Abstract
The importance of iron to bacteria is shown by the presence of numerous iron-scavenging and transport systems and by many genes whose expression is tightly regulated by iron availability. We have taken a global approach to gene expression analysis of Salmonella enterica serovar Typhimurium in response to iron by combining efficient, high-throughput methods with sensitive, luminescent reporting of gene expression using a random promoter library. Real-time expression profiles of the library were generated under low- and high-iron conditions to identify iron-regulated promoters, including a number of previously identified genes. Our results indicate that approximately 7% of the genome may be regulated directly or indirectly by iron. Further analysis of these clones using a Fur titration assay revealed three separate classes of genes; two of these classes consist of Fur-regulated genes. A third class was Fur independent and included both negatively and positively iron-responsive genes. These may reflect new iron-dependent regulons. Iron-responsive genes included iron transporters, iron storage and mobility proteins, iron-containing proteins (redox proteins, oxidoreductases, and cytochromes), transcriptional regulators, and the energy transducer tonB. By identifying a wide variety of iron-responsive genes, we extend our understanding of the global effect of iron availability on gene expression in the bacterial cell.
Salmonella enterica serovar Typhimurium is a facultative, gram-negative intestinal pathogen that is a major cause of acute gastroenteritis worldwide. Salmonella encounters a range of environments and gene regulation is tightly controlled to adapt to the requirements of the bacterial cell, including changes in nutrient availability. Iron is an essential element, acting as a cofactor for numerous enzymes, and is involved in electron transport and redox reactions in the cell. The absolute requirement for iron is compounded by the ability of Fe3+ to generate free radicals that are capable of damaging the cell through the Fenton and Haber-Weiss reactions, resulting in oxidative stress (11). The Salmonella serovar Typhimurium genome contains a number of iron-responsive genes that allow for the uptake and storage of iron, with regulation mediated primarily through the ferric uptake regulator (Fur). Fur is a well-characterized transcriptional repressor that regulates gene expression in response to iron (18-20). Iron regulation in this bacterium has primarily focused on the identification and characterization of Fur-responsive elements, including 14 genes identified by a Fur titration assay (47) and a Salmonella-specific iron transport system that is required for virulence (23, 54). However, Salmonella contains a number of iron-containing proteins and iron-responsive elements (15, 30) that have not been identified in Fur-dependent screens and therefore may be regulated through other mechanisms. As pathogens encounter environments of both limiting and replete iron, it is important to characterize the response of the entire genome to iron availability. This will aid in a better understanding of the mechanisms of gene regulation and adaptation in the host.
The availability of complete bacterial genome sequences has permitted a shift in the focus of microbial gene expression studies from the analysis of a few transcriptional units to the examination of expression patterns on a whole-genome scale. The technological advances accompanying genomic studies have led to the development of a variety of approaches for gathering large-scale gene expression data. Several methods for analysis of differential gene expression exist, including microarrays, LuxArray, and differential fluorescence induction. Complications of microarray analysis include its reproducibility and standardization (9, 10, 21, 24, 27, 44). Differential fluorescence induction uses flow cytometry to select bacteria containing transcriptionally active regulatory regions cloned upstream of a promoterless green fluorescence protein (GFP) reporter gene (29, 43, 48-50); however, there are technical challenges associated with sorting bacteria. The LuxArray system of genome-wide transcriptional analysis uses specific bioluminescent reporter strains arrayed on solid-phase media (51). Only three published studies have examined the impact of iron on the global response of bacteria, specifically Pasteurella multocida (39), Mycobacterium tuberculosis (42), and Pseudomonas aeruginosa (37), all using DNA microarrays.
In this study, we present a high-throughput approach based on the construction of a random promoter library for differential gene expression profiling in Salmonella serovar Typhimurium in response to iron. This method uses a sensitive, luminescent reporter-detection system that readily monitors ranges in promoter activity and accurately reflects real-time changes in gene expression in response to iron. This global gene expression profile provides greater insight into the complex genetic regulation associated with iron availability.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The Salmonella serovar Typhimurium strain used in this study for library construction was ATCC 14028 (American Type Culture Collection, Manassas, Va.). Salmonella serovar Typhimurium and Escherichia coli strains were cultured aerobically at 37°C in Luria-Bertani (LB) medium (Invitrogen Canada Inc., Burlington, Ontario, Canada) or M9 minimal medium (Becton Dickinson Canada Inc., Mississauga, Ontario, Canada) supplemented with 0.1% Casamino Acids (Becton Dickinson Canada Inc.) and 0.5% glycerol. For analysis of the vector controls and flagellum promoter, RP437 (obtained from J. S. Parkinson), a chemotactic E. coli K-12 derivative, was grown aerobically at 30°C in tryptone broth (per liter: 10 g of tryptone, 8 g of NaCl). Kanamycin (KAN) was included in the culture media or agar plates at a concentration of 50 μg/ml as required. Iron-limiting and iron-rich conditions were created by the addition of 200 μM 2,2′-dipyridyl and 100 μM FeCl3, respectively, to LB medium (unless otherwise stated). E. coli donor strain 1808 (MC4100Δasd, recA::RP4-2Tc::Mu Km) was used for conjugations with the iron-responsive library subset. This strain was grown aerobically at 37°C in LB medium supplemented with 0.4% glucose and diaminopimelic acid (100 μg/ml). KAN was added at a concentration of 50 μg/ml, and tetracycline (TET) was added at a concentration of 15 or 30 μg/ml when appropriate.
Construction of the Salmonella serovar Typhimurium random promoter library.
