Abstract
Proteins of the Tol-Pal (Tol-OprL) system play a key role in the maintenance of outer membrane integrity and cell morphology in gram-negative bacteria. Here we describe an additional role for this system in the transport of various carbon sources across the cytoplasmic membrane. Growth of Pseudomonas putida tol-oprL mutant strains in minimal medium with glycerol, fructose, or arginine was impaired, and the growth rate with succinate, proline, or sucrose as the carbon source was lower than the growth rate of the parental strain. Assays with radiolabeled substrates revealed that the rates of uptake of these compounds by mutant cells were lower than the rates of uptake by the wild-type strain. The pattern and amount of outer membrane protein in the P. putida tol-oprL mutants were not changed, suggesting that the transport defect was not in the outer membrane. Consistently, the uptake of radiolabeled glucose and glycerol in spheroplasts was defective in the P. putida tol-oprL mutant strains, suggesting that there was a defect at the cytoplasmic membrane level. Generation of a proton motive force appeared to be unaffected in these mutants. To rule out the possibility that the uptake defect was due to a lack of specific transporter proteins, the PutP symporter was overproduced, but this overproduction did not enhance proline uptake in the tol-oprL mutants. These results suggest that the Tol-OprL system is necessary for appropriate functioning of certain uptake systems at the level of the cytoplasmic membrane.
Two membranes separate the cytoplasm from the external environment in gram-negative bacteria. The outermost membrane contains pore-forming proteins that allow the entry of small molecules by passive diffusion (38), whereas the cytoplasmic membrane houses proteins that link across the periplasmic space with the outer membrane to provide the energy required for the uptake across the outer membrane of certain nutrients, such as iron siderophores and vitamin B12 (7). The cytoplasmic membrane also houses the transport systems that promote the entry of compounds from the periplasm to the cytoplasm, in addition to energy-generating systems. In Escherichia coli and Pseudomonas sp. maintenance of the structure of the cellular envelope involves the products of the tol-pal (tol-oprL in Pseudomonas) operons (4, 27, 32, 43). The transcriptional organization of the tol-oprL gene cluster of Pseudomonas putida is shown in Fig. 1 (33). The Tol-OprL (or Tol-Pal [peptidoglycan-associated lipoprotein]) system is made up of seven proteins. The E. coli Tol-Pal system is organized into two protein complexes: a cytoplasmic membrane complex composed of the TolQ, TolR, and TolA proteins, which interact with each other via their transmembrane domains (12, 17, 18, 24, 25); and an outer membrane complex made up of TolB and Pal, which also interact with Lpp, OmpA, and the peptidoglycan layer (6, 10, 25).
FIG. 1.
Physical and transcriptional organization of the tol-oprL cluster of P. putida KT2440. The solid arrows represent the different tol genes, their relative sizes, and the directions of transcription. The positions of the P1 and PL promoters are indicated (33).
TolQ is an integral cytoplasmic membrane protein that contains three transmembrane domains. The TolR and TolA proteins are anchored to the cytoplasmic membrane by a single transmembrane-spanning segment near the N terminus, which leaves most of the protein exposed to the periplasm (31, 37).
TolB is a periplasmic protein (23), whereas Pal is an outer membrane peptidoglycan-associated lipoprotein (30). The latter protein is anchored to the outer membrane by its N-terminal lipid moiety and strongly interacts with the peptidoglycan layer through its C-terminal region (5, 27). The C-terminal region of Pal also interacts with TolB (41). Interactions of Pal with TolB and the peptidoglycan appear to be mutually exclusive since the TolB-Pal complex is not strongly associated with the peptidoglycan (5, 10).
The cytoplasmic membrane TolQRA and outer membrane TolB-Pal complexes have been shown to be associated through the interactions of TolA with Pal (9) and TolB (14, 52). The TolA-Pal interactions require the proton motive force and the TolQ and TolR proteins (9). These interactions imply that the complexes form a link between the cytoplasmic and outer membranes, confirming the previous observation which indicated that Tol proteins are preferentially located in the contact regions between the cytoplasmic and outer membranes (19).
How the Tol-Pal (Tol-OprL) system plays its role in the maintenance of outer membrane integrity is unknown. It has been hypothesized that TolA could be involved in the energy-dependent passage of synthesized outer membrane components across the periplasm (34). This hypothesis is consistent with the observed in vitro interactions of TolB and TolA with trimeric outer membrane porins (13, 42). Furthermore, TolA was found to be required for surface expression of the O-antigen-containing lipopolysaccharide (16).
