Abstract
Background
Currently available vaccines against leptospirosis are of low efficacy, have an unacceptable side-effect profile, do not induce long-term protection, and provide no cross-protection against the different serovars of pathogenic leptospira. The current major focus in leptospirosis research is to discover conserved protective antigens that may elicit longer-term protection against a broad range of Leptospira. There is a need to screen vaccine candidate genes in the genome of Leptospira interrogans.
Results
Bioinformatics, comparative genomic hybridization (CGH) analysis and transcriptional analysis were used to identify vaccine candidates in the genome of L. interrogans serovar Lai strain #56601. Of a total of 4727 open reading frames (ORFs), 616 genes were predicted to encode surface-exposed proteins by P-CLASSIFIER combined with signal peptide prediction, α-helix transmembrane topology prediction, integral β-barrel outer membrane protein and lipoprotein prediction, as well as by retaining the genes shared by the two sequenced L. interrogans genomes and by subtracting genes with human homologues. A DNA microarray of L. interrogans strain #56601 was constructed for CGH analysis and transcriptome analysis in vitro. Three hundred and seven differential genes were identified in ten pathogenic serovars by CGH; 1427 genes had high transcriptional levels (Cy3 signal ≥ 342 and Cy5 signal ≥ 363.5, respectively). There were 565 genes in the intersection between the set encoding surface-exposed proteins and the set of 307 differential genes. The number of genes in the intersection between this set of 565 and the set of 1427 highly transcriptionally active genes was 226. These 226 genes were thus identified as putative vaccine candidates. The proteins encoded by these genes are not only potentially surface-exposed in the bacterium, but also conserved in two sequenced L. interrogans. Moreover, these genes are conserved among ten epidemic serovars in China and have high transcriptional levels in vitro.
Conclusion
Of the 4727 ORFs in the genome of L. interrogans, 226 genes were identified as vaccine candidates by bioinformatics, CGH and transcriptional analysis on the basis of the theory of reverse vaccinology. The proteins encoded by these genes might be useful as vaccine candidates as well as for diagnosis of leptospirosis.
Background
Leptospirosis is a globally important zoonotic disease caused by pathogenic Leptospira species[1]. Leptospires are thin, helically coiled, motile bacteria, classified into 17 genomospecies (including the saprophyte Leptospira biflexa and the pathogen Leptospira interrogans) on the basis of DNA-DNA hybridization studies, or serologically classified into more than two hundred pathogenic serovars on the basis of structural heterogeneity in the carbohydrate component of the lipopolysaccharide[2,3]. Currently available vaccines, based on inactivated whole bacteria or membrane preparations from pathogenic leptospires, are of low efficacy, have an unacceptable side-effect profile, require annual booster immunizations and do not confer cross-protective immunity against different serovars [4-6]. Because of these concerns, the current major focus in leptospirosis research is to discover cross-species-conserved or cross-serovar-conserved protective antigens that may elicit longer-term protection against a broad range of Leptospira[5,7]. New vaccine development strategies are thus needed for preventing this zoonosis. Reverse vaccinology, which based on the genomic approach, has been applied to some bacteria, and novel vaccine candidate sequences have been identified [8-11]. The genome projects of two Leptospira strains give us intensive knowledge on the whole genome level [12-14]. Although many efforts have been made to identify the surface-exposed proteins of leptospires, finding perfect vaccine candidate antigens that provide cross-protection against different serovars of pathogenic L. interrogans still requires further work[7,15-17].
In our current study, we identified 226 potential candidate vaccine genes against L. interrogans using in silico analysis, comparative genomic hybridization (CGH) and transcriptional analysis, based on a genome-wide DNA microarray comprising 3528 open reading frames (ORFs) derived from the original annotation of L. interrogans strain #56601. These candidate genes not only encode surface-exposed proteins of L. interrogans strain #56601, but also have high transcription levels in vitro. Moreover, the proteins encoded by these genes are conserved in two sequenced L. interrogans and ten epidemic pathogenic serovars in China.
Results
In silico analysis for identification of genes encoding surface-exposed proteins
In 4727 ORFs of L. interrogans strain #56601, 1282 proteins were predicted to be surface-exposed using P-CLASSIFIER, 654 proteins had signal peptides, 813 were predicted to have no more than four α-helices with transmembrane topology, 96 were predicted to have β-barrel topology implying that they are integral β-barrel outer membrane proteins, and 158 were predicted have a lipoprotein signal peptide using SpLiP. The number of genes in the intersection between the set of surface-exposed proteins identified by P-CLASSIFIER and the set of proteins characterized by at least one of the four characteristic topologies is 688. We calculated the similarity of proteins between serovar Lai and serovar Copenhageni as well as between serovar Lai and human (cut-off value: similarity >70% and E value = 1e-10 for two serovars, E value = 1e-10 for serovar Lai and human) using BLASTP. We found 3672 orthologs between the two serovars, and 605 proteins that are similar in serovar Lai and human. Finally, 616 genes were yielded by the bioinformatics study by retaining the orthologs between the two serovars and subtracting the genes that were similar in serovar Lai and human.
Comparative genomic hybridization
We prepared a gene chip microarray corresponding to the complete genome sequence of L. interrogans strain #56601. The chips were hybridized to labelled total DNA extracted from strain Fiocruz L1–130 and ten pathogenic serovars. On the basis of test hybridizations of strain Fiocruz L1–130 vs. the reference sample, we considered genes that gave hybridization ratios between 1.0 and 3.0 to be present in both strains and greater than 10.0 to be absent from the test strain. Ambiguous values between 3.0 and 10.0 may have been due to highly divergent genes or hybridization to paralogous genes. The CGH results revealed that 307 genes of L. interrogans strain #56601 were absent or highly divergent in at least one strain tested. After subtracting these 307 differential genes, we were left with 565 genes, which not only encode presumably surface-exposed proteins but also are conserved in the ten pathogenic serovars.