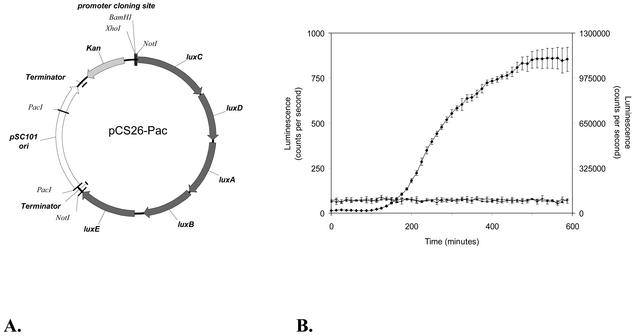
Genomic DNA was isolated (2) and partially digested with Sau3AI (Invitrogen Canada Inc.). Fragments of 1 to 2 kb were obtained by sucrose density gradient centrifugation (2) and ligated using T4 DNA ligase (Invitrogen Canada Inc.) into the BamHI-digested site of pCS26-Pac (Fig. 1A). All transformations were carried out according to standard procedures (2) and occurred by electroporation using a Gene Pulser (Bio-Rad Laboratories Inc., Hercules, Calif.). To increase efficiency of library construction, ligation products were first introduced into ElectroMAX DH10B cells (Invitrogen Canada Inc). Each electroporation yielded six pools of 150 μl, and each pool consisted of ∼8,000 transformants. Plasmid DNA was prepared from each pool and introduced into Salmonella serovar Typhimurium TN2540 (hsd r− m+). Pools of plasmid DNA were prepared and moved into Salmonella serovar Typhimurium 14028. Transformants were picked and transferred to LB medium in black 384-well microtiter plates (3710 Costar; Corning Incorporated, Corning, N.Y.) using a colony-picking robot (Norgren Systems, Palo Alto, Calif.) and incubated at 37°C for 6 h. The clones were assayed for luminescence in a Wallac Victor2 1420 multilabel counter (Perkin-Elmer Life Sciences, Boston, Mass.) and transferred into minimal medium by using a 384-pin manual plate replicator (catalog no. VP 386; V&P Scientific, San Diego, Calif.). Following incubation at 37°C, both sets of plates were assayed for promoter activity at 20 h. Clones exhibiting promoter activity were rearrayed (using Norgren Systems software) and grown in LB medium in 384-well plates. These selected clones exhibiting promoter activity represent the random promoter library.
FIG. 1.
(A) The reporter plasmid pCS26-Pac. The salient features of this pZS derivative (28) include low copy number, strong transcriptional terminators, unique promoter cloning sites, and luxCDABE of Photorhabdus luminescens (33). pCS26-Pac was developed by introducing PacI sites adjacent to the pSC101 origin of replication to facilitate transfer of the reporter cassette to other vectors. (B) The mean luminescence activity in counts per second was measured in triplicate for all strains grown in tryptone broth. The growth curves were within 5%, and no significant differences in growth were observed between these strains and the parent strain (data not shown). RP437/pCGNot21 (open square) contains the vector lacking the lux reporter, RP437/pCS26 (closed triangle) contains a promoterless vector, and RP437/pCS16 (closed diamond) contains the flgB promoter in pCS26. RP437/pCS16 luminescence is plotted on the secondary y axis.
Library screening method.
To screen for genes responsive to iron, the random promoter library was cultured overnight in low-iron medium supplemented with KAN. The library was then inoculated into the appropriate medium for the screen (low or high iron) with a 384-pin plate replicator. Inoculated plates were incubated at 37°C and luminescence readings were measured in the Wallac Victor2 1420 multilabel counter. When performing the initial screens for iron-responsive clones readings were taken at 6 and 20 h. A subset of clones with differential expression of threefold or more were identified and rearrayed as a separate promoter set for further study. To determine and characterize genuine versus false iron-responsive promoters, additional screens with this subset were performed. When rescreening smaller subsets, the overnight cultures were diluted 1:300 in the appropriate media in 96-well clear-bottom black plates (9520 Costar; Corning Incorporated) and were assayed for both luminescence and absorbance over the desired time course. For this study, raw luminescence data was not normalized, to allow comparison of absolute expression levels between different clones.
DNA sequencing and sequence analysis.
Iron-responsive promoters were PCR amplified using the pZE.05 (5′CCAGCTGGCAATTCCGA-3′) and pZE.06 (5′AATCATCACTTTCGGGAA-3′) primers, flanking the BamHI site of pCS26-Pac. PCR products were sequenced (QIAGEN Genomics Inc. Sequencing Services, Bothell, Wash.) using pZE.06 and the DNA sequences obtained were compared with the GenBank database by using the NCBI standard nucleotide-nucleotide BLAST program blastN and further analyzed using Vector NTI (InforMax).
Fur regulation.
For examination of Fur regulation of the iron-responsive clones, we used a variation of the Fur titration experiment (45) utilizing a high- copy conjugal plasmid containing a Fur binding site. The E. coli MG1655 fepA promoter (∼612,091 to 611,671 bp) (5) was cloned into the conjugal donor plasmid pEX18Tc (22) and this construct was moved into E. coli conjugal donor strain 1808 by electroporation. The donor was grown overnight and the cells were harvested by centrifugation in a Beckman JA-20 rotor in a Beckman J2-21 centrifuge for 5 min at 5,000 rpm at 4°C. Cells were washed twice and resuspended in LB medium at 1/30 the original culture volume. The recipient cells (the iron-responsive clones) were grown from frozen stock cultures overnight. LB agar plates containing KAN (75 μg/ml) and TET (30 μg/ml) were heavily inoculated with the donor strain and the recipients were stamped onto this. Plates were incubated at 37°C for 2 days. Conjugants were stamped into fresh liquid LB medium containing KAN (75 μg/ml) and TET (30 μg/ml) and grown overnight. Screens were performed as described above. Time points for assay were 2, 4, 6, 8, and 24 h. A second assay, including time points at 14, 16, and 18 h, was done to ensure that delays in promoter expression were not being overlooked due to slow growth of the conjugants.
Expression data analysis.