In the present study we identified an unexpected new role for the Tol-Pal (Tol-OprL) system. We found that tol-oprL mutants of P. putida do not grow on a number of carbon sources because of limited uptake of these nutrients across the cytoplasmic membrane. This observation was corroborated for the well-defined tol mutants of E. coli and hypothetical tol mutants of Pseudomonas aeruginosa. In this paper we discuss the hypothesis that the Tol-OprL system is required for proper functioning of certain transport systems in the cytoplasmic membrane.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture media, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. Bacterial strains were grown by using routine methods in liquid Luria-Bertani (LB) medium or in M9 minimal medium with benzoic acid (5 mM) as the sole carbon source (44). To isolate outer membranes, cells were grown in LB medium or BM2 minimal medium with glucose as a carbon source (20). Pseudomonas putida was incubated at 30°C, and P. aeruginosa and E. coli strains were incubated at 37°C on an orbital platform operating at 200 strokes per min.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference(s) or source |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR1 recA1 endA1 gyrA96 thi1 relA1 | 44 |
| HB101 | supE44 hsdS20 recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 44 |
| 1292 | supE hsdS met gal lac Y fhuA | 4, 6, 23 |
| JC7782 | 1292 tolA (stop after codon 40) | 4 |
| JC7752 | 1292 tolB (stop after codon 363) Δpal | 6 |
| JC3417 | 1292 tolB (stop after codon 329) | 4, 23 |
| JC8031 | 1292 ΔtolRA | 12 |
| GM1 | ara Δ(lac-pro) thi/F′ [lac-pro] | 48 |
| TPS13 | GM1 tolQ (stop after codon 36) | 48, 4949 |
| TPS300 | GM1 tolR::Cmr | 49 |
| P. aeruginosa strains | ||
| H103 | Wild-type PAO1 | 20 |
| H636 | H103 oprF::ΩSmr | 55 |
| H729 | H103 oprD::Kmr | 22 |
| PAO1403 | PAO1 tol-1, aeruginocin-tolerant mutant, requires tryptophan | 21 |
| PAO1408 | PAO1 tol-2, aeruginocin-tolerant mutant, requires tryptophan | 21 |
| PAO1419 | PAO1 tol-4, aeruginocin-tolerant mutant, requires methionine | 21 |
| PAO1654 | PAO1, tol-6, aeruginocin-tolerant mutant | 21 |
| P. putida strains | ||
| KT2440 | hsdR1 | 32 |
| AX | KT2440 tolA::xylE | 32 |
| AΩ | KT2440 tolA::ΩKm | 32 |
| BX | KT2440 tolB::xylE | 32 |
| BΩ | KT2440 tolB::ΩKm | 43 |
| CRR216 | KT2440 putP | Our laboratory |
| DOT-OX2 | KT2440 oprL::xylE | 32 |
| oprB | KT2440 with plasmid pCHESI-B inserted into the oprB gene (insertion after codon 112), Kmr | This study |
| QX | KT2440 tolQ::xylE | 32 |
| QΩ | KT2440 tolQ::ΩKm | 32 |
| RX | KT2440 tolR::xylE | 32 |
| RΩ | KT2440 tolR::ΩKm | 32 |
| Plasmids | ||
| pBBR1MCS-5 | Gmr, oriT RK2 | 26 |
| pBBRPutP | pBBR1MCS-5 carrying a 1.5-kb chromosomal fragment containing the P. putida KT2440 putP gene obtained by PCR as an HindIII-Xbal fragment | This study |
| pCHESIΩkm | Apr Kmr, pUNφ18 with the HindIII insert from pHP45Ωkm (Ω-Km interposon) at the HindIII site, oriT RP4 | Rodríguez-Herva and Marquésb |
| pCHESI-B | pCHESIΩkm carrying, at the EcoRI site, a 338-bp chromosomal fragment from the P. putida KT2440 oprB gene obtained by PCR as an EcoRI-EcoRI fragment | This study |
| pHP45Ω-Km | Apr Kmr, ori ColE1, source of the Ω-Km interposon | 15 |
| pRK600 | Cmr, helper plasmid, ori ColE1 mobRK2 traRK2 | 32 |
| pUNφ18 | Apr, pUC18 with oriT RP4 to allow replication in Pseudomonas | 35 |
Apr, Cmr, Gmr, Kmr, and Smr, resistance to ampicillin, chloramphenicol, gentamicin, kanamycin, and streptomycin, respectively.
J. J. Rodríguez-Herva and S. Marqués, unpublished data.
When required, antibiotics were used at the following final concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 30 μg ml−1; gentamicin, 10 μg ml−1 (for E. coli) or 30 μg ml−1 (for Pseudomonas sp.); kanamycin, 25 μg ml−1 (for E. coli), 50 μg ml−1 (for P. putida), or 300 μg ml−1 (for P. aeruginosa); and streptomycin, 50 μg ml−1 (for E. coli), 100 μg ml−1 (for P. putida), or 500 μg ml−1 (for P. aeruginosa).
General molecular biology methods.
Standard molecular biology techniques were used for DNA manipulations (44). Southern blot analyses, PCR, and nucleotide sequencing were done as previously described (32, 33).
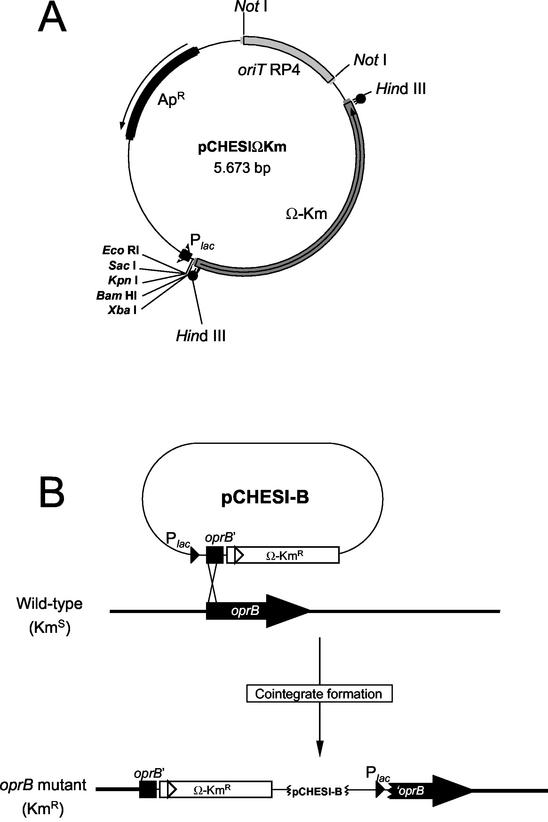
Construction of a P.putida oprB mutant strain.
A mutant strain bearing an inactivated chromosomal oprB gene was constructed as follows. Plasmid pCHESIΩKm is a pUC18 derivative containing the oriT origin of transfer of RP4 and the Ω-Km interposon of plasmid pHP45ΩKm (15) cloned as a HindIII fragment (Fig. 2A). To generate the oprB mutation, a 338-bp fragment of the P. putida oprB gene was amplified by PCR by using primers with EcoRI sites and was subsequently cloned in the EcoRI site of pCHESIΩKm in the same transcriptional direction as the Plac promoter. The resulting plasmid, pCHESI-B (Fig. 2B), was mobilized from E. coli DH5α into P. putida KT2440 by triparental mating by using the E. coli HB101(pRK600) helper strain (32) P. putida transconjugants bearing a cointegrate of the plasmid in the host chromosome were selected on M9 minimal medium with benzoic acid (10 mM) as the sole C source and 50 μg of kanamycin per ml. A few Kmr clones were chosen for Southern blot hybridization to confirm that pCHESI-B integrated into and disrupted the oprB gene. All of the clones analyzed contained an inactivated oprB gene, and a single random clone was chosen and designated P. putida oprB.
FIG. 2.
Strategy used to construct an oprB mutant of P. putida. (A) Physical map of pCHESIΩKm. Single restriction sites are indicated by boldface type. (B) Physical map of pCHESI-B and cointegration into the host chromosome to obtain an oprB mutant. Details of the cloning and cointegration procedures are described in the text.
Outer membrane isolation and immunodetection.
Outer membranes were prepared from cultures grown to a turbidity (measured at 660 nm [OD660]) of ∼0.8 by sucrose gradient density centrifugation (20). Protein profiles were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (20). Immunoblotting procedures were performed as previously described (32, 40).
Uptake assays.
(i) Assays with whole cells.