Transcriptome analysis
Microarray analysis of the mRNA extracted from in vitro grown leptospires revealed that the fluorescence signals of Cy3 and Cy5 ranged from 10.5 to 51,707 (see Figure 1); 1427 genes were expressed above the median level (Cy3 signal ≥ 342 and Cy5 signal ≥ 363.5) in the microarray and therefore as genes with high transcriptional levels. The intersection between the sets of 565 and 1427 genes contained 226 genes. Among them, 8.0% (18/226) were located extracellularly, 53.1% (120/226) in the outer membrane, 16.4% (37/226) in the periplasmic space and 22.6% (51/226) in the inner membrane according to predictions. These vaccine candidates were classified further according their gene names and clusters of orthologous groups (COGs) [18,19](Table 1, 2, 3, 4); 60.6% (137/226) of the candidates had COG annotations.
Figure 1.

Identification of highly expressed genes in L. interrogans by microarray. Bacteria were grown in EMJH medium at 37°C and were collected when the culture reached mid-exponential-phase. RNA was purified and labelled with either Cy3 or Cy5 and hybridized with the microarray of L. interrogans strain #56601 (3528 genes). Transcription analysis revealed that 1427 genes were highly expressed (cy3 signal ≥ 342 and cy5 signal ≥ 363.5).
Table 1.
The result of vaccine candidates according to localization sites: extracellular
| gene | Cy3 signal | Cy5 signal | COG | product |
| LA0074 | 402.2 | 574.7 | - | hypothetical protein |
| LA0322 | 1,118 | 760.5 | - | hypothetical protein |
| LA0444 | 1,699 | 1,024 | COG1196D COG4254S | hypothetical protein |
| LA0587 | 3,246 | 2,998 | COG1075R | Lactonizing lipase |
| LA0617 | 672 | 487.5 | - | hypothetical protein |
| LA1433 | 1,946 | 1,552 | - | hypothetical protein |
| LA1508 | 559.2 | 853 | - | putative outermembrane protein |
| LA1569 | 755.3 | 391.3 | COG5651N | putative lipoprotein |
| LA2471 | 354.5 | 572.5 | COG0457R | putative outermembrane protein |
| LA2823 | 1,466 | 884 | - | putative lipoprotein |
| LA2975 | 478.8 | 454.2 | - | hypothetical protein |
| LA2992 | 663.8 | 535.3 | COG0419L | hypothetical protein |
| LA3210 | 410.7 | 847.7 | - | hypothetical protein |
| LA3338 | 899.8 | 795.8 | - | putative lipoprotein |
| LA3394 | 392.3 | 652 | - | hypothetical protein |
| LA3779 | 374.8 | 431.7 | - | hypothetical protein |
| LA3848 | 395.5 | 368.8 | - | putative lipoprotein |
| LB225 | 798 | 1,069 | - | hypothetical protein |
Table 2.
The result of vaccine candidates according to localization sites: outermembrane
| gene | Cy3 signal | Cy5 signal | COG | product |
| LA0049 | 471 | 478.2 | COG0840NT COG2202T | aerotaxis sensor receptor, flavoprotein |
| LA0099 | 1,081 | 853.5 | - | hypothetical protein |
| LA0166 | 2,149 | 1,113 | COG1196D | hypothetical protein |
| LA0178 | 1,224 | 953.8 | COG0706U | 60Kd inner membrane protein |
| LA0241 | 554.5 | 905.7 | COG1999R | SCO1/SenC family protein |
| LA0253 | 755.8 | 559.2 | COG2849S | hypothetical protein |
| LA0272 | 462.2 | 734.8 | - | hypothetical protein |
| LA0301 | 787 | 771.8 | COG2885M | outer membrane protein OmpA family |
| LA0330 | 554 | 397.3 | COG2366R | Penicillin G acylase precursor |
| LA0339 | 1,060 | 1,185 | COG0584C | Glycerophosphoryl diester phosphodiesterase |
| LA0365 | 1,290 | 1,959 | - | hypothetical protein |
| LA0370 | 720.7 | 952.8 | - | hypothetical protein |
| LA0378 | 692.5 | 560.3 | COG0457R | TPR-repeat-containing proteins |
| LA0379 | 1,134 | 957 | - | hypothetical protein |
| LA0410 | 3,451 | 3,604 | COG2834M | hypothetical protein |
| LA0423 | 553 | 712.8 | COG2931Q | hypothetical protein |
| LA0505 | 6,317 | 7,727 | COG1409R | probable glycosyl hydrolase |
| LA0532 | 736.5 | 684.7 | - | hypothetical protein |
| LA0568 | 434.8 | 435.3 | COG2067I | hypothetical protein |
| LA0635 | 1,131 | 712.8 | - | S-layer-like array protein |
| LA0678 | 973.3 | 952.5 | COG0840NT | Methyl-accepting chemotaxis protein mcpB |
| LA0811 | 478.8 | 770.5 | - | hypothetical protein |
| LA0818 | 481.3 | 946.8 | - | hypothetical protein |
| LA0878 | 886.7 | 555.7 | COG0266L | DshA protein |
| LA0940 | 1,467 | 1,026 | - | hypothetical protein |
| LA0957 | 2,798 | 1,542 | COG1538MU | outer membrane efflux protein |