Promoter expression data were analyzed with Cluster Software (12) hierarchical clustering. The expression data was adjusted in Cluster first by mean centering each gene and then by normalizing the expression magnitudes of the genes. Unweighted hierarchical clustering using uncentered correlation as a similarity measure and average linkage clustering was performed. Cluster analysis was visualized with Treeview software (12).
RESULTS
Optimizing library construction.
Prior to construction of an entire genomic library, the methods were optimized with a partial E. coli MG1655 random promoter library. The method consisted of ligating Sau3A-digested DNA fragments into the BamHI site of pCS26-Pac (Fig. 1A) and transforming the ligation products into the appropriate strain background. An important feature of this vector is the luxCDABE reporter (33) that does not require the addition of substrate or further manipulations to produce light from an active promoter. The absence of endogenous promoter activity from the construct, which has strong transcriptional terminators, ensures that the strain containing the vector is nonluminescent (Fig. 1B), allowing promoters even with very low basal activity to be detected. Positive promoter clones were identified by light production (as a measure of their expression levels) under different assay conditions and compiled to form the library. Here the term promoter refers to the regulatory region(s) that controls gene expression in addition to the RNA polymerase binding site.
This strategy has the limitation that only the promoters that are active under the assay conditions used for library construction will be included in the final promoter library. To investigate this, we examined the MG1655 library for activity under a variety of conditions, including LB medium, M9 minimal medium supplemented with glycerol, LB medium supplemented with various carbon sources, LB medium under anaerobic conditions, LB medium at pH 5.5, and LB medium with 300 mM NaCl (data not shown). Surprisingly, the two assay media LB and M9-glycerol enabled nearly comprehensive promoter clone identification (96% of the positives identified under all conditions). Of the single conditions, only anaerobic screening contributed significantly to the data set (∼3% of total positives).
Construction of the Salmonella serovar Typhimurium random promoter library.
Construction of the random promoter library involved generating 111,360 random clones and screening them for light production in LB and minimal media at two different time points. Statistics associated with library construction are summarized in Table 1. Promoters displaying expression levels greater than or equal to three times the median of assayed activity from all random clones were included in the library, with a final total of 6,528 clones. With estimations of one promoter for every two kilobases of DNA and a genome size for Salmonella serovar Typhimurium of 4,857 kb (32), a library this size represents approximately 2.7-fold coverage of predicted promoter regions. Analysis of sequenced clones demonstrated the random distribution of the fragments throughout the genome (data not shown).
TABLE 1.
Statistics and result summary for the generation of the Salmonella serovar Typhimurium 14028 random promoter library
| Characteristic | Result for Salmonella serovar Typhimurium |
|---|---|
| Genome size (kb) | 4,857 |
| No. of clones screened | 111,360 |
| No. of predicted promoters | 2,430 |
| Size of library | 6,528 |
| Estimated coveragea (fold) | 2.7 |
| No. of positives (initial screen) with differential expression of >3-fold | 628 |
| No. (%) of false positives | 148 (23.6) |
| No. (%) of positives (rescreen) with differential expression of >3-fold | |
| High iron | 182 (37.9) |
| Low iron | 298 (62.1) |
Coverage with respect to predicted promoters.
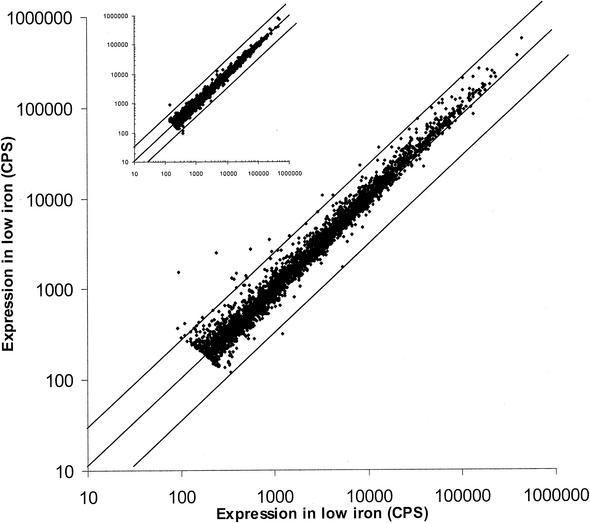
For screening the library, a simple protocol was followed that requires minimal automation. Multiple cultures were inoculated into selected media in 384-well plates using a 384-pin replicator. This approach allows the screens to be set up rapidly and without the requirement for automated pipetting. To determine if this approach is consistent and reproducible, the library was screened under duplicate conditions. The results for each condition were plotted against each other and visualized in a scatter plot (Fig. 2). The expression levels lie on the diagonal with a slope of one, indicating that the growth of the replica cultures is the same in duplicate wells and that manual stamping is a reliable means for subculturing a library under screen conditions. Importantly, the reproducibility extends to promoters with moderate to low activity. The small degree of variability observed (i.e., points off the diagonal) can most likely be attributed to minor growth effects, as differences in growth were not accounted for in this assay; variability decreases as the time course proceeds (Fig. 2, inset).
FIG. 2.
Scatter plot analysis of duplicate screens under low-iron conditions. Luminescence readings were taken at 6 and 20 h (inset). The lines demarcate the threefold differences in expression level. The linear trend for the duplicate data points indicates that plate-to-plate variability is minimal. Points falling outside the threefold range (3% of data points) can most likely be attributed to growth effects arising from variation in inoculum. Promoter clones with expression levels below or equal to background were not included in the data set.
Screening the library with iron.