Cells of the wild-type strain P. putida KT2440 and different mutant strains were grown to an OD660 of ∼0.5 to 0.7 in M9 minimal medium supplemented with the appropriate carbon source. Cells were washed once and resuspended in substrate-free M9 medium to an OD660 of ∼0.3. This cell density ensured that uptake remained linear throughout the experiment. Radiolabeled substrates, added at a final concentration of 250 μM, contained the radiolabeled compound at a concentration of 1 μM. The specific activity was 160 mCi/mmol for [14C]glycerol, 306 mCi/mmol for [14C]glucose, and 242 mCi/mmol for [14C]proline. The cells were incubated at 30°C. After radiolabeled substrates were added, samples were taken at 0, 10, 20, and 30 min. One milliliter was removed, filtered through 0.45-μm-pore-size filters (Millipore) by using a Millipore filter manifold, and washed twice with 3 ml of 0.1 M LiCl. Background radioactivity due to unspecific binding to bacterial cells and filters was assessed by using cells incubated on ice. Filters were placed in scintillation vials, and 4 ml of scintillation liquid was added. Samples were allowed to equilibrate for 12 h before scintillation counting.
(ii) Assays with spheroplasts.
Cells were converted to spheroplasts by treatment with lysozyme and EDTA. The cells were grown to an OD660 of ∼0.5 to 0.7 in M9 minimal medium supplemented with the appropriate carbon source. Cultures (50 to 100 ml) were centrifuged (10,000 × g, 5 min, 4°C), and the cells were resuspended in 6.3 ml of 0.75 M sucrose-10 mM Tris-HCl (pH 7.8) and treated with 0.1 mg of lysozyme per ml for 2 to 5 min on ice. Then 13.2 ml of a 1.5 mM EDTA (pH 7.5) solution was added. The cell suspension was incubated for 15 min at 30°C and observed under a phase-contrast microscope. More than 95% of the cells appeared to be spheroplasts. For uptake assays, the OD660 of the sample was adjusted to ∼0.4, and for the uptake of radiolabeled substrates we used the procedure described above.
Determination of Δψ.
Generation of a membrane potential (Δψ) in spheroplasts of the wild-type strain and tol-oprL mutants was monitored by using the cationic dye 3,3′-diethylthiadicarbocyanine iodide [DiSC2(5)], which translocates into the lipid bilayer of hyperpolarized membranes, resulting in quenching of its fluorescence. Each reaction mixture (total volume, 1 ml) contained buffer A (125 mM HEPES, 0.9% [wt/vol] NaCl, 1 mM KCl, 1 mM MgCl2, 0.4% [wt/vol] glucose), about 106 spheroplasts, and 0.08 μM DiSC2(5). The fluorescence emitted by DiSC2(5) was measured at 670 nm with excitation at 650 nm by using an SLM Aminco SPF-500C fluorimeter. To dissipate the Δψ, valinomycin (final concentration, 2 μM) was added to the reaction mixture. The Δψ was determined by determining the difference between the fluorescence quenching of DiSC2(5) when it accumulated in the cytoplasmic membrane and the fluorescence of DiSC2(5) when it was released upon addition of valinomycin.
RESULTS
Growth of P. putida tol-oprL mutants on different carbon sources.
The role of the proteins of the Tol-OprL system in maintaining the integrity of the outer membrane is well established. However, the involvement of these proteins in other cellular processes is less clear. We observed that the turbidity of overnight cultures of P. putida tol-oprL mutants grown on M9 minimal medium with glucose was significantly lower (OD660, ∼0.5) than that of the wild-type strain (OD660, >3) (Table 2). This prompted us to analyze the growth of P. putida KT2440 and the different tol-oprL mutant strains in M9 minimal medium with different carbon sources, including arginine (10 mM), benzoate (10 mM), fructose (10 mM), glucose (10 mM), glycerol (20 mM), proline (10 mM), succinate (10 mM), and sucrose (10 mM). All P. putida tol-oprL mutants grew as well as the wild-type strain in minimal medium with benzoate (Table 2); however, the turbidities of tol mutant cultures were significantly lower with the other carbon sources. In particular, growth of the tol mutants was negligible with arginine, fructose, and glycerol (Table 2). When proline was used as the sole carbon source, we observed that any polar mutation in the tolQRAB cluster (strains QΩ, RΩ, AΩ, and BΩ) impeded or seriously limited growth (Table 2). In contrast, mutant strains with nonpolar mutations in the tolQ (strain QX), tolR (strain RX), and tolA (strain AX) genes grew relatively well (the lack of a polar effect in xylE was confirmed by reverse transcription-PCR assays performed with the appropriate primers [data not shown]), although inactivation of tolB, the last gene in the cluster, produced a mutant which showed limited growth on proline. Given that the P. putida tolQRAB genes form an operon (33), insertion of the Ω-Km interposon into tolQ, tolR, or tolA seems to exert a polar effect on expression of the tolB gene. The inability of these polar mutants and of the BX mutant, in which the tolB gene is inactivated by an xylE insertion, to grow on M9 minimal medium with proline seems to be mainly due to the lack of the TolB protein in the cell envelope. In marked contrast, growth on succinate was compromised in all mutants except those that lacked the tolB gene product (the BX and BΩ mutants), and with sucrose as the sole carbon source, growth of all mutants was slower than growth of the parental strain.
TABLE 2.
Growth of the P. putida tol-oprL mutants with different carbon sources
| Carbon source | Growth of P. putida strainsa
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| KT2440 | QΩ | RΩ | AΩ | BΩ | QX | RX | AX | BX | DOT-OX2 | |
| Arginine | ++ | − | − | − | − | − | − | − | − | − |
| Benzoate | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Fructose | +++ | − | − | − | − | − | − | − | − | − |
| Glucose | +++ | + | + | + | + | + | ++ | ++ | + | ++ |
| Glycerol | +++ | − | − | − | − | − | − | − | − | − |
| Proline | +++ | − | − | + | + | ++ | ++ | ++ | + | ++ |
| Succinate | ++ | + | − | + | ++ | + | + | + | ++ | + |
| Sucrose | ++ | + | − | + | + | − | − | + | + | + |
Culture cell density was determined after 23 h of incubation at 30°C in M9 minimal medium supplemented with the different carbon sources. +++, OD660 ≥ 2; ++, 0.7 ≤ OD660 < 2; +, 0.2 ≤ OD660 < 0.7; −, OD660 < 0.2. The initial OD660 of the cultures were 0.06 to 0.1.
Growth limitation of the tol mutants of P. putida is not due to defects in the synthesis of porins.