| LA1009 | 764.8 | 887.7 | COG5009M | Penicillin-binding protein 1A |
| LA1010 | 1,124 | 829 | - | putative outermembrane protein |
| LA1087 | 376.5 | 536 | - | hypothetical protein |
| LA1099 | 2,388 | 1,500 | COG3103T | hypothetical protein |
| LA1100 | 2,901 | 2,796 | COG1538MU | outer membrane efflux protein |
| LA1161 | 474.2 | 403 | COG2067I | long-chain fatty acid transport protein |
| LA1174 | 615 | 423 | COG0834ET | amino acid ABC transporter, periplasmic amino acid-binding protein |
| LA1192 | 616.3 | 545.3 | - | putative outermembrane protein |
| LA1404 | 1,377 | 977 | - | putative outermembrane protein |
| LA1412 | 1,034 | 640.3 | - | hypothetical protein |
| LA1495 | 1,920 | 2,172 | - | putative outermembrane protein |
| LA1501 | 558.2 | 545.7 | COG4775M COG5009M | hypothetical protein |
| LA1507 | 1,615 | 1,747 | - | hypothetical protein |
| LA1690 | 744.7 | 471.7 | COG0449M | hypothetical protein |
| LA1733 | 1,418 | 1,557 | - | hypothetical protein |
| LA1912 | 873.8 | 745 | - | putative outermembrane protein |
| LA1917 | 595.5 | 535.3 | - | hypothetical protein |
| LA1931 | 941.5 | 1,540 | - | putative outermembrane protein |
| LA1987 | 909.3 | 994.5 | - | putative outermembrane protein |
| LA1996 | 556.8 | 674 | - | hypothetical protein |
| LA2024 | 2,594 | 2,079 | - | hypothetical protein |
| LA2063 | 1,463 | 1,967 | - | hypothetical protein |
| LA2094 | 548.2 | 380.3 | COG1716T | FHA-domain containing protein |
| LA2126 | 1,223 | 979.7 | COG0616OU | Putative signal peptide peptidase sppA |
| LA2215 | 1,045 | 679.2 | COG1196D COG1360N | Chemotaxis motB protein |
| LA2238 | 420 | 464.7 | COG0726G | polysaccharide deacetylase |
| LA2266 | 367.3 | 364.5 | - | putative outermembrane protein |
| LA2267 | 886.2 | 1,542 | COG0457R | putative outermembrane protein |
| LA2268 | 971.7 | 1,074 | - | putative outermembrane protein |
| LA2295 | 4,445 | 6,689 | COG0532J COG4254S | LipL45 protein |
| LA2318 | 813.2 | 673.8 | COG4775M | Predicted outer membrane protein |
| LA2368 | 347.8 | 585 | COG1555L COG3156U COG0477GEPR COG0075E | type II secretion pathway related protein etpK-like protein |
| LA2375 | 1,255 | 2,047 | COG1450NU | General secretory pathway protein D |
| LA2377 | 377.5 | 418 | COG0739M | peptidase, M23/M37 family protein |
| LA2395 | 847.3 | 1,736 | COG2815S | putative outermembrane protein |
| LA2413 | 540.7 | 381.2 | COG0791M | Probable lipoprotein nlpC precursor |
| LA2464 | 362 | 435.7 | COG3225N | gliding motility protein GldG |
| LA2468 | 3,653 | 6,205 | COG1196D | hypothetical protein |
| LA2510 | 1,230 | 846 | COG1452M | hypothetical protein |
| LA2537 | 1,329 | 1,304 | - | hypothetical protein |
| LA2538 | 624.2 | 606.5 | - | hypothetical protein |
| LA2612 | 532.8 | 574 | COG3190N | flagellar protein required for flagellar formation |
| LA2617 | 656.5 | 697.8 | - | hypothetical protein |
| LA2656 | 1,128 | 637.2 | COG2968S | hypothetical protein |
| LA2664 | 905.3 | 867.8 | COG1706N | flagellar P-ring protein precursor |
| LA2672 | 662.3 | 1,116 | - | hypothetical protein |
| LA2741 | 1,649 | 916.7 | - | hypothetical protein |
| LA2742 | 814.8 | 524.2 | - | hypothetical protein |
| LA2755 | 4,175 | 2,808 | COG0768M | probable penicillin-binding protein |
| LA2757 | 1,213 | 1,270 | COG1792M | rod shape-determining protein mreC |
| LA2800 | 665 | 1,591 | - | hypothetical protein |
| LA2818 | 681.7 | 440.8 | - | hypothetical protein |
| LA2857 | 506.7 | 516.5 | COG0596R | Predicted hydrolase or acyltransferase, alpha/beta hydrolase superfamily |
| LA2949 | 407.5 | 460 | COG0265O | heat shock protein, HtrA1 |
| LA3069 | 1,221 | 786.3 | - | hypothetical protein |
| LA3091 | 995.5 | 879.7 | - | hypothetical protein |
| LA3118 | 771.7 | 1,239 | COG0466O | hypothetical protein |
| LA3149 | 608.3 | 421.8 | COG1629P | Hemin receptor |
| LA3165 | 749.2 | 454.7 | COG4642S | conserved hypothetical protein with MORN repeat |
| LA3353 | 432.2 | 698.2 | - | hypothetical protein |
| LA3403 | 391.3 | 388.8 | - | hypothetical protein |
| LA3434 | 724.7 | 625.7 | COG0860M | N-acetylmuramoyl-L-alanine amidase |
| LA3440 | 915.5 | 864.3 | COG0237H | hypothetical protein |
| LA3468 | 618 | 584.8 | COG1629P | probable TonB-dependent receptor |
| LA3469 | 658.2 | 685.7 | COG3487P | iron-reglulated protein A |