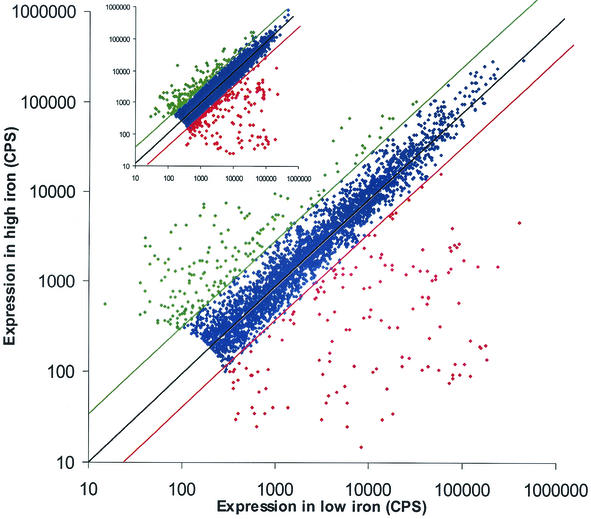
The random promoter library was screened for differential responses to replete and limiting iron conditions (Fig. 3). The closeness of the fit of the expression data to the diagonal linear trend line indicates the level of differential activity of a clone under the two assay conditions. Promoters with differences in expression of threefold or more are indicated in color. Differences in expression levels of >1,000-fold were observed between the two conditions for some of the iron-responsive genes.
FIG. 3.
Scatter plot analysis of library screens under low- versus high-iron conditions. Luminescence readings were taken at 6 and 20 h (inset). The colored data points represent promoter clones exhibiting >3-fold differences in expression between the two conditions; red represents promoters active under low-iron conditions, and green represents promoters active under high-iron conditions. Promoter clones with expression levels below or equal to background were not included in the data set.
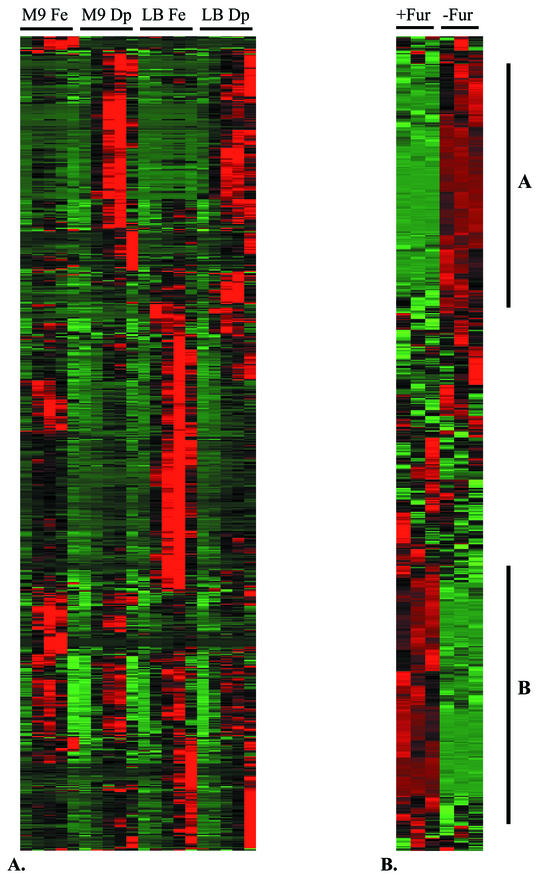
Rescreening 628 putative iron-regulated promoter clones over an 8-h time course (Fig. 4A) resulted in the identification of false positives (n = 148) and 480 differentially expressed clones (Table 1). Real-time expression profiles for promoters, under a variety of conditions, were generated concurrently with measurements of bacterial growth. The severalfold induction levels for all promoters in response to iron are listed in Table 2. Expression data generated over a 24-h time course in minimal and LB media with high and low iron levels are presented in Fig. 4A. We utilized hierarchical clustering of the promoter expression data with the software Cluster (12). Clustering the temporal profiles from various medium conditions allows for the refined definition of coregulated genes, showing numerous clusters responding to different combinations of medium conditions in high or low iron.
FIG. 4.
Hierarchical clustering of iron-responsive expression data. Expression levels above the mean center for each gene are indicated by red, below the mean center by green, and around the mean center in black. Clones are clustered on the y axis and the time course is shown on the x axis. (A) Temporal expression data for promoter clones in supplemented M9 minimal (M9) or LB (LB) media containing 200 μM FeCl3 (Fe) or 200 μM 2,2′-dipyridyl (Dp). Luminescence was measured at 2, 4, 6, 8, and 24 h. Clustering illustrates differential regulation of promoter sets in response to various medium conditions. (B) +Fur, library strains containing pEX18Tc. −Fur, library strains containing fepA-pEX18Tc. Each data set is shown as a time course through 14, 16, and 18 h of the growth curve in high-iron medium (400 μM FeCl3). Highlighted clusters refer to promoters with increased expression (class A) or decreased expression (class B) when Fur availability is reduced. The remaining promoters (class C) appear to be regulated independently of Fur (not highlighted).
TABLE 2.