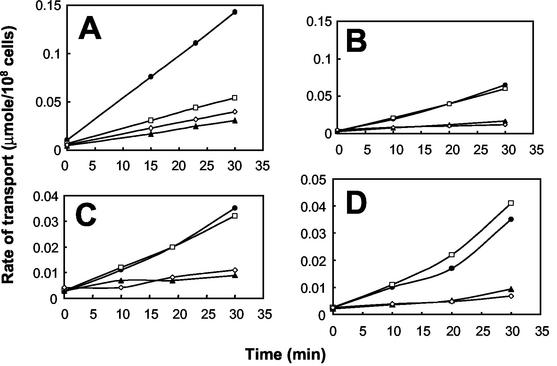
The Tol-Pal protein complex of E. coli has been proposed to be involved in various steps in the biogenesis of porins and in their assembly in the outer membrane (27, 34). Thus, the growth deficiencies in the P. putida tol-oprL mutants may be due to defects in the assembly or synthesis of porins involved in the entry of the substrates tested into the periplasm. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis showed that the pattern of outer membrane proteins in the P. putida tol-oprL mutants was similar to that in the wild-type strain (Fig. 3A)
FIG. 3.
Uptake of different carbon sources by P. putida KT2440 and its isogenic mutants. Uptake of glucose (A and B) and glycerol transport (C and D) by whole cells (A and C) or spheroplasts (B and D) of the P. putida wild-type strain (•) and P. putida QX (tolQ mutant) (⋄), BX (tolB mutant) (▴), and oprB (□) mutant strains. Cells were grown in minimal glucose medium until the exponential growth phase was reached, and uptake was analyzed as described in Materials and Methods.
In P. aeruginosa, OprD has been shown to facilitate the diffusion into the periplasm of arginine and small peptides containing basic amino acids (50). The OprB porin was shown to contain a binding site for glucose and other compounds, such as glycerol and fructose (1, 56). We analyzed the presence of these two outer membrane porins in P. putida tol-oprL strains by Western blotting. Cells were grown in BM2 minimal medium with glucose to induce OprB expression (45) or in LB medium to detect OprD. The amounts of OprD and OprB in the outer membranes of all mutant strains were similar to the amounts in the wild-type strain (data not shown).
We examined growth of the wild-type P. putida strain and the oprB mutant with glucose, benzoate, glycerol, or fructose as the sole carbon source, and we found that the doubling times in the exponential phase of the mutant strain with these carbon sources were similar to those of the wild-type strain with the same carbon sources (data not shown). Together, these results suggest that the poor growth of the P. putida tol-oprL mutants on certain carbon sources is not the result of defective assembly of the porins for these compounds in the outer membrane.
Uptake of glucose, glycerol, and proline in intact cells and spheroplasts.
To determine whether transport of glucose, glycerol, and proline through the cell envelope was affected in the P. putida tol-oprL mutants, we used radiolabeled substrates to analyze the uptake of these compounds by intact cells. For the glucose and glycerol uptake assays, cells were grown on M9 minimal medium with glucose, whereas for the proline uptake assays, cells were grown on LB medium until an OD660 of ∼0.5 was reached, washed once, and incubated in minimal medium with proline for 3 h. The rates of glucose, glycerol, and proline uptake across the cell envelope in the tolQ and tolB mutants were between 5- and 10-fold lower than the rates of uptake in the wild-type strain (Fig. 3A and C and 4). In contrast, the loss of OprB resulted in a decrease in glucose transport across the cell envelope (Fig. 3A) but not in a decrease in glycerol transport (Fig. 3C).
FIG. 4.
Uptake of proline by whole cells of different strains of P. putida. The strains used were P. putida KT2440 (squares), P. putida CRR216 (putP mutant) (circles), P. putida BX (tolB mutant) (triangles), P. putida QX (tolQ mutant) (diamonds), and P. putida QΩ (tolQ mutant) (rectangles) bearing pBBR1MCS-5 as a negative control (open symbols) or pBBRPutP (solid symbols). Cells were grown in LB medium until an OD660 of ∼0.5 was reached, washed once, resuspended in M9 minimal medium, and incubated for 3 h. Uptake was measured as described in Materials and Methods.
Since the amounts of outer membrane proteins appeared to be unaffected in the tol-oprL mutants, the reduced uptake of various carbon sources might have been due to a defect in transport across the cytoplasmic membrane. To test this possibility, the uptake of glucose and glycerol was measured in spheroplasts, in which the outer membrane barrier had been lost. The rates of uptake of these compounds by spheroplasts of the wild-type strain and the oprB mutants were similar (Fig. 3B and D). This result was expected, since the transport defect of the oprB mutant was related to outer membrane transport. The incorporation rates in these spheroplasts were between 10- and 20-fold higher than the rates determined for spheroplasts of the tolQ and tolB mutants (Fig. 3B and D). These results therefore suggest that in the tol mutants, glucose and glycerol uptake is affected at the level of cytoplasmic membrane transport.
Proline transport in cells that overproduce the PutP cytoplasmic membrane protein.
The reduced uptake of certain carbon sources across the cytoplasmic membrane of the P. putida tol-oprL mutants might have been a result of the absence of or reduced amounts of specific transport proteins in the cytoplasmic membrane. To test this hypothesis, we investigated whether the tol mutations could be complemented by overproduction of these transporters from a constitutive promoter. The uptake of proline in P. putida is mediated by the cytoplasmic membrane-located PutP protein, which functions as an Na+ symporter. The wild-type putP gene of P. putida KT2440 (51) was subcloned into plasmid pBBR1MCS-5 to obtain pBBRPutP. In this plasmid, the putP gene is expressed from the Plac promoter, which is constitutively expressed in Pseudomonas sp.; thus, all cells contain PutP. The rate of proline uptake was analyzed in P. putida KT2440 and in QX (tolQ mutant), QΩ (tolQ mutant), BX (tolB mutant), and putP mutant strains bearing or not bearing pBBRPutP. Figure 4 shows that overproduction of the PutP symporter partially restored the ability of the P. putida putP mutant strain to transport proline, which shows that the putP gene was functionally expressed. However, plasmid pBBRPutP did not enhance the uptake of proline in the QX, QΩ, and BX mutant strains. The rates of transport of proline in the P. putida BX strain, which had a nonpolar mutation in the tolB gene, and the P. putida QΩ strain, which had a polar mutation in the tolQ gene, were lower than the rate of transport of proline in the P. putida QX strain, which had a nonpolar mutation in the tolQ gene (Fig. 4). This finding is in agreement with the fact that the P. putida tolQRA polar mutants, as well as the P. putida BX and BΩ mutants, were unable to grow with proline as the carbon source, whereas leaky growth of the P. putida tolQRA nonpolar mutants was observed.
Determination of ΔΨ in P. putida tol-oprL mutant strains.