| LA3506 | 1,407 | 1,266 | COG0840NT | Methyl-accepting chemotaxis protein |
| LA3552 | 1,652 | 2,900 | - | hypothetical protein |
| LA3632 | 1,028 | 1,337 | COG1413C | PBS lyase HEAT-like repeat containing protein |
| LA3681 | 463.5 | 459 | - | phage-related-like protein |
| LA3744 | 1,318 | 840.2 | - | hypothetical protein |
| LA3862 | 537.3 | 761.3 | COG0532J | hypothetical protein |
| LA3872 | 385.3 | 648.8 | COG0616OU | Putative signal peptide peptidase sppA |
| LA3938 | 589 | 1,026 | COG0457R | hypothetical protein |
| LA3970 | 532.7 | 398.5 | - | hypothetical protein |
| LA4070 | 9,764 | 5,630 | - | hypothetical protein |
| LA4212 | 1,678 | 1,684 | - | hypothetical protein |
| LA4227 | 2,465 | 1,953 | COG5621R | hypothetical protein |
| LA4232 | 509.3 | 612.2 | COG2982M | hypothetical protein |
| LA4261 | 485.7 | 612.5 | COG0451MG | UDP-glucose 4-epimerase |
| LA4263 | 1,012 | 1,290 | - | hypothetical protein |
| LA4285 | 726.3 | 791.2 | COG3858R | hypothetical protein |
| LA4341 | 1,009 | 1,146 | COG0739M | Peptidase family M23/M37 |
| LB018 | 1,549 | 1,589 | COG1635H | hypothetical protein |
| LB025 | 371.8 | 382.5 | - | hypothetical protein |
| LB050 | 344 | 533.3 | - | hypothetical protein |
| LB056 | 443.5 | 523.8 | COG0457R | TPR-repeat-containing protein |
| LB061 | 550.2 | 769.7 | COG3211R | hypothetical protein |
| LB191 | 344.3 | 410 | COG1629P COG4771P | putative TonB-dependent outer membrane receptor protein |
| LB199 | 917.3 | 925.2 | COG1629P | putative outermembrane protein |
| LB258 | 552.5 | 1,082 | COG4870O | Cysteine protease |
| LB277 | 1,634 | 984.3 | - | hypothetical protein |
| LB279 | 1,115 | 804.3 | COG1629P | hypothetical protein |
| LB328 | 1,591 | 2,672 | COG1360N COG2885M | outer membrane protein OmpA |
| LB362 | 1,246 | 7,69 | - | hypothetical protein |
Table 3.
The result of vaccine candidates according to localization sites: periplasmic
| gene | Cy3 signal | Cy5 signal | COG | product |
| LA0430 | 2,614 | 2,094 | COG1830G | hypothetical protein |
| LA0011 | 1472.2 | 2164 | - | putative lipoprotein |
| LA0093 | 963.2 | 539.3 | - | hypothetical protein |
| LA0107 | 476 | 466.3 | - | hypothetical protein |
| LA0222 | 9,873 | 18,863 | COG2885M | outer membrane protein OmpA family |
| LA0312 | 526.2 | 366.7 | COG0739M | M23/M37 family protein |
| LA0413 | 505.3 | 544.2 | - | hypothetical protein |
| LA0494 | 551 | 1,165 | - | hypothetical protein |
| LA0569 | 404.2 | 366.3 | - | hypothetical protein |
| LA0616 | 8,877 | 7,462 | COG0457R | outer membrane lipoprotein lipL41 |
| LA1118 | 610.2 | 614.3 | - | putative outermembrane protein |
| LA1136 | 636.5 | 1,301 | COG2834M | hypothetical protein |
| LA1155 | 534.3 | 563.8 | COG1613P | sulfate-binding protein precursor |
| LA1312 | 1,514 | 1,070 | - | hypothetical protein |
| LA1448 | 1,090 | 1,857 | COG1464P | putative outermembrane protein |
| LA1998 | 676 | 700.8 | COG0726G | polysaccharide deacetylase |
| LA2023 | 622 | 405 | COG2010C | cytochrome c |
| LA2208 | 2,252 | 2,334 | COG3858R | hypothetical protein |
| LA2277 | 609.5 | 391.3 | - | hypothetical protein |
| LA2316 | 633.3 | 707.2 | - | putative outermembrane protein |
| LA2372 | 1,427 | 2,257 | COG2165NU | General secretory pathway protein G |
| LA2531 | 1,177 | 894.5 | COG1196D | hypothetical protein |
| LA2637 | 51,707 | 37,602 | - | LipL32 protein |
| LA2748 | 714.5 | 537.3 | COG1613P | Sulfate-binding protein precursor |
| LA2820 | 691.3 | 525.5 | - | hypothetical protein |
| LA2950 | 373.8 | 661 | COG0265O | HtrA2 |
| LA2993 | 349 | 433.8 | - | hypothetical protein |
| LA3507 | 1,360 | 721.7 | COG2010C | putative cytochrome c |
| LA3535 | 541.2 | 659.8 | - | hypothetical protein |
| LA3571 | 607.2 | 492.8 | COG2010C | putative cytochrome c |
| LA3576 | 595.8 | 594.5 | COG1360N | flagellar motor protein |
| LA3780 | 1,365 | 1,432 | - | hypothetical protein |
| LA3839 | 664 | 618.3 | COG1881R | Phosphatidylethanolamine-binding family protein |
| LA3944 | 507.3 | 595.2 | - | hypothetical protein |
| LA4262 | 355 | 515.8 | - | hypothetical protein |
| LB047 | 506.3 | 2,137 | COG2849S | hypothetical protein |
| LB098 | 735.5 | 507.3 | COG0726G | Predicted xylanase/chitin deacetilase |
Table 4.