Classification of annotated iron-responsive promoters with respect to Fur regulation
| Classe | Gene | Maximum fold inductiona | Function(s) | Reference(s) |
|---|---|---|---|---|
| A | iroBc,d | −8,900 | Putative glycosyl transferase, related to UDP-glucuronosyltransferase/putative ATP binding cassette (ABC) transporter | 3, 4 |
| entCc,d | −1,805 | Isochorismate synthetase, enterochelin biosynthesis | 6, 7, 26 | |
| cirAc | −620 | Outer membrane porin, receptor for colicin I, requires TonB | 17, 18, 20, 53 | |
| fepAc,d | −569b | Outer membrane porin, receptor for ferric enterobactin (enterochelin) and colicins B and D | 47 | |
| ydiEd | −353 | Putative hemin uptake factor | 38 | |
| fhuFc | −241 | Ferric hydroxamate transport, involved in reduction of ferric iron in cytoplasmic ferrioxamine B | 19, 35, 36 | |
| sitAc,d | −142b | Salmonella iron transporter | 54 | |
| yqjA | −86 | Putative DedA family, membrane protein | 32 | |
| bfd | −81b | Regulatory or redox component complexing with Bfr in iron storage and mobility | 32, 37 | |
| yajHd | −58b | Putative iron chelator utilization protein | 38 | |
| STM0363d | −55b | Putative transcription regulator; AraC family | 32 | |
| STM4305 | −52b | Putative anaerobic dimethyl sulfoxide reductase, subunit A | 32 | |
| STM1586 | −46 | Putative periplasmic protein | 32 | |
| tonBc | −22 | Energy transducer: uptake of iron, cyanocobalamin: sensitivity to phages, colicins | 20, 40 | |
| fhuAc,d | −21 | Outer membrane protein receptor or transporter for ferrichrome, colicin M, and phages T1, T5, and phi80 | 20, 47 | |
| clpB | −20b | ATP-dependent protease, Hsp 100, part of novel multichaperone system with DnaK, DnaJ, and GrpE | 32 | |
| copA | −12 | Cu(I)-translocating P-type ATPase | 32 | |
| yafA | −11b | Putative hemolysin | 32 | |
| prsA | −2b | Phosphoribosylpyrophosphate synthetase | 32 | |
| holC | 3 | DNA polymerase [III], chi subunit | 32 | |
| invH | 6 | Invasion protein | 32 | |
| ytfK | 6b | Putative cytoplasmic protein | 32 | |
| rfaL | 7 | O-antigen ligase | 32 | |
| narKd | 17 | MFS superfamily, nitrite extrusion protein | 32 | |
| fdnG | 26b | Putative molybdopterin oxidoreductase | 32 | |
| rfaJ | 28 | UDP-d-glucose:(galactosyl)lipopolysaccharide glucosyltransferase | 32 | |
| STM3820 | 29b | Putative cytochrome c peroxidase | 32 | |
| cobD | 36 | Putative aminotransferase in cobalamin synthesis | 32 | |
| pduA | 274b | Propanediol utilization: polyhedral bodies | 32 | |
| pduQ | 363b | Propanediol utilization: propanol dehydrogenase | 32 | |
| B | adhE | −42b | Iron-dependent alcohol dehydrogenase of the multifunctional alcohol dehydrogenase AdhE | 32 |
| treF | −15 | Cytoplasmic trehalase | 32 | |
| xapAd | −13 | Xanthosine phosphorylase (purine nucleoside phosphorylase) | 32 | |
| yhgl | −8b | Putative thioredoxin-like proteins and domain | 32 | |
| STM1554 | −4b | Putative coiled-coil protein | 32 | |
| ycfC | −2b | Membrane-associated protein of unknown function | 32 | |
| fimZ | 4 | Fimbrial protein Z, putative transcriptional regulator (LuxR/UhpA family) | 32 | |
| pykF | 4 | Pyruvate kinase 1 (formerly F), fructose stimulated | 32 | |
| STM1330 | 4 | Putative DNA/RNA nonspecific endonuclease | 32 | |
| yhcGd | 4b | Putative cytoplasmic protein | 32 | |
| fumAc | 5b | Fumarase A (fumarate hydratase class I), aerobic isozyme | 30 | |
| STM1239 | 5 | Putative cytoplasmic protein | 32 | |
| udhA | 5 | Soluble pyridine nucleotide transhydrogenase | 32 | |
| sopA | 6 | Secreted effector protein of Salmonella serovar Dublin | 32 | |
| yhfK | 9 | Putative inner membrane protein | 32 | |
| ysaA | 9 | Paral putative oxidoreductase | 32 | |
| sicA | 10 | Surface presentation of antigens: secretory proteins | 32 | |
| STM3595 | 10 | Putative phosphatase | 32 | |
| rhaA | 16b | l-Rhamnose isomerase | 32 | |
| moaAd | 19 | Molybdopterin biosynthesis, protein A | 32 | |
| ftnB | 21 | Ferritin-like protein | 32 | |
| pepT | 24 | Putative peptidase T (aminotripeptidase) | ||
| STM0989 | 24 | MukF protein (killing factor KicB) | 32 | |
| frdAd | 28 | Fumarate reductase, anaerobic, flavoprotein subunit | 32 | |
| glpA | 30b | sn-glycerol-3-phosphate dehydrogenase (anaerobic), large subunit | 32 | |
| STM4510 | 59b | Putative aspartate racemase | 32 | |
| dmsAd | 88 | Anaerobic dimethyl sulfoxide reductase, subunit A | 32 | |
| yqhC | 104 | Putative transcriptional regulator (AraC/XylS family) | 32 | |
| C | yfcZ | −18 | Putative cytoplasmic protein | 32 |
| STM1851 | −14b | Putative cytoplasmic protein | 32 | |
| yhcO | −10 | Putative cytoplasmic protein | 32 | |
| STM4448 | −8 | Putative periplasmic protein/putative phosphotransferase system mannitol/fructose-specific IIA domain (Ntr type) | 32 | |
| yciG | −6 | Putative cytoplasmic protein | 32 | |
| ydhI | −4 | Putative inner membrane protein | 32 | |
| nagB | −3 | Glucosamine-6-phosphate deaminase | 32 | |
| narW | −3 | Nitrate reductase 2, delta subunit, assembly function | 32 | |
| STM1537 | 4b | Putative Ni/Fe-hydrogenase 1 b-type cytochrome subunit | 32 | |
| STM4067 | 4 | Putative ADP-ribosylglycohydrolase | 32 | |
| cybC | 5b | Cytochrome b (562) | 32 | |
| yeaA | 6b | Putative domain frequently associated with peptide methionine sulfoxide reductase | 32 | |
| nirB | 7b | Nitrite reductase, large subunit | 32 | |
| yfbE | 8 | LexA regulated, putative SOS response | 32 | |
| sifB | 9 | Salmonella translocated effector: translocated by SP1-2 | 32 | |
| yjdE | 10 | Putative APC family, putrescine/ornithine transport protein, cryptic | 32 | |
| yliG | 10 | Putative Fe-S oxidoreductase family 1 | 32 | |
| ais | 12 | Aluminum-inducible protein | 32 | |
| yfiR | 14 | Putative periplasmic protein | 32 | |
| aspAd | 15b | Aspartate ammonia-lyase (aspartase) | 32 | |
| STM1255 | 15b | Putative ABC transporter periplasmic binding protein | 32 | |
| fucR | 22b | Positive regulator of the fuc operon (DeoR family) | 32 | |
| STM3138d | 23 | Putative methyl-accepting chemotaxis protein | 32 | |
| araE | 28 | MFS family, l-arabinose: proton symport protein (low-affinity transporter) | 32 | |
| eutS | 28 | Putative carboxysome structural protein, ethanol utilization | 32 | |
| yhbU | 29 | Putative protease | 32 | |
| STM4519 | 41b | Putative NAD-dependent aldehyde dehydrogenase | 32 | |
| mgsA | 78b | Methylglyoxal synthase | 32 |
Indicates approximate maximum induction in LB medium between high-iron (positive numbers) and low-iron (negative numbers) conditions ± 1.