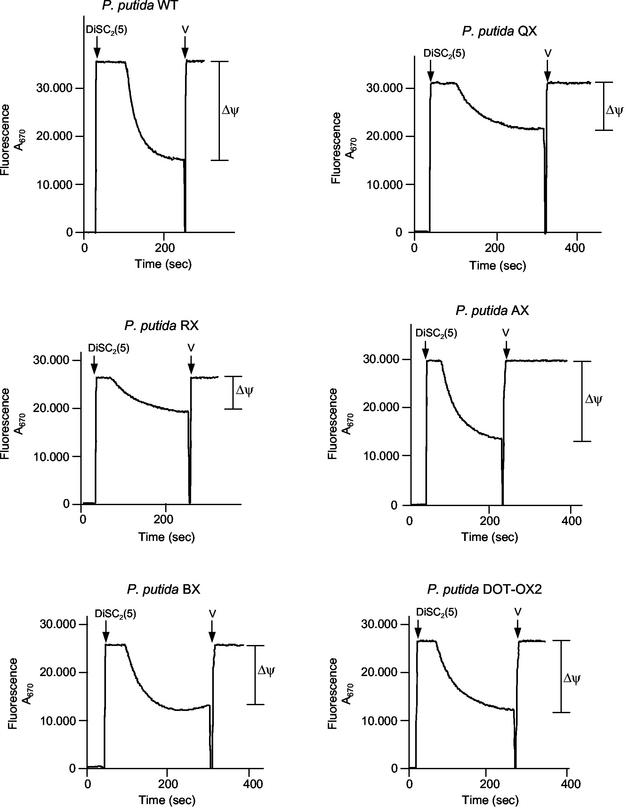
The reduced activities of certain cytoplasmic membrane transporters in the P. putida tol-oprL mutants might have been due to a reduction in the proton motive force. To test this possibility, we measured ΔΨ in the P. putida tol-oprL mutant strains using the fluorochrome DiSC2(5). Cells were grown on LB medium, and generation of the proton motive force in spheroplasts was monitored by monitoring the fluorescence quenching of DiSC2(5). The proton motive forces in the P. putida AX (tolA mutant), BX (tolB mutant), and DOT-OX2 (oprL mutant) strains were similar (about 93.7, 83.4, and 87.5%, respectively) to the proton motive force produced by the wild-type strain (Fig. 5). Although the ΔΨ was found to be lower in the P. putida QX (tolQ mutant) and RX (tolR mutant) strains, it was not completely dissipated (Fig. 5), and therefore, it seems highly unlikely that the reduced uptake of various carbon sources in the tol mutants could have been a result of the reduced proton motive force.
FIG. 5.
Determination of Δψ in P. putida KT2440 (WT) and tol::xylE mutant strains (P. putida QX, RX, AX, BX, and DOT-OX2). Cells were grown in LB medium, harvested in the exponential phase of growth (OD660, ∼0.4), and converted to spheroplasts as described in Materials and Methods. Generation of the proton motive force by spheroplasts was monitored by monitoring the fluorescence quenching of DiSC2(5) as described in Materials and Methods. The arrows indicate the times when the fluorescence probe and valinomycin (V) were added.
Growth of P. aeruginosa and E. coli tol-oprL (tol-pal) mutants with different carbon sources.
The reduced uptake of various carbon sources observed in the P. putida mutants represents a new phenotype for tol-oprL mutations. Previously, a number of P. aeruginosa and E. coli mutants with mutations in the tol-pal (tol-oprL) system were isolated (Table 1), but their growth on various carbon sources was not investigated systematically. Therefore, we decided to determine whether these mutants exhibited limited growth with different carbon sources. To do this, growth of the wild-type and tol mutant strains was assayed in M9 minimal medium supplemented with arginine (10 mM), fructose (10 mM), glycerol (20 mM), glucose (10 mM), and proline (10 mM) in the case of P. aeruginosa and with glucose (10 mM), glycerol (20 mM), and maltose (10 mM) in the case of E. coli. P. aeruginosa tol-1 and tol-2 mutant strains were not able to grow or grew poorly with arginine, glycerol, praline, or succinate (Table 3). Growth of the P. aeruginosa tol-4 mutant was compromised with arginine, glucose, and glycerol and was less notably compromised with proline and succinate (Table 3). Growth of the P. aeruginosa tol-6 mutant was affected only with arginine and glucose as the carbon sources (Table 3).
TABLE 3.
Growth of the P. aeruginosa tol mutants with different carbon sources
| Carbon source | Growth of P. aeruginosa strains
|
||||
|---|---|---|---|---|---|
| H103 | tol-1 | tol-2 | tol-4 | tol-6 | |
| Arginine | ++ | − | − | + | + |
| Glucose | +++ | − | + | + | ++ |
| Glycerol | ++ | + | − | + | ++ |
| Proline | ++ | − | + | ++ | ++ |
| Succinate | ++ | − | + | ++ | ++ |
aCulture cell density was determined after 20 h of incubation at 37°C in M9 minimal medium supplemented with different carbon sources. +++, OD660 ≥ 2; ++, 0.7 ≤ OD660 < 2; +, 0.2 ≤ OD660 < 0.7; −, OD660 < 0.2. The initial OD660 of the cultures were 0.06 to 0.1.
For E. coli we used two parental strains, GM1 and 1292, and their corresponding isogenic mutants. Growth of GM1 and growth of the isogenic tolQ TPS13 and tolR TPS300 mutants were similar in glucose and glycerol; however, in maltose growth of the E. coli mutant strains was seriously compromised (Table 4). Growth of a tolB mutant derivative of E. coli 1292 (i.e., strain JC3417) in glucose was similar to growth of the wild-type strain, but its growth was compromised in glycerol and maltose (Table 4). E. coli JC7782 (ΔtolA), JC8031 (ΔtolRA), and JC7752 (ΔtolBpal) were not able to grow or grew very poorly with glucose, glycerol, or maltose as the sole carbon source. Hence, reduced uptake of certain carbon sources, first observed for the P. putida tol-oprL mutants, appears to be a characteristic of many, although not all, P. aeruginosa and E. coli tol-oprL (tol-pal) mutants.
TABLE 4.
Growth of the E. coli tol-pal mutants with different carbon sources
| Carbon source | Growth of E. coli strainsa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| GM1 | TPS13 (tolQ) | TPS300 (tolR) | 1292 | JC3417 (tolB) | JC7752 (ΔtolBpal) | JC7782 (tolA) | JC8031 (ΔtolRA) | |
| Glucose | ++ | ++ | ++ | ++ | ++ | + | + | + |
| Glycerol | ++ | ++ | ++ | ++ | + | − | − | − |
| Maltose | +++ | + | + | +++ | − | − | − | − |
Culture cell density was determined after 20 h of incubation at 37°C in M9 minimal medium supplemented with the different carbon sources. +++, OD660 ≥ 2; ++, 0.7 ≤ OD660 ≤ 2; +, 0.2 ≤ OD660 < 0.7; −, OD660 < 0.2. The initial OD660 of the cultures were 0.06 to 0.1.