The result of vaccine candidates according to localization sites: innermembrane
| gene | Cy3 signal | Cy5 signal | COG | product |
| LA0238 | 662.5 | 433.2 | COG1612O | cytochrome-c oxidase assembly factor ctaA |
| LA0250 | 651.2 | 738.8 | COG4956R | TRAM family protein |
| LA0314 | 577.2 | 368 | COG0168P | Trk system potassium uptake protein trkH |
| LA0550 | 1,353 | 886.5 | COG0841V | NolG efflux transporter |
| LA0639 | 858.2 | 469.7 | - | hypothetical protein |
| LA0650 | 870.7 | 628 | COG0705R | Rhomboid family protein |
| LA0680 | 530.2 | 707.5 | COG0004P | hypothetical protein |
| LA0960 | 760.7 | 452 | - | hypothetical protein |
| LA1056 | 702.7 | 607.8 | COG0840NT | hypothetical protein |
| LA1143 | 4,027 | 4,074 | COG0341U | Preprotein translocase subunit SecF |
| LA1191 | 1,014 | 790.7 | COG0840NT | Methyl-accepting chemotaxis protein |
| LA1283 | 902.2 | 1,162 | COG0845M | hypothetical protein |
| LA1284 | 415.7 | 543 | COG4591M | Lipoprotein releasing system transmembrane protein lolC |
| LA1321 | 374.8 | 860.8 | COG4232OC | thiol:disulfide interchange protein DsbD |
| LA1397 | 722.8 | 672.3 | COG1033R | putative Protein export membrane protein SecD/SecF |
| LA1435 | 612.2 | 524.3 | COG0392S | hypothetical protein |
| LA1451 | 415.2 | 435.2 | COG1183I | Phosphatidylglycerophosphate synthase |
| LA1471 | 3,360 | 7,809 | COG3808C | Pyrophosphate-energized vacuolar membrane proton pump |
| LA1477 | 566.7 | 436.8 | COG1519M | 3-deoxy-D-manno-octulosonic-acid transferase |
| LA1535 | 521.5 | 685.5 | - | hypothetical protein |
| LA1554 | 498.7 | 398.2 | COG1502I | hypothetical protein |
| LA1695 | 4,493 | 2,360 | - | CrcB-like protein |
| LA1958 | 2,663 | 1,551 | COG0526OC | putative outermembrane protein |
| LA1979 | 483.8 | 657.8 | COG0463M | Putative glycosyl transferase |
| LA1982 | 342.5 | 452 | COG3307M | hypothetical protein |
| LA2050 | 411.3 | 848.8 | COG0707M | UDP-N-acetylglucosamine:LPS N-acetylglucosamine transferase |
| LA2250 | 10,742 | 9,624 | - | Nuclease S1 |
| LA2275 | 1,415 | 1,071 | COG0586S | dedA protein |
| LA2320 | 1,319 | 1,496 | COG0811U | biopolymer transport protein, putative |
| LA2604 | 464.3 | 448.7 | - | hypothetical protein |
| LA2737 | 3,813 | 2,157 | COG0204I | putative acyltransferase |
| LA2891 | 5,229 | 3,140 | COG1055P | hypothetical protein |
| LA3072 | 1,970 | 1,665 | COG0477GEPR | hypothetical protein |
| LA3110 | 1,262 | 2,371 | COG2156P | potassium-transporting ATPase, C chain |
| LA3146 | 877 | 523.2 | COG2076P | hypothetical protein |
| LA3577 | 1,618 | 1,198 | COG1291N | motility protein A |
| LA3586 | 2,348 | 1,746 | COG4270S | hypothetical protein |
| LA3754 | 667.3 | 449.7 | COG0681U | Signal peptidase I |
| LA3777 | 497.3 | 539 | COG0239D | Protein crcB homolog |
| LA3806 | 2,116 | 2,869 | COG0004P | Probable ammonium transporter |
| LA3916 | 5,518 | 5,510 | - | hypothetical protein |
| LA3926 | 967.8 | 1,802 | COG0841V | transmembrane efflux pump protein |
| LA4062 | 1,326 | 2,138 | - | hypothetical protein |
| LA4154 | 638.7 | 759 | COG3225N | hypothetical protein |
| LA4155 | 1,140 | 1,015 | COG1277R | probable permease of ABC transporter |
| LA4172 | 411 | 392.5 | - | hypothetical protein |
| LA4228 | 559.5 | 627.8 | COG4174R | Dipeptide transport system permease protein dppB |
| LA4233 | 409 | 985.7 | COG1172G | hypothetical protein |
| LA4269 | 1,907 | 2,240 | COG2207K COG0477GEPR | transcriptional regulator, AraC family |
| LB174 | 2,150 | 3,440 | COG0501O | heat shock protein HtpX |
| LB281 | 5,026 | 2,708 | COG0811U | transport protein ExbB |
Discussion
Vaccines composed of whole cells or outer membrane envelope are available in some countries to prevent human leptospirosis, and clinical trials have been reported [20-23]. In view of their disadvantages, especially their inability to elicit longer-term protection against different serovars of pathogenic leptospires, efforts have been focused on developing subunit vaccines[24]. During recent years, Hap1[25] (also known as LipL32[26]), LipL41, OmpL1[27] and Lig[28,29] proteins have been identified as promising vaccine candidates for preclinical trials.
