverse of the dimensional covariance matrix Ξ. Unfortunately, there is apparently no general algebraic solution to this maximization problem using an arbitrary, symmetric, dimensional weight matrix (2, 11, 16). Koschat and Swayne have proposed a solution for an arbitrary diagonal D × D weight matrix using a computationally intensive, iterative numerical algorithm assuming that Σ = I (16). However, the Koschat–Swayne algorithm is reported to have poor convergence properties and is sensitive to the seed values (16).
In our ML Procrustes treatment, the dimensional weight matrix is not arbitrary; rather, we weight specifically by the inverse of the dimensional covariance matrix (i.e., Ξ̂U−1). In this case, there is a much simpler, analytic solution to maximizing the estimated ML target for the optimal rotation. Here the dimensionally weighted Procrustes problem reduces conveniently (see Appendix III) to maximizing
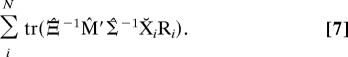 |
The rotations that maximize Eq. 7 can be found with the usual Procrustes singular value decomposition (SVD) solution (11). Let the SVD of an arbitrary matrix D be UΛV′. Then the maximizing rotations R̂i are given by
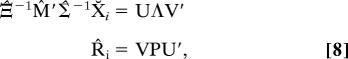 |
where rotoinversions can be avoided by constraining the determinant of R̂i to be 1 by using P = I if |V||U| = 1 or P = diag(1, …, 1, −1) if |V||U| = −1.
ML Estimation of the Scaling Factors
When superposing and comparing the shapes of different sized objects, scaling factors must be estimated to remove differences due to size alone. The unconstrained ML estimates of the scaling parameters βi are given by
However, unlike the rotations which are generally arbitrary with real data, it may be reasonable to model the scaling factors themselves as random variables drawn from a relevant probability distribution. For instance, the log-normal distribution arises as the limiting distribution of many small, multiplicative random effects, as may be expected for random deviations in size (17). Many biological entities subject to random variations in growth naturally fit a log-normal distribution. Assuming that βi ≈ LN (θ, θ), where
then the probabilities P(βi) = P(1/βi), and the mode of the distribution is 1 as required. The ML estimate β̂i for the lognormally distributed scaling factors is the positive root of (Appendix IV)
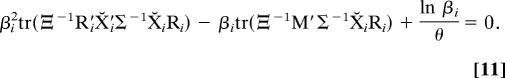 |
The (ln βi)/θ term in Eq. 11 can be thought of as a “penalty term” that concentrates the β̂ estimates around unity; otherwise it is identical to the nonparametric solution (Eq. 9).
ML Estimate of the Mean Form
Maximizing the likelihood with respect to the mean form M gives the familiar average (11)
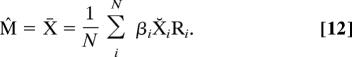 |
However, the average X̄ is an asymptotically biased estimate of the mean form M (it is too large) for constant K when superpositioning without scaling (i.e., setting βi = 1 for all i) (1, 7, 18). For the case when Ξ and Σ are diagonal (i.e., no correlations), we offer the following easily computable, bias-corrected ML Procrustes estimator M̃ of the mean (Appendix V)
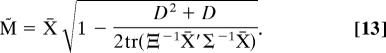 |
This bias-corrected estimate is consistent (for constant K as N → ∞) provided that the errors are not very large, specifically when tr (Ξ−1X̄′Σ−1X̄) ≥ (D2 + D)/2. For three-dimensional forms, for example, this estimator is valid when the landmark variances are less than K/6 times the squared radius of gyration of the average form.
An Iterative Algorithm for ML Procrustes Analysis
In practice, the ML solutions given above must be solved simultaneously via numerical methods, because each of the unknown parameters is a function of the others. We have developed the following iterative algorithm (11, 13, 19) for determining solutions to each of the Procrustes ML estimators.
Initialize. Set Σ̂ = I, Ξ∼ = I, and βi = 1 for all i; choose one of the Xi to serve as M̂.
Translate. Obtain each X̆i by centering Xi according to footnote †.
Rotate. Calculate each R̂i according to Eq. 8, and set each Xi = X̆iR̂i.