DISCUSSION
The results presented here demonstrate that mutations in the tol-oprL (tol-pal) genes of P. putida, P. aeruginosa, and E. coli lead to organisms that are defective in the utilization of a number of carbon sources and support the hypothesis that the Tol system of P. putida influences the transport of certain carbon sources across the cytoplasmic membrane, although the molecular mechanisms behind this phenotype(s) is still unknown. These results are surprising because functions previously associated with the Tol-Pal (Tol-OprL) system were all related to the correct biogenesis of the outer membrane, the assembly of porins, and transfer of lipopolysaccharide.
Lazzaroni et al. (28) found that the level of expression of E. coli OmpF and LamB outer membrane porins was lower in an E. coli tolA mutant and suggested that the TolA protein exerted positive indirect control at the transcriptional level over the ompF and lamB genes. In contrast, our immunodetection analysis of P. putida demonstrated that the levels of the OprD, OprB, and OprF porins of the outer membrane in the tol-oprL mutants were similar to the levels in the wild-type strain. Furthermore, immunofluorescence assays showed that at least the OprF protein seemed to be correctly exposed on the surface of the outer membrane of the P. putida tol-oprL mutants (data not shown). This is in agreement with the fact that in the E. coli tol-pal mutants, the outer membrane porins are correctly processed and assembled in the outer membrane (11, 27). These results suggest that the P. putida Tol-OprL system is not involved in the export or assembly of the porins in the outer membrane in this microorganism. However, we cannot rule out the possibility that some components of the outer membrane are not functional in the tol-oprL mutants, since the stability of the membrane is seriously affected in these strains (32).
It was surprising to find that the carbon sources that are not taken up by the P. putida tol-pal mutants are transported by transport systems that are very different. Thus, glycerol uptake involves facilitated diffusion down a concentration gradient (53, 54), whereas the uptake of proline is mediated by the PutP protein, which is an energy-dependent Na+ symporter (8, 51). For the transport of glucose there are two separate inducible systems in P. aeruginosa (36); one is a low-affinity transport system which involves the extracellular oxidation of glucose to gluconate or to 2-ketogluconate prior to transport into the cytoplasm, and the second is a high-affinity system which transports glucose directly into the cytoplasm via a periplasmic binding protein-dependent ABC transport system that requires ATP as an energy source (2). The existence in P. putida of a periplasmic glucose binding protein is suspected because (i) the rate of glucose transport across the spheroplasts was lower than the rate of glucose transport across whole cells of the wild-type strain (Fig. 3A and B) and (ii) a polyclonal antibody against the P. aeruginosa glucose binding protein cross-reacted with a protein of P. putida (data not shown). We identified in the P. putida KT2440 genome a gene similar to the P. aeruginosa gltK gene (1) which encodes one of the cytoplasmic membrane components of the glucose ABC transport system. In P. aeruginosa, the uptake of arginine is also mediated by a periplasmic binding protein-dependent ABC transport system (39). Although the transport of glucose and arginine might have been influenced by the generation of a proton motive force, the tol-pal mutants of P. putida were not particularly defective in the generation of a proton motive force, which eliminated the possibility that the reduced transport rates in the tol-oprL mutants were due to a low proton motive force. Consistently, the transport of glycerol, which was taken up through a facilitated diffusion process, was also affected in the P. putida tol-oprL mutants. In E. coli, the transport and catabolism of glycerol are mediated by the components of the glp regulon. The glpFK operon encodes a membrane diffusion facilitator for glycerol and a cytoplasmic glycerol kinase (53). In P. aeruginosa, glycerol was also found to be transported by a facilitated diffusion system (54) that involved a GlpFK system which was 80% identical to the E. coli glycerol diffusion facilitator and to cytoplasmic glycerol kinase (46, 47). Our results with P. putida suggest that glycerol transport is not associated with a periplasmic binding protein, since the rates of glycerol transport were similar in whole cells and spheroplasts (Fig. 3C and D). Furthermore, the P. putida KT2440 genome contains the glpFK operon, and the proteins exhibit strong homology to the E. coli and P. aeruginosa GlpF and GlpK proteins. The data available for P. aeruginosa and P. putida are consistent with transport of glycerol by a facilitated diffusion system, which does not require energy. The rate of glycerol uptake by the P. putida tol-oprL mutants is as low as the rate of glycerol uptake by a mutant lacking the glpFK genes that encode the transport system (46). This reinforces the hypothesis that the Tol system influences glycerol uptake at the cytoplasmic membrane level.
Why are tol-oprL mutants defective in the uptake of certain carbon sources? One possibility is that the Tol-OprL system can directly or indirectly influence the correct insertion or functioning of certain transport systems in the cytoplasmic membrane. To our knowledge, this is the first time that such a role has been attributed to the Tol-Pal (Tol-OprL) system, although reduced utilization of some carbon sources has been reported previously for E. coli tol-pal mutants. This phenotype in E. coli was ascribed to loss of the periplasmic binding proteins of the corresponding transporters (3, 29). However, in this work we demonstrated that this hypothesis is not correct since the P. putida tol-oprL mutants are affected in transport at the level of the cytoplasmic membrane. Although the P. putida tol-oprL mutants release periplasmic proteins into the extracellular medium (32), the amounts of periplasmic glucose binding protein that remain in the periplasm of the mutants are similar to the amount found in the wild-type strain (data not shown). Moreover, not only the transport systems that do not depend on a periplasmic binding protein are affected in these mutants, as described above.
In P. putida the polar mutations in tolQRAB had more dramatic effects on the range and levels of utilization of different carbon sources than individual knockouts, as assessed by culture turbidity (Table 2). In this regard, the tol-1 and tol-2 mutants of P. aeruginosa exhibited very limited or no growth with several carbon sources, whereas utilization of carbon sources was less affected in the tol-4 and tol-6 mutants. It would therefore be interesting to determine the specific nature of the mutations in these P. aeruginosa tol mutants to establish a closer link with the mutations in P. putida. Our results with P. putida suggest that while TolB is the most critical protein affecting proline transport, this protein is not essential for growth on succinate, which is affected more in mutants lacking the other components of the Tol system. On the other hand, in E. coli, growth defects were more evident with maltose than with any other sugar, and double mutants showed greater impairment than single mutants. The growth defects with maltose could be partially due to the known leakiness of the outer membrane and the loss of MalE from the periplasm, although it should be noted that not only periplasmic binding protein-dependent transport systems are affected in the E. coli tol-pal mutants since these mutants are also unable to grow in glycerol. The defects in growth of these mutants seem to be similar to that of P. putida, for which a cytoplasmic membrane transport defect has been demonstrated. The weak phenotype of the tolQ and tolR mutants of E. coli could be explained by the presence of the ExbB and ExbD proteins, which might take over the functions of TolQ and TolR to some extent (27).