The availability of complete genome sequence information for many pathogens and the development of sophisticated computer programs have led to a new paradigm in vaccine development. Now it is possible to screen potential vaccine candidate genes in a reverse manner starting from the genome. This reverse vaccinology was first applied to MenB[30] and is now applied routinely in vaccine development, as in the search for vaccines against S. pneumoniae, Streptococcus agalactiae, Staphylococcus aureus, Porphyromonas gingivalis, Chlamydia pneumoniae and other microorganisms[10]. Bioinformatics analysis is the first important strategy of reverse vaccinology. Gram-negative bacteria have five subcellular location sites: cytoplasm, inner membrane, outer membrane, periplasm and extracellular space. The surface-exposed proteins, i.e. those located in sites other than the cytoplasm, are the most suitable vaccine candidates because they are more susceptible to antibody recognition and can therefore elicit protective immune responses. Many sophisticated computer programs have been developed to predict the subcellular locations of putative proteins in the whole genome [31-33]. Analyzing the gene transcription profile using DNA microarrays provides a second vaccine candidate selection strategy in reverse vaccinology. A gene having a fluorescent signal above the median value corresponds to an expression level higher than 5–10 mRNA copies per genome[34]. Those highly expressed genes could be potential vaccine candidates[34]. Finally, other approaches such as proteomic technology can be used to screen vaccine candidates. Using combined these strategies, genes encoding potential vaccine antigens can eventually be identified.
In our preliminary selection, all genes in L. interrogans strain #56601 were searched using P-CLASSIFIER, a system for predicting the subcellular locations of proteins on the basis of amino acid subalphabets and a combination of multiple support vector machines[33]. Moreover, four topologies were predicted by the corresponding programs. Proteins predicted to be surface-exposed and having any of these four topologies were screened as preliminary vaccine candidates. All proteins with more than four predicted transmembrane spanning regions were removed from the list of candidates, not only because they are likely to be completely embedded in the cell membrane and therefore inaccessible to antibodies, but also because they are difficult to express in E. coli[34]. We retained the genes shared by the two sequenced serovars and subtracted genes that had human homologues. The reason we subtracted human homologues is they are likely to cause problems of autoimmunity[35]. Finally, we narrowed the list of vaccine candidates to 616 genes in the genome of L. interrogans strain #56601.
In order to explore vaccine candidates that could generate cross-protection against the diverse serovars of leptospires, we applied CGH to identify genes that are conserved among the ten pathogenic strains involved in most infections[36]. This approach allowed us to refine the vaccine candidate shortlist further by eliminating antigens that were not conserved among these serovars. The 565 vaccine candidates not only presumably surface-exposed but also conserved among the ten prevalent serovars in China were identified as the result of this approach.
Transcriptome analysis was performed using DNA microarrays of L. interrogans in order to assess the transcription levels of all genes in the genome. A graph of the signal obtained for each gene gave a diagonal distribution reflecting the expression level of that gene. After subtracting genes with transcriptional levels below the median, we were left with 226 genes as vaccine candidates.
Applying the theory of reverse vaccinology, 226 genes had been identified as potential vaccine candidates against L. interrogans combined bioinformatics, CGH and transcriptional analysis. Among them, 60.6% (137/226) have COG annotations; thus, nearly 40% either have an unknown function or have no COG annotation. This group of gene products offers great promise as it comprises a pool of previously unexploited vaccine targets. To evaluate our results, we compared our candidates with those identified by others. Gamberini et al. (2005) found approximately 20% potential surface proteins using in silico approach, and sixteen proteins were recognized by antibodies present in human sera[15]. However, only three of them (LA0222, LA2637 and LA2741) appear in our final set. This is not unexpected, since 206 genes encoding hypothetical or unknown proteins were selected from approximately 20% of the genome for cloning and expression. Nally et al. (2005) characterized 32 proteins in outer membrane vesicles of L. interrogans serovar Copenhageni by two-dimensional gel electrophoresis, including previously-described outer membrane proteins (OMPs); in addition, unknown, hypothetical and putative OMPs were also identified[17]. Interestingly, only two proteins (LA0222 and LA2637) are represented among the sixteen proteins found by Gamberini and co-workers. There is an overlap of eight genes between our result and that of Nally et al. (2005) (LA0222, LA0505, LA0616, LA1495, LA2024, LA2295, LA2637 and LA3091). The reasons responsible for the discrepancies among the results may be due to differing methodologies. Genomics, transcriptional profiling and proteomics have emerged in the post genomic era with potential to speed up the vaccine discovery research process. It should be pointed out that those methods have their respective advantages and limitations, and can be complementally utilized in the development of the novel vaccines. Genomics involves the use of various softwares to predict sublocalization of proteins. However, some algorithms have limited accuracy. Although transcriptome analysis uses gene chip array to measure gene expression but suffers from the fact that mRNA levels may not reflect protein levels. Expression of a transcribed gene may be regulated at the level of translation. It is believed that the proteome maps of microorganisms are important to understand cellular status at the protein level, which cannot be deciphered from genome or transcriptome analysis[37]. Proteomics of outer membrane can rapidly identify almost all proteins in outer membrane. However, some of the proteins identified in membrane preparations are in fact typical cytoplasmic proteins[10,38]. Moreover, one of the major disadvantages of subproteomic studies by 2-D gel electrophoresis and mass spectrometry is the potential for contamination via leaky fractionation or lysis[39]. Nally et al. (2005) also revealed that outer membrane vesicles contain small amounts of inner membrane or cytoplasmic proteins in their proteomic study[17]. It is worth mentioning here that mainly surface-exposed proteins such as LipL32 (LA2637)[26,40], LipL41 (LA0616)[27,40], LipL45 (LA2295)[41] and LipL21 (LA0011)[42] have higher transcriptional levels in our results; this suggests that the genes with higher transcriptional levels identified in our current research may be preferable for development as vaccine candidates.