Scale. For lognormally distributed scale factors, first find the lognormal shape/location parameter θ with Eq. 19. Newton–Raphson can then be used to find the lognormal scale estimates with an initial guess equal to the arbitrary scale factor estimate Eq. 9 or to the scale factor from the previous iteration.
Estimate the mean. Find the new average M̂ according to Eq. 12 or 13. Return to step 3 and loop to convergence.
Estimate the inverse gamma distributed eigenvalues. Calculate Σ̂U from Eq. 3. The Σ̂U covariance matrix must be decomposed, and the positive eigenvalues fit iteratively to an inverse gamma distribution. The rank of the sample covariance matrix is min(ND − D − 3, K − 3). At convergence, the rank is reduced by two due to the collinearity induced by the linear constraints from the scale and shape parameters of the inverse gamma regularization (see Eq. 6). The rank is further reduced (by one) due to additional collinearity from centering the form matrices (6). Thus, the maximally obtainable number of positive eigenvalues in our hierarchical treatment is K − 3; this rank deficiency is inherent in the Procrustes sample covariance matrix and cannot be eliminated by increasing the number of forms. Thus, we treat all zero eigenvalues as missing data using an Expectation-Maximization algorithm and include only the full-rank, positive eigenvalues of the sample covariance matrix when estimating the parameters of the inverse gamma distribution. If the covariance matrix is assumed to be diagonal, one must include only the K − 3 largest variances in the inverse gamma fit, as the smallest three are known a priori to be a zero-valued eigenvalues. Estimation of the regularized eigenvalues thus cycles until convergence between two steps: (i) fit the current estimates of the full-rank eigenvalues to an inverse gamma distribution, and (ii) obtain updated estimates of the eigenvalues by modification of the sample eigenvalues analogous to Eq. 6. λIγ,j = (NDλU,j + 2α)/(2(1 + γ) + ND).
Estimate the landmark covariance matrix. Calculate Σ̂Iγ from Eq. 6 using the α̂ and γ̂ inverse gamma parameters determined in the previous step.
Estimate the dimensional covariance matrix. Calculate Ξ∼ from Eq. 5. Return to step 6 and loop to convergence.
Align the superposition with Ξ∼. Let the spectral decomposition of Ξ∼ be VΔV′. Rotate the entire superposition, including the average form M̂, with the matrix V′ of left eigenvectors (i.e. set M̂ = M̂V′) so that Ξ∼ is equivalent to the diagonal matrix Δ.
Loop. Return to step 2 until convergence (e.g., stop if the relative difference between rotation matrices between iterations fall below a specified tolerance, such as 10−7).
We have implemented this algorithm in THESEUS, a UNIX C program intended for likelihood analysis of biological macromolecules (25). Because of the complexity of the problem, the efficiency of the algorithm is difficult to determine generally. If Σ is assumed to be diagonal, then the algorithm is very efficient (O(NK)). In our experience, even with large three-dimensional problems, when assuming a diagonal landmark covariance matrix (e.g., K = 200 and N = 500), the algorithm converges rapidly after a few dozen iterations (from milliseconds to a few seconds on a 500 MHz G4 laptop). When using full, arbitrary covariance matrices on a low-dimensional problem, the algorithm is O(K3), as the limiting step is the inversion of the landmark covariance matrix Σ̂; in this case, the calculation may take a few minutes.
Comparison with Previous Analyses
Our likelihood analysis offers several ML Procrustes estimators, including the covariance matrices (Ξ∼ and Σ̂), a bias-corrected average form (M̃), lognormally distributed scaling factors (β̂i), and simplified estimates of the rotations (R̂i) when weighting the dimensions. Several of our solutions differ significantly from corresponding solutions to similar problems proposed previously in the Procrustes literature (1, 7, 8, 10, 11, 16). Unlike the unconstrained covariance estimators reported in Goodall and recounted by others (1, 7, 8, 11), each of our covariance estimators is itself a function of the other covariance matrix. Equations 10.2 and 10.3 of Goodall (11) are only valid when either Ξ or Σ is constrained to be equal to the identity matrix. Additionally, to find the optimal rotations when weighting with a factored dimensional covariance matrix, Goodall proposes a method using the Koschat–Swayne algorithm (16). Goodall's method aims to maximize numerically the same estimated likelihood target as our method, corresponding to the first term of Eq. 2. In contrast to the Goodall and Koschat–Swayne treatments, our analytic solution (Eq. 8) is shown to be valid for a nondiagonal dimensional weight matrix Ξ−1 and for Σ ≠ I. Finally, we present estimates of the scaling factors that differ from that reported in Goodall (10). When X̆iRi is identical to the mean form M, the scaling factor must necessarily equal unity, since no scaling is required when the mean form is superposed on itself. Unlike the scale factor estimator given in Goodall (equation 2.4), both of our ML estimators (Eqs. 9 and 11) conform to this logically necessary condition.