In summary, the results presented here suggest a new physiological role for the Tol-Pal (Tol-OprL) system, namely, an effect on the uptake of a number of carbon sources at the level of the cytoplasmic membrane.
Acknowledgments
We thank E. A. Worobec for providing the polyclonal antibody against P. aeruginosa GBP and OprB and Hendrik Adams for advice concerning Δψ measurements. We thank M. M. Ochs and S. W. Farmer for support, M. M. Fandila and C. Lorente for secretarial assistance, and K. Shashok for improving the language of the manuscript.
M. A. Llamas was the recipient of a fellowship from the Spanish Ministry of Education and Culture to work at the University of British Columbia and the recipient of a short-term EMBO fellowship to visit the University of Utrecht. The work in Spain was supported by grants from the European Commission (grants QLK3-CT-2000-0170 and QLK3-CT-2001-00435) and the Junta de Andalucía and Ministerio de Ciencia y Tecnología (grant BIO 2000-0964). R. E. W. Hancock was supported by a grant from the Canadian Institute for Health Research.
REFERENCES
- 1.Adewoye, L. O., L. Tschetter, J. O'Neil, and E. A. Worobec. 1998. Channel specificity and secondary structure of the glucose-inducible porins of Pseudomonas sp. J. Bioenerg. Biomembr. 30:257-267. [DOI] [PubMed] [Google Scholar]
- 2.Adewoye, L. O., and E. A. Worobec. 2000. Identification and characterization of the gltK gene encoding a membrane-associated glucose transport protein of Pseudomonas aeruginosa. Gene 253:323-330. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. J., J. M. Wilson, and D. L. Oxender. 1979. Defective transport and other phenotypes of a periplasmic “leaky” mutant of Escherichia coli K-12. J. Bacteriol. 140:351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernadac, A., M. Gavioli, J. C. Lazzaroni, S. Reina, and R. Lloubès. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 180:4872-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouveret, E., H. Bénédetti, A. Rigal, E. Loret, and C. Lazdunski. 1999. In vitro characterization of peptidoglycan-associated lipoprotein (PAL)-peptidoglycan and PAL-TolB interactions. J. Bacteriol. 181:6306-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouveret, E., R. Derouiche, A. Rigal, R. Lloubès, C. Lazdunski, and H. Bénédetti. 1995. Peptidoglycan-associated lipoprotein-TolB interaction. J. Biol. Chem. 270:11071-11077. [DOI] [PubMed]
- 7.Braun, V. 1995. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16:295-307. [DOI] [PubMed] [Google Scholar]
- 8.Cairney, J., C. F. Higgins, and I. R. Booth. 1984. Proline uptake through the major transport system of Salmonella typhimurium is coupled to sodium ions. J. Bacteriol. 160:22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cascales, E., M. Gavioli, J. N. Sturgis, and R. Lloubès. 2000. Proton motive force drives the interaction of the inner membrane TolA and outer membrane Pal proteins in Escherichia coli. Mol. Microbiol. 38:904-915. [DOI] [PubMed] [Google Scholar]
- 10.Clavel, T., P. Germon, A. Vianney, R. Portalier, and J. C. Lazzaroni. 1998. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol. Microbiol. 29:359-367. [DOI] [PubMed] [Google Scholar]
- 11.de Cock, H., M. Pasveer, J. Tomassen, and E. Bouveret. 2001. Identification of phospholipids as new components that assist in the in vitro trimerization of a bacterial pore protein. Eur. J. Biochem. 268:865-875. [DOI] [PubMed] [Google Scholar]
- 12.Derouiche, R., H. Bénédetti, J. C. Lazzaroni, C. Lazdunski, and R. Lloubès. 1995. Protein complex within Escherichia coli inner membrane. J. Biol. Chem. 270:11078-11084. [DOI] [PubMed] [Google Scholar]
- 13.Derouiche, R., M. Gavioli, H. Bénédetti, A. Prilipov, C. Lazdunski, and R. Lloubès. 1996. TolA central domain interacts with Escherichia coli porins. EMBO J. 15:6408-6415. [PMC free article] [PubMed] [Google Scholar]
- 14.Dubuisson, J. F., A. Vianney, and J. C. Lazzaroni. 2002. Mutational analysis of the TolA C-terminal domain of Escherichia coli and genetic evidence for an interaction between TolA and TolB. J. Bacteriol. 184:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 16.Gaspar, J. A., J. A. Thomas, C. L. Marolda, and M. A. Valvano. 2000. Surface expression of O-specific lipopolysaccharide in Escherichia coli requires the function of the TolA protein. Mol. Microbiol. 38:262-275. [DOI] [PubMed] [Google Scholar]
- 17.Germon, P., T. Clavel, A. Vianney, R. Portalier, and J. C. Lazzaroni. 1998. Mutational analysis of the Escherichia coli K-12 TolA N-terminal region and characterization of its TolQ-interacting domain by genetic suppression. J. Bacteriol. 180:6433-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Germon, P., M. C. Ray, A. Vianney, and J. C. Lazzaroni. 2001. Energy-dependent conformational change in the TolA protein of Escherichia coli involves its N-terminal domain, TolQ, and TolR. J. Bacteriol. 183:4110-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guihard, G., P. Boulanger, H. Bénédetti, R. Lloubès, M. Besnard, and L. Letellier. 1994. Colicin A and the Tol proteins involved in its translocation are preferentially located in the contact sites between the inner and outer membranes of Escherichia coli cells. J. Biol. Chem. 269:5874-5880. [PubMed] [Google Scholar]
- 20.Hancock, R. E. W., and A. M. Carey. 1979. Outer membrane of Pseudomonas aeruginosa: heat- and 2-mercaptoethanol-modifiable proteins. J. Bacteriol. 140:902-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holloway, B. W., H. Rossiter, D. Burgess, and J. Dodge. 1973. Aeruginocin tolerant mutants of Pseudomonas aeruginosa. Genet. Res. Camb. 22:239-253. [DOI] [PubMed] [Google Scholar]
- 22.Huang, H., and R. E. W. Hancock. 1993. Genetic definition of the substrate selectivity of outer membrane protein OprD of Pseudomonas aeruginosa. J. Bacteriol. 175:7793-7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isnard, M., A. Rigal, J. C. Lazzaroni, C. Lazdunski, and R. Lloubès. 1994. Maturation and localization of the TolB protein required for colicin import. J. Bacteriol. 176:6392-6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Journet, L., A. Rigal, C. Lazdunski, and H. Bénédetti. 1999. Role of TolR N-terminal, central, and C-terminal domains in dimerization and interaction with TolA and TolQ. J. Bacteriol. 181:4476-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koebnik, R. 1995. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol. Microbiol. 16:1269-1270. [DOI] [PubMed] [Google Scholar]