This is the first time that CGH and transcription analysis have been used to identify potential candidates for vaccines against L. interrogans. Our present work corroborates previous studies, showing the advantages of reverse vaccinology[8,11]. The next step following our present research is to verify whether the selected vaccine candidates are surface-exposed and to evaluate the protective activities of these proteins. Such studies will lead to the development of safe and effective new vaccines against leptospirosis in the future.
Conclusion
We have performed high-throughput in silico and microarray-based processes that are useful for determining potential vaccine candidates against leptospirosis. In total, 226 genes were identified in the genome of L. interrogans serovar Lai type strain #56601 using bioinformatics, CGH and transcriptional analysis. The proteins encoded by these genes are not only potentially surface-exposed in the bacterium, but also conserved in two sequenced L. interrogans. Moreover, these genes are conserved among ten epidemic serovars in China and have high transcriptional levels in vitro. These proteins might therefore be useful for vaccine candidates as well as for the diagnosis of leptospirosis. Further research, including verification that these vaccine candidates are surface-exposed and evaluation their protective activities, will aid in the study of vaccines against leptospirosis in the future.
Methods
Bacteria strains and growth condition
Ten strains of L. interrogans were used in this study (Table 5). All the strains were obtained from the Institute for Infectious Disease Control and Prevention (IIDC), Beijing, China. Leptospires were maintained by serial passages in guinea pigs for preservation of virulence and were cultured in liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium at 28°C or 37°C with shaking under aerobic conditions. Culture conditions were then developed to ensure that only mid-exponential-phase bacterial cultures at a mean density of 106/ml were used in further experimentation. The cells were harvested by centrifugation at 10,000 g for 10 min at 4°C.
Table 5.
Bacterial strains used in the study
| serogroup | serovar | strain |
| Icterohaemorrhagiae | Lai | Lai(56601) |
| Canicola | Canicola | Lin |
| Pyrogenes | Pyrogenes | 4 |
| Autumnalis | Autumnalis | Lin 4 |
| Australis | Australis | 65-9 |
| Pomona | Pomona | Luo |
| Grippotyphosa | Linhai | Lin 6 |
| Hebdomadis | Hebdomadis | P 7 |
| Bataviae | Paidjan | L 37 |
| Sejroe | Wolffi | L 183 |
The L. interrogans serogroup Icterohaemorrhagiae serovar Lai type strain #56601 (strain Lai) was used to construct the DNA microarray. The genomic DNA of strain Fiocruz L1–130 was kindly provided by the Centro de Pesquisas Goncalo Moniz.
In silico analysis
Genes and protein data for human and for the sequenced L. interrogans (serovar Lai and serovar Copenhageni) were downloaded from NCBI. P-CLASSIFIER[33] was applied to predict the subcellular locations of proteins in L. interrogans strain #56601. Signal peptide prediction was carried out using SignalP 3.0[43]. α-Helix transmembrane topology prediction was carried out using TMHMM[44]. BOMP was used to predict β-barrel outer membrane proteins[45]. Putative lipoproteins were predicted by SpLiP[46]. To identify proteins orthologous between serovar Lai and serovar Copenhageni as well as between serovar Lai and human, all predicted proteins were searched against each other locally using BLASTP[47].
Comparative genomic hybridization
DNA microarrays of L. interrogans strain #56601 consisting of 3528 annotated ORFs longer than 250bp were prepared as previously described [48]. The genomic DNA of L. interrogans strain #56601 was used for reference in the double-fluorescence hybridization, and the genomic DNA of strain Fiocruz L1–130 was used as a control. A CGH microarray analysis of strain Lai and strain Fiocruz L1–130 was performed first. The qualified threshold determined in this control experiment was used to identify gene deletions in other strains. Reference or test DNA was fluorescently labelled through direct incorporation of Cy3-dCTP or Cy5-dCTP (Amersham Pharmacia Biotech) respectively by a randomly primed polymerization reaction. Unincorporated nucleotides and random primers were removed using QIAquick Nucleotide Removal columns (QIAGEN) according to the manufacturer's instructions.
Hybridizations were conducted in a hybridization chamber at 42°C overnight. Slides were washed at 55°C with 1 × SSC containing 0.2% SDS for 10 min and then at 55°C with 0.1 × SSC containing 0.2% SDS for 20 min and finally at room temperature with 0.1 × SSC for 3 min. Competitive hybridization was performed twice for each strain. In the first experiment, L. interrogans strain #56601 reference DNA and the sample DNA were labelled with Cy3 and Cy5, respectively. In the second hybridization, the dyes for labelling were interchanged.
Microarrays were scanned using a Chipreader laser scanner GenePix 4000B AXON (Axon Instruments, Union City, CA) according to the manufacturer's recommendations. Spot quantification, signal normalization and data visualization were performed using the programs GeneSpring 5.0.2 (Silicon Genetics) and Microsoft Excel.
Transcriptome analysis
L. interrogans was grown in EMJH medium at 37°C under aerobic conditions for transcriptome analysis. Only mid-log-phase cultures at a mean density of 106/ml in 100 ml were used in transcriptional experiments.