Several authors have reported that the Procrustes estimate of the mean form (corresponding to the estimated ML estimator, Eq. 12) is inconsistent (1, 7), being too large as the number of forms approaches infinity. The inconsistency of the average form is a result of ignoring the uncertainty introduced by using estimates of the rotations and translations in Eq. 12, rather than using their true values, which are unknown. However, we note that Eq. 23 implies that the average X̄ is in fact a consistent estimator of the mean M (assuming nonisotropic and uncorrelated errors) in the limit as both the numbers of landmarks (K) and forms (N) increase together, since then the bias factor approaches 1 in probability.
Performance of ML Procrustes Analysis with Simulated Data
We conducted simulations to investigate the ability of our ML Procrustes method to correctly infer known parameters of a family of forms. One common application of Procrustes methods in structural biology is superposing the atomic coordinates of multiple protein structures, each adopting a similar yet variable molecular conformation. To create an artificial data set, 400 perturbed protein structures were generated randomly, assuming a matrix Gaussian error distribution with a known mean form and known covariance matrices. At this stage of the simulation, the resulting family of perturbed structures consists of a known superposition, since the structures have not yet been subjected to arbitrary rotations and translations. As the final step in creating our test set of forms, we randomly translated and rotated each of the perturbed structures.
We then applied our ML Procrustes procedure to this simulated data set, obtaining estimates of the coordinates of the mean structure, the covariance matrices, and the original superposition. To illustrate the robustness of the ML method to violations of the assumptions, in this superposition analysis we assumed a diagonal landmark covariance matrix, even though the true covariance matrix contains large correlations. For comparison, we also performed a standard LS Procrustes analysis on the same data set. Results of a representative simulation are shown in Fig. 1.
Fig. 1.
ML and LS superposition of simulated protein structure data. (A) The “true” superposition, generated from a known mean structure with known covariance matrices, before arbitrary translations and rotations have been applied. In generating the simulated data, the set of alpha carbon atoms from model 1 of Protein Data Bank entry 2SDF (www.pdb.org) was used as the mean form (67 atoms/landmarks, squared radius of gyration = 152 Å2). The nondiagonal 67 × 67 landmark covariance matrix was based on values calculated from the superposition given in 2SDF, with variances ranging from 0.01 to 80 Å2 and correlations ranging from 0 to 0.99 (see Data Sets 1 and 2). The known dimensional covariance matrix had eigenvalues of 0.16667, 0.33333, and 0.5 corresponding to the x, y, and z axes of 2SDF model 1, respectively. (B) An ordinary LS superposition of the simulated data. (C) A ML superposition of the simulated data, assuming a diagonal landmark covariance matrix Σ (i.e., no correlations), Ξ = 1, and inverse gamma distributed variances.
Visual inspection clearly shows the remarkably close correspondence between the ML superposition and the known “true” superposition (Fig. 1). In contrast, the LS superposition is largely inaccurate, with a marked excess of positional variability in the regions of the structure with the least true variance. A residuals plot of the LS superpositions shows the tell-tale signs of extremely heteroscedastic data, indicating that the LS assumption of homoscedastic data has been violated (Fig. 2, which is published as supporting information on the PNAS web site). In contrast, a residuals plot of the ML superposition shows a very even spread for each landmark, as would be expected if the covariance matrices were estimated accurately. Similarly, probability plots of the residuals indicate large deviations from normality for the LS method, whereas ML recovers the true Gaussian distribution very well (Fig. 3, which is published as supporting information on the PNAS web site). A χ2 analysis comparing the inferred covariance matrices to the known covariance matrices also indicates the superior accuracy of ML vs LS superpositioning (reduced χ2 = 0.83, P = 1.0 vs. χ2 = 14,293, P = 0.0, respectively). Finally, both methods recovered the mean structure with good accuracy, although the ML method was slightly better than LS (0.074 vs 0.11 Å root mean squared error, respectively). Simulations with a large set of forms (400 here) should reflect the asymptotic behavior of the methods. However, qualitatively similar results are also obtained when superposing much smaller numbers of simulated structures (even as few as N = 2, see Data Sets 1 and 2, which are published as supporting information on the PNAS web site).