Promoter maximally expressed in supplemented M9 minimal medium.
Promoters known to be Fur regulated.
Promoter represented more than once.
Class A represents promoters that are negatively regulated by Fur. Class B represents promoters that are positively regulated by Fur. Class C represents promoters that appear to be regulated independently of Fur.
False positives represent promoters that may have been selected as iron responsive initially due to a variety of factors (small growth effects, luminescence cross talk in the multiwell plate) but in fact are not shown as iron responsive once a more detailed screen is performed. Fifty-one clones that were not affected by iron were included as negative controls and their expression falls within the threefold expression difference used as the threshold for regulated clone selection (data not shown).
Analysis of Fur dependence.
The negative transcriptional regulator Fur is responsible for the regulation of a number of iron-responsive genes in Salmonella (4, 15, 47). In order to characterize the iron-responsive clones with respect to Fur regulation, a variation of the Fur titration assay (45, 47) was carried out and expression data were clustered as shown in Fig. 4B. Three groups were identified, consisting of promoters with increased expression when Fur was titrated (class A) and decreased expression when Fur was titrated (class B) and promoters that appeared to be regulated independently of Fur (class C).
Characterization and sequence analysis of selected clones.
Nucleotide sequence data were produced for 125 of 480 iron-responsive clones (Table 2). We identified 12 promoters for genes that have previously been shown or postulated to respond to iron. In addition, many other genes of known (47%; 35 of 74) or unknown (53%; 39 of 74) function not previously identified as iron responsive were shown to be regulated by iron in this study. We estimate 2.7-fold coverage of the genome and we found that 16 promoters were represented more than once, with 5 promoters represented more than twice. For example, the iron-regulated promoter fhuA is represented by two nonidentical clones that have overlapping sequences but different 5′ and 3′ termini and possess identical expression profiles (data not shown).
We have identified sixteen fragments containing active promoters that, based on their chromosomal location, were not anticipated. These orphan promoters are located in positions that would not drive the expression of annotated open reading frames (data not shown).
DISCUSSION
The procedure for cloning random DNA fragments into promoterless vectors to identify promoters displaying condition-dependent gene regulation is a well-established method (29, 32, 43, 48-50). We adapted this approach and incorporated the luxCDABE reporter to develop sensitive methods amenable to real-time resolution of gene expression. The result is that the construction and screening of random promoter libraries is highly applicable to global gene expression studies.
We demonstrate the efficacy of this approach with the high-throughput expression profiling of a Salmonella serovar Typhimurium random promoter library in response to iron. One-quarter of iron-responsive promoter clones were sequenced, and as expected for a library with nearly threefold coverage, 16 promoters were represented more than once. Ten previously characterized iron-regulated genes were identified, including iroB, fhuA, fhuF, entC, fepA, cirA, sitA, tonB, bfd, and fumA. In addition, we observed iron regulation of genes coding for iron storage and mobility proteins and iron-containing proteins. We also identified putative transcriptional regulators, cytoplasmic, periplasmic and membrane-associated proteins, transporters, a protease, and oxidoreductases. By generating a global iron response profile, it may be possible to identify uncharacterized iron transport and utilization systems, as well as iron-containing proteins that may exist in Salmonella.
We observed the expression of several virulence genes in response to iron. The fragment containing the upstream regulatory region for sitA, a component of the Salmonella pathogenicity island 1-encoded iron uptake system (54), was identified as an iron-responsive promoter in this screen in addition to several other virulence-associated genes (fimZ, invH, sicA, sifB, and sopA). While expression of the sitA transporter is induced in low iron, the remaining genes were optimally expressed in high iron. The ability to respond to changes in iron levels is important for the establishment of pathogens (41). Traditionally, the host has been portrayed as a low-iron environment (14, 41). In contrast, recent work suggests that bacteria experience gradients of iron in the gastrointestinal tract between the lumen and epithelial cell surface (8). Moreover, the Salmonella-containing vacuole in the macrophage is not iron restricted (13). The recognition that iron levels are variable in the host indicates that bacterial gene regulation in response to iron through the course of an infection may be more complex than previously thought.
Fur is a well-established negative transcriptional regulator that responds to iron availability and class A expression profiles are consistent with traditional Fur regulation. Several gene products of sequenced promoters are involved in Fur-regulated iron scavenging systems. Interestingly, two promoters, ydiE (putative hemin uptake factor) and yqjH (putative iron chelator utilization protein), predicted to be Fur regulated using a comparative genomic approach (38), were also identified as such in this screen.