- 26.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 27.Lazdunski, C., E. Bouveret, A. Rigal, L. Journet, R. Lloubès, and H. Bénédetti. 1998. Colicin import into Escherichia coli cells. J. Bacteriol. 180:4993-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazzaroni, J. C., N. Fognini-Lefebvre, and R. C. Portalier. 1986. Effects of lkyB mutations on the expression ompF, ompC and lamB porin structural genes in Escherichia coli K-12. FEMS Microbiol. Lett. 33:235-239. [Google Scholar]
- 29.Lazzaroni, J. C., and R. C. Portalier. 1981. Genetic and biochemical characterization of periplasmic leaky mutants of Escherichia coli K-12. J. Bacteriol. 145:1351-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazzaroni, J. C., and R. C. Portalier. 1992. The excC gene of Escherichia coli K-12 required for cell envelope integrity encodes the peptidoglycan-associated lipoprotein (PAL). Mol. Microbiol. 6:735-742. [DOI] [PubMed] [Google Scholar]
- 31.Levengood, S. K., W. F. Beyer, Jr., and R. E. Webster. 1991. TolA: a membrane protein involved in colicin uptake contains an extended helical region. Proc. Natl. Acad. Sci. USA 88:5939-5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Llamas, M. A., J. L. Ramos, and J. J. Rodríguez-Herva. 2000. Mutations in each of the tol genes of Pseudomonas putida reveal that they are critical for the maintenance of outer membrane stability. J. Bacteriol. 182:4764-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llamas, M. A., J. L. Ramos, and J. J. Rodríguez-Herva. 2003. Transcriptional organization of the Pseudomonas putida tol-oprL genes. J. Bacteriol. 185:184-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloubès, R., E. Cascales, A. Walburger, E. Bouveret, C. Lazdunski, A. Bernadac, and L. Journet. 2001. The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152:523-529. [DOI] [PubMed] [Google Scholar]
- 35.Marqués, S., M. T. Gallegos, M. Manzanera, A. Holtel, K. N. Timmis, and J. L. Ramos. 1998. Activation and repression of transcription at the double tandem divergent promoters for the xylR and xylS genes of the TOL plasmid of Pseudomonas putida. J. Bacteriol. 180:2889-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Midgley, M., and E. A. Dawes. 1973. The regulation of transport of glucose and methyl α-glucoside in Pseudomonas aeruginosa. Biochem. J. 132:141-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller, M. M., A. Vianney, J. C. Lazzaroni, R. E. Webster, and R. C. Portalier. 1993. Membrane topology of the Escherichia coli TolR protein required for cell envelope integrity. J. Bacteriol. 175:6059-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikaido, M. 1992. Porins and specific channels of bacterial outer membranes. Mol. Microbiol. 6:435-442. [DOI] [PubMed] [Google Scholar]
- 39.Nishijyo, T., S. M. Park, C. D. Lu, Y. Itoh, and A. T. Abdelal. 1998. Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J Bacteriol. 180:5559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rawling, E. G., N. L. Martin, and R. E. W. Hancock. 1995. Epitope mapping of the Pseudomonas aeruginosa major outer membrane porin protein OprF. Infect. Immun. 63:38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ray, M. C., P. Germon, A. Vianney, R. Portalier, and J. C. Lazzaroni. 2000. Identification by genetic suppression of Escherichia coli TolB residues important for TolB-Pal interaction. J. Bacteriol. 182:821-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rigal, A., E. Bouveret, R. Lloubès, C. Lazdunski, and H. Bénédetti. 1997. The TolB protein interacts with the porins of Escherichia coli. J. Bacteriol. 179:7274-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodríguez-Herva, J. J., and J. L. Ramos. 1996. Characterization of an OprL null mutant of Pseudomonas putida. J. Bacteriol. 178:5836-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Saravolac, E. G., N. F. Taylor, R. Benz, and R. E. W. Hancock. 1991. Purification of glucose-inducible outer membrane protein OprB of Pseudomonas putida and reconstitution of glucose-specific pores. J. Bacteriol. 173:4970-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schweizer, H. P., R. Jump, and C. Po. 1997. Structure and gene-polypeptide relationships of the region encoding glycerol diffusion facilitator (glpF) and glycerol kinase (glpK) of Pseudomonas aeruginosa. Microbiology 143:1287-1297. [DOI] [PubMed] [Google Scholar]
- 47.Schweizer, H. P., and C. Po. 1996. Regulation of glycerol metabolism in Pseudomonas aeruginosa: characterization of the glpR repressor gene. J. Bacteriol. 178:5215-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun, T.-P., and R. E. Webster. 1986. fii, a bacterial locus required for filamentous phage infection, and its relation to colicin-tolerant tolA and tolB. J. Bacteriol. 165:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, T.-P., and R. E. Webster. 1987. Nucleotide sequence of a gene cluster involved in entry of E colicins and single-stranded DNA of infecting filamentous bacteriophages into Escherichia coli. J. Bacteriol. 169:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trias, J., and H. Nikaido. 1990. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J. Biol. Chem. 265:15680-15684. [PubMed] [Google Scholar]
- 51.Vílchez, S., L. Molina, C. Ramos, and J. L. Ramos. 2000. Proline catabolism by Pseudomonas putida: cloning, characterization, and expression of the put genes in the presence of root exudates. J. Bacteriol. 182:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walburger, A., C. Lazdunski, and Y. Corda. 2002. The Tol/Pal system function requires an interaction between the C-terminal domain of TolA and the N-terminal domain of TolB. Mol. Microbiol. 44:695-708. [DOI] [PubMed] [Google Scholar]
- 53.Weissenborn, D. L., N. Wittekindt, and T. J. Larson. 1992. Structure and regulation of the glpFK operon encoding glycerol diffusion facilitator and glycerol kinase of Escherichia coli K-12. J. Biol. Chem. 267:6122-6131. [PubMed] [Google Scholar]
- 54.Williams, S. G., J. A. Greenwood, and C. W. Jones. 1994. The effect of nutrient limitation on glycerol uptake and metabolism in continuous cultures of Pseudomonas aeruginosa. Microbiology 140:2961-2969. [DOI] [PubMed] [Google Scholar]
- 55.Woodruff, W. A., and R. E. W. Hancock. 1988. Construction and characterization of Pseudomonas aeruginosa protein F-deficient mutants after in vitro and in vivo insertion mutagenesis of the cloned gene. J. Bacteriol. 170:2592-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wylie, J. L., and E. A. Worobec. 1995. The OprB porin plays a central role in carbohydrate uptake in Pseudomonas aeruginosa. J. Bacteriol. 177:3021-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]