The Procrustes likelihood analysis described here should be of widespread applicability for quantitative comparison of shape. Unlike LS, our ML analysis results in approximately Gaussian distributed residuals, opening the possibility for the application of standard statistical analyses that rely on Gaussian errors for their validity. Additionally, because of the greatly improved estimation of the covariance matrices, it should be possible to accurately analyze major modes of covariation in a family of forms via principal components analysis (18).
Supplementary Material
Acknowledgments
We thank Meredith Betterton, Ian Dryden, Colin Goodall, Subhash Lele, and Norm Pace for helpful comments and suggestions. This work was supported by the Arnold and Mable Beckman Foundation and National Institutes of Health Grant GM59414. D.L.T. was supported by Postdoctoral Fellowship Grant PF-04-118-01-GMC from the American Cancer Society.
Abbreviations
- LS
least squares
- ML
maximum likelihood.
Appendices
In the following derivations, all log-likelihoods with respect to a given parameter are stated up to an arbitrary constant. Likelihoods are maximized by standard application of matrix derivatives (20), by finding the first derivative of the log-likelihood function with respect to the parameter of interest, setting it to zero, and solving for the parameter.
Appendix I: ML Estimates of the Factored Covariance Matrices.
We first define the error displacement matrix Ei as the matrix of differences from the mean M for each translated, scaled, and rotated form Xi
The log-likelihood for the row-wise covariance matrix Σ is
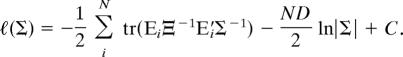 |
Taking the derivative with respect to Σ and setting it equal to zero gives
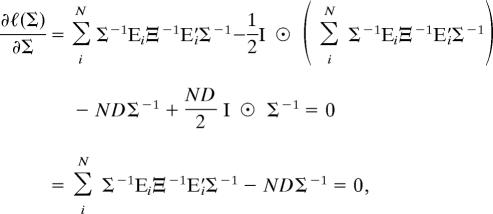 |
where A ⊙ B denotes the Hadamard (element-wise) product of two matrices A and B. Thus, after simplification and multiplication by Σ twice, we have Σ̂ = (1/ND) ΣiN EiΞ−1E′i, which is equivalent to Eq. 3. Derivation of the ML estimator of Ξ proceeds similarly.
Appendix II: Inverse Gamma Distributed Variances.
The probability distribution function for an inverse gamma distribution of the eigenvalues of the row-wise covariance matrix is
The corresponding log-likelihood of the eigenvalues is
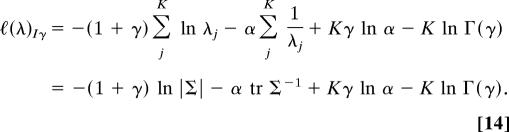 |
The unrestricted log-likelihood for Σ can be written as (compare Appendix I)
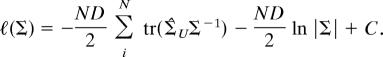 |
The Procrustes extended log-likelihood (5, 15) assuming inverse gamma distributed eigenvalues ℓ(Σ)Iγ is simply the sum ℓ(λ)Iγ + ℓ(Σ). Taking the derivative of ℓ(Σ)Iγ with respect to Σ and setting it equal to zero gives
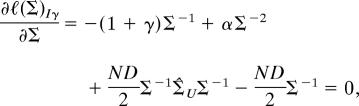 |
which, after pre- and postmultiplication by Σ−1 and rearrangement, gives Eq. 6.
Appendix III: ML Rotations with Dimensional Weighting by Ξ−1.