Our screens have established bfd, a bacterioferritin-associated ferredoxin, as a class A gene. bfd was also shown to be iron responsive in P. aeruginosa, and a candidate Fur box was identified (37). Fur regulation of bfd suggests that when iron is low in the cell, bfd would be derepressed and its product would function to mobilize iron from bacterial storage proteins. In contrast, bacterioferritin (Bfr), an iron storage protein and the second gene in an operon with bfd (16, 30), is up-regulated in a Fur-dependent manner in high iron through regulation by the small RNA RyhB (30). Production of RyhB is repressed by Fur in the presence of iron, leading indirectly to positive Fur regulation in the cell. When iron is low, Bfr levels would be down-regulated by RyhB, halting the production of iron storage proteins in the absence of available iron. Under high-iron conditions, significant basal expression of bfd (data not shown) was observed, suggesting incomplete repression of the operon by Fur and, subsequently, expression of bfr. The differential expression of this operon is an example of the complex regulation of Salmonella serovar Typhimurium in response to iron.
Where class A genes are negatively regulated by Fur in the classical manner, class B genes appear to be positively regulated by Fur. Small regulatory RNA molecules such as RyhB provide a mechanism for positive Fur regulation; it is likely that RyhB, dependent on the Hfq RNA-binding protein for function, regulates the target gene by terminating transcription postinitiation (30, 31, 34). Many of our random clones contain not only the promoter regions but also a significant portion of the gene and may include the target sites for small RNAs. In this study, ftnB and fumA were positively regulated by Fur under high-iron conditions. Ferritin (ftnA) from E. coli was shown to be regulated through RyhB, suggesting that ftnB may be similarly regulated. fumA was also identified as a RyhB-regulated gene (1, 30). There are several additional class B genes encoding iron-containing or iron-dependent proteins, including dmsA, frdA, cybC, fdnG, and adhE, which may be similarly regulated. These results illustrate that this approach may be suitable for measuring posttranscriptional effects at the level of RNA stability. Alternative mechanisms for regulation in this class may involve other small RNA-dependent mechanisms (52), Fur regulation of other regulators, and indirect induction of oxidative stress in the cell (46). Remaining promoters in class C may be subject to more complex or new mechanisms of iron regulation and are currently under investigation. These classes are the result of examining global effects of iron changes in the environment that may act directly (Fur) or indirectly (oxidative stress) on gene expression.
Some of the difficulties encountered with other methods of whole-genome expression profiling are overcome with this random promoter library approach. The reporter-detection system readily monitors low promoter activity to allow for a more thorough evaluation of genes being expressed and accurately reflects real-time changes in gene expression. One complication of this random approach is the nonspecific cloning of DNA with promoter activity that may be a product of several regulatory effects. However, a significant advantage is the potential discovery of unannotated promoters and regulatory regions. There is also some likelihood that promoters may be missed, yet the redundancy of sequenced clones that include the same promoter is indicative of good genome-wide coverage. The screening method is versatile and a wide range of conditions can be assayed in liquid, biofilm, solid medium, and mixed-community assays.
Temporal expression profiling that is readily achieved using promoter reporter fusions can reveal subtleties in gene regulatory networks that may be difficult to resolve using other approaches (25), allowing the inference of internal physiological states. This strategy for transcriptional analysis has the potential to generate quantitative high-resolution global expression patterns. Such information along with proteome and metabolome data will ultimately lead to the development of predictive models of gene regulation and microbial physiology.
Acknowledgments
J. Bjarnason and C. M. Southward contributed equally to this work.
We thank Tony Schryvers for helpful comments on the manuscript and members of the Surette laboratory for insightful discussions.
This work was supported by the Canadian Institutes of Health Research.
REFERENCES
- 1.Abdul-Tehrani, H., A. J. Hudson, Y. S. Chang, A. R. Timms, C. Hawkins, J. M. Williams, P. M. Harrison, J. R. Guest, and S. C. Andrews. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. Greene Publishing Associates, New York, N.Y.
- 3.Baumler, A. J., F. Heffron, and R. Reissbrodt. 1997. Rapid detection of Salmonella enterica with primers specific for iroB. J. Clin. Microbiol. 35:1224-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumler, A. J., R. M. Tsolis, A. W. van der Velden, I. Stojiljkovic, S. Anic, and F. Heffron. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183:207-213. [DOI] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Brickman, T. J., B. A. Ozenberger, and M. A. McIntosh. 1990. Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J. Mol. Biol. 212:669-682. [DOI] [PubMed] [Google Scholar]
- 7.Buss, K., R. Muller, C. Dahm, N. Gaitatzis, E. Skrzypczak-Pietraszek, S. Lohmann, M. Gassen, and E. Leistner. 2001. Clustering of isochorismate synthase genes menF and entC and channeling of isochorismate in Escherichia coli. Biochim. Biophys. Acta 1522:151-157. [DOI] [PubMed] [Google Scholar]
- 8.Conrad, M. E., and J. N. Umbreit. 2002. Pathways of iron absorption. Blood Cells Mol. Dis. 29:336-355. [DOI] [PubMed] [Google Scholar]
- 9.Conway, T., and G. K. Schoolnik. 2003. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol. Microbiol. 47:879-889. [DOI] [PubMed] [Google Scholar]
- 10.Diehn, M., and D. A. Relman. 2001. Comparing functional genomic datasets: lessons from DNA microarray analyses of host-pathogen interactions. Curr. Opin. Microbiol. 4:95-101. [DOI] [PubMed] [Google Scholar]
- 11.Earhart, C. F. 1996. Uptake and metabolism of iron and molybdenum, p. 1075-1090. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 12.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 14.Faraldo-Gomez, J. D., and M. S. Sansom. 2003. Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 4:105-116. [DOI] [PubMed] [Google Scholar]