If all structures have been centered, the likelihood equations simplify. For centered form matrices, the log-likelihood with respect to the rotations is
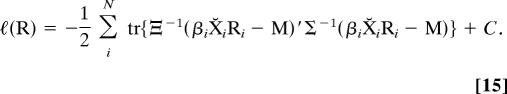 |
The nuisance parameters Ξ, Σ, and M are in general unknown, and as a consequence the likelihood given in Eq. 15 cannot be maximized directly. Therefore, as in Goodall (11), we use the method of estimated likelihood (5, 21, 22) to construct a modified target likelihood function by replacing these nuisance parameters with their ML estimates (Ξ̂, Σ̂, and M̂, respectively). To simplify the derivation here, we choose Σ to be the normalized member of the factored covariance matrices. After substitution with ML estimates, Eq. 15 can be expanded as the estimated likelihood ℓe(R) = ℓ(R, Ξ̂, Σ̂, M̂)
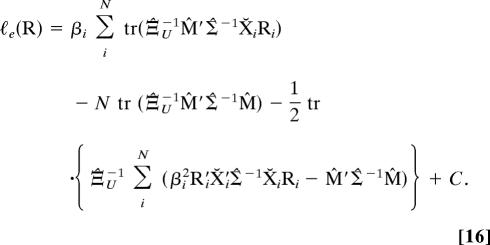 |
After substitution with Eq. 12 for the estimate of the mean form M̂, the ML estimate of the dimensional covariance matrix Ξ̂U can be expressed as
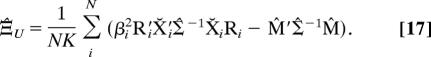 |
Substitution of Eq. 17 into Eq. 16 gives
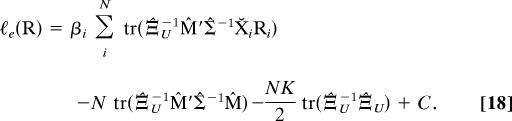 |
In the final term of Eq. 18 the two matrices cancel reducing to NKD/2. Hence, when maximizing the estimated likelihood ℓe(R) with respect to the rotations (holding other nuisance parameters fixed), both the last two terms of Eq. 18 are constant. Maximization of the first term alone (as given in Eq. 8) provides a ML estimate of the rotation Ri.
Appendix IV: ML Procrustes Estimate of Lognormally Distributed Scale Factors.
In the lognormal distribution given in Eq. 11, the shape and location parameter both equal θ to constrain the mode of the distribution to 1. Given a sample of N scale factors βi, the ML estimate of θ is
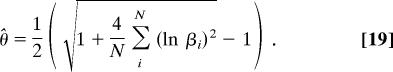 |
The log-likelihood for the scale factors β given a lognormal LN:(θ,θ) distribution is
The Procrustes extended log-likelihood ℓP,LN (5, 15) assuming lognormally distributed scale factors is then given by the sum of ℓ(βi)LN and ℓP (from Eq. 2). With respect to βi the joint log-likelihood is
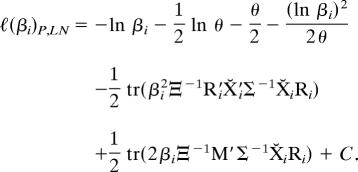 |
Taking the derivative of ℓ(β)P,LN with respect to βi, setting it equal to zero, and multiplying through by −βi yields Eq. 11.
Appendix V: Bias-Corrected ML Procrustes Estimate of the Mean Form.
In the following, we assume that Ξ and Σ are both diagonal, and that βi = 1 for all i. The sum of squared distances from the mean for each landmark (Gj) in a set of optimally superimposed forms is given by
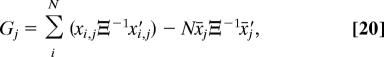 |
where xi,j is the jth 1 × D row vector in the optimally translated and rotated form matrix X̆iRi, such that X̆iRi = [x′i,1 …x′i,j]′. It can be shown from standard multivariate analysis that (12)
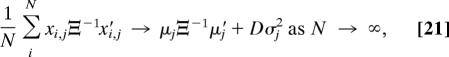 |
where similarly μj is the jth row vector in the mean form matrix M. Assuming that Gj is approximately χ2 distributed (1, 23, 24), then Gj ≈ σj2χFj2 with Fj degrees of freedom. For each of the K landmark row vectors in the N forms there are D degrees of freedom. However, for each of the N forms, D degrees of freedom are lost due to translational estimation, and ½ D(D − 1) degrees are lost due to rotational estimation (1, 24). Thus, by distributing the lost degrees of freedom over each of the K row vectors in proportion to the contribution of each landmark to the minimization criterion (wj = μjΞ−1μ′j/[σj2 tr(Ξ−1M′Σ−1M)]), we have
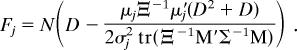 |
It then follows trivially that (17)
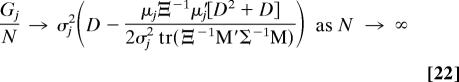 |
Combining Eqs. 20–22 yields the asymptotic result as N → ∞
The final factor on the right hand side of Eq. 23 is an estimate of the asymptotic bias of the average. Setting μj = Bx̄j (implying M = BX̄) and solving for B gives Eq. 13.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Row-wise weighted centering can be applied to an uncentered structure Xi by X̆i = Xi + 1Kt̂i, where t̂i is the ML estimate of the translation. The ML translation is independent of the rotations Ri, the scaling factors βi, and the dimensional covariance matrix Ξ, and it is given by −(1′KΣ−1Xi/1′KΣ−11K), where Xi is an uncentered structure (11).
References
- 1.Dryden IL, Mardia KV. Statistical Shape Analysis. New York: Wiley; 1998. [Google Scholar]
- 2.Gower JC, Dijksterhuis GB. Procrustes Problems. Vol 30. Oxford: Oxford Univ Press; 2004. [Google Scholar]
- 3.Lele S, Richtsmeier JT. An Invariant Approach to Statistical Analysis of Shapes. Boca Raton, FL: Chapman and Hall/CRC; 2001. [Google Scholar]
- 4.Mardia K, Goodall C. In: Multivariate Environmental Statistics. Patil G, Rao C, editors. Vol 6. New York: North-Holland; 1993. pp. 347–385. [Google Scholar]
- 5.Pawitan Y. In All Likelihood: Statistical Modeling and Inference Using Likelihood. Clarendon, Oxford: Oxford Science Publications; 2001. [Google Scholar]
- 6.Lele S, Richtsmeier JT. Syst Zool. 1990;39:60–69. [Google Scholar]
- 7.Lele S. Math Geol. 1993;25:573–602. [Google Scholar]
- 8.Glasbey C, Horgan G, Gibson G, Hitchcock D. Biometrical J. 1995;37:481–495. [Google Scholar]
- 9.Goodall C. J Roy Stat Soc B Met. 1991;53:334–339. [Google Scholar]
- 10.Goodall C. In: Mardia KV, Gill CA, editors. Proceedings in Current Issues in Statistical Shape Analysis; Leeds, UK: Leeds Univ Press; 1995. pp. 18–33. [Google Scholar]
- 11.Goodall C. J Roy Stat Soc B Met. 1991;53:285–321. [Google Scholar]
- 12.Arnold SF. The Theory of Linear Models and Multivariate Analysis. New York: Wiley; 1981. [Google Scholar]
- 13.Dutilleul P. J Stat Comput Sim. 1999;64:105–123. [Google Scholar]
- 14.Broemeling L. Bayesian Analysis of Linear Models. New York: Dekker; 1985. [Google Scholar]
- 15.Bjornstad JF. J Am Stat Assoc. 1996;91:791–806. [Google Scholar]
- 16.Koschat MA, Swayne DF. Psychometrika. 1991;56:229–239. [Google Scholar]
- 17.Evans M, Hastings N, Peacock JB. Statistical Distributions. 3rd Ed. New York: Wiley; 2000. [Google Scholar]
- 18.Kent JT, Mardia KV. J Roy Stat Soc B Met. 1997;59:281–290. [Google Scholar]
- 19.Dempster AP, Laird NM, Rubin DB. J Roy Stat Soc B Met. 1977;39:1–38. [Google Scholar]
- 20.Dwyer PS. J Am Stat Assoc. 1967;62:607–625. [Google Scholar]
- 21.Royall R. Statistical Evidence: A Likelihood Paradigm. Vol 71. New York: Chapman and Hall; 1991. [Google Scholar]
- 22.Royall R, Tsou T-S. J Am Stat Assoc. 1995;90:316–320. [Google Scholar]
- 23.Langron SP, Collins AJ. J Roy Stat Soc B Met. 1985;47:277–284. [Google Scholar]
- 24.Sibson R. J Roy Stat Soc B Met. 1979;41:217–229. [Google Scholar]
- 25.Theobald DL, Wuttke DS. Bioinformatics. 2006;22:2171–2172. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