- 15.Foster, J. W., and H. K. Hall. 1992. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J. Bacteriol. 174:4317-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garg, R. P., C. J. Vargo, X. Cui, and D. M. Kurtz, Jr. 1996. A [2Fe-2S] protein encoded by an open reading frame upstream of the Escherichia coli bacterioferritin gene. Biochemistry 35:6297-6301. [DOI] [PubMed] [Google Scholar]
- 17.Griggs, D. W., B. B. Tharp, and J. Konisky. 1987. Cloning and promoter identification of the iron-regulated cir gene of Escherichia coli. J. Bacteriol. 169:5343-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hantke, K. 1984. Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol. Gen. Genet. 197:337-341. [DOI] [PubMed] [Google Scholar]
- 19.Hantke, K. 2002. Members of the Fur protein family regulate iron and zinc transport in E. coli and characteristics of the Fur-regulated fhuF protein. J. Mol. Microbiol. Biotechnol. 4:217-222. [PubMed] [Google Scholar]
- 20.Hantke, K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182:288-292. [DOI] [PubMed] [Google Scholar]
- 21.Heller, M. J. 2002. DNA microarray technology: devices, systems, and applications. Annu. Rev. Biomed. Eng. 4:129-153. [DOI] [PubMed] [Google Scholar]
- 22.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 23.Janakiraman, A., and J. M. Slauch. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35:1146-1155. [DOI] [PubMed] [Google Scholar]
- 24.Jenssen, T. K., M. Langaas, W. P. Kuo, B. Smith-Sorensen, O. Myklebost, and E. Hovig. 2002. Analysis of repeatability in spotted cDNA microarrays. Nucleic Acids Res. 30:3235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalir, S., J. McClure, K. Pabbaraju, C. Southward, M. Ronen, S. Leibler, M. G. Surette, and U. Alon. 2001. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science 292:2080-2083. [DOI] [PubMed] [Google Scholar]
- 26.Kwon, O., M. E. Hudspeth, and R. Meganathan. 1996. Anaerobic biosynthesis of enterobactin Escherichia coli: regulation of entC gene expression and evidence against its involvement in menaquinone (vitamin K2) biosynthesis. J. Bacteriol. 178:3252-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucchini, S., A. Thompson, and J. C. Hinton. 2001. Microarrays for microbiologists. Microbiology 147:1403-1414. [DOI] [PubMed] [Google Scholar]
- 28.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marra, A., J. Asundi, M. Bartilson, S. Lawson, F. Fang, J. Christine, C. Wiesner, D. Brigham, W. P. Schneider, and A. E. Hromockyj. 2002. Differential fluorescence induction analysis of Streptococcus pneumoniae identifies genes involved in pathogenesis. Infect. Immun. 70:1422-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masse, E., N. Majdalani, and S. Gottesman. 2003. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 6:120-124. [DOI] [PubMed] [Google Scholar]
- 32.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 33.Meighen, E. A., and R. B. Szittner. 1992. Multiple repetitive elements and organization of the lux operons of luminescent terrestrial bacteria. J. Bacteriol. 174:5371-5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moll, I., D. Leitsch, T. Steinhauser, and U. Blasi. 2003. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 4:284-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller, K., B. F. Matzanke, V. Schunemann, A. X. Trautwein, and K. Hantke. 1998. FhuF, an iron-regulated protein of Escherichia coli with a new type of [2Fe-2S] center. Eur. J. Biochem. 258:1001-1008. [DOI] [PubMed] [Google Scholar]
- 36.Niehaus, F., K. Hantke, and G. Unden. 1991. Iron content and FNR-dependent gene regulation in Escherichia coli. FEMS Microbiol. Lett. 68:319-323. [DOI] [PubMed] [Google Scholar]
- 37.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 38.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2001. Comparative analysis of FUR regulons in gamma-proteobacteria. Nucleic Acids Res. 29:5195-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paustian, M. L., B. J. May, D. Cao, D. Boley, and V. Kapur. 2002. Transcriptional response of Pasteurella multocida to defined iron sources. J. Bacteriol. 184:6714-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Postle, K. 1990. Aerobic regulation of the Escherichia coli tonB gene by changes in iron availability and the fur locus. J. Bacteriol. 172:2287-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider, W. P., S. K. Ho, J. Christine, M. Yao, A. Marra, and A. E. Hromockyj. 2002. Virulence gene identification by differential fluorescence induction analysis of Staphylococcus aureus gene expression during infection-simulating culture. Infect. Immun. 70:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoolnik, G. K. 2002. Microarray analysis of bacterial pathogenicity. Adv. Microb. Physiol. 46:1-45. [DOI] [PubMed] [Google Scholar]
- 45.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 46.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 47.Tsolis, R. M., A. J. Baumler, I. Stojiljkovic, and F. Heffron. 1995. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J. Bacteriol. 177:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valdivia, R. H., and S. Falkow. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367-378. [DOI] [PubMed] [Google Scholar]
- 49.Valdivia, R. H., and S. Falkow. 1998. Flow cytometry and bacterial pathogenesis. Curr. Opin. Microbiol. 1:359-363. [DOI] [PubMed] [Google Scholar]
- 50.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 51.Van Dyk, T. K., E. J. DeRose, and G. E. Gonye. 2001. LuxArray, a high-density, genomewide transcription analysis of Escherichia coli using bioluminescent reporter strains. J. Bacteriol. 183:5496-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wassarman, K. M. 2002. Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell 109:141-144. [DOI] [PubMed] [Google Scholar]
- 53.Worsham, P. L., and J. Konisky. 1981. Use of cir-lac operon fusions to study transcriptional regulation of the colicin Ia receptor in Escherichia coli K-12. J. Bacteriol. 145:647-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, D., W. D. Hardt, and J. E. Galan. 1999. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect. Immun. 67:1974-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]