Total RNA was isolated from leptospires using Trizol reagent (Invitrogen) according to the manufacturer's protocol. Contaminating DNA was digested with RQ1 RNase-free DNase (Promega Corp.). The treated RNA was purified with a QIAGEN RNeasy Kit (QIAGEN).
RNA (10 μg) was labelled with Cy3 by reverse transcription using Superscript α (Invitrogen). Unincorporated dye was removed using a QIAquick Nucleotide Removal Kit (QIAGEN) as specified in the manufacturer's protocol. Samples were hybridized under cover slides to the microarray slides overnight at 42°C, and then washed as usual. The hybridization slides were processed by Tiffsplit (Agilent) and data were further analyzed using Genespring software 5.0.2 and normalized using mean values combined with Microsoft Excel software. Microarrays were used to assay relative RNA abundance. Flagged spots or SN<2 spots were excluded for intrachip and interchip reproducibility analysis. We calculated the coefficients of three spots in same chip for each gene to estimate intrachip reproducibility using Microsoft Excel. The signal values from the experiments represent average mRNA abundances. As in the CGH experiments, the dyes for labelling Cy3 and Cy5 were interchanged in the second hybridization.
Figure 2 is a scheme of the procedure we used to identify the vaccine candidates as described above (the numbers in parentheses are the results after the corresponding procedure step).
Figure 2.
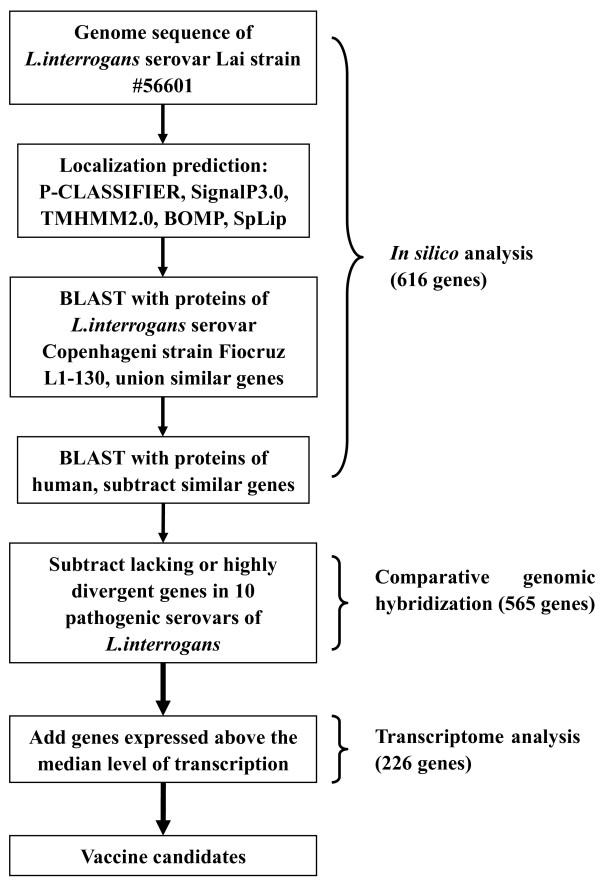
Schematic representation of general procedure to identify the vaccine candidates in the genome of L. interrogans (the numbers in parentheses are the results after the corresponding procedure step).
Authors' contributions
HLY and XKG designed the research project. HLY and YZZ carried out the bioinformatics analysis. PH and HLY completed the CGH. JHQ and HLY coordinated the transcriptome analysis. HLY and XKG drafted the manuscript. XCJ and GPZ participated in the design of the study and helped to draft the manuscript. All authors contributed to the writing and preparation of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Bao-Yu Hu and Yang Yang (Department of Microbiology and Parasitology, Shanghai Jiao Tong University School of Medicine) for help in bacterial culture preparation. This work was supported in part by grants from the National Natural Science Foundation of China (No.30370071 & 30670102), the National High Technology Research and Development Program of China and Shanghai Leading Academic Discipline Project (T0206). Moreover, we are grateful to the editors and anonymous reviewers for thoughtful comments on the manuscript.
Contributor Information
Hong-Liang Yang, Email: yanghongliang@sjtu.edu.cn.
Yong-Zhang Zhu, Email: zhuzhu198022@126.com.
Jin-Hong Qin, Email: jinhongq@hotmail.com.
Ping He, Email: hpatsh@hotmail.com.
Xu-Cheng Jiang, Email: xjiang@shsmu.edu.cn.
Guo-Ping Zhao, Email: gpzhao@sibs.ac.cn.
Xiao-Kui Guo, Email: microbiology@sjtu.edu.cn.
References
- Faine S, Alder B, Bolin C, Perolat P. Leptospira and Leptospirosis. Melbourne , Medisci; 1999. [Google Scholar]
- Human leptospirosis:guidance for diagnosis,surveillance and control. World Health Organization; 2003. [Google Scholar]
- de la Pena-Moctezuma A, Bulach DM, Kalambaheti T, Adler B. Comparative analysis of the LPS biosynthetic loci of the genetic subtypes of serovar Hardjo: Leptospira interrogans subtype Hardjoprajitno and Leptospira borgpetersenii subtype Hardjobovis. FEMS Microbiol Lett. 1999;177:319–326. doi: 10.1111/j.1574-6968.1999.tb13749.x. [DOI] [PubMed] [Google Scholar]
- Sonrier C, Branger C, Michel V, Ruvoen-Clouet N, Ganiere JP, Andre-Fontaine G. Evidence of cross-protection within Leptospira interrogans in an experimental model. Vaccine. 2000;19:86–94. doi: 10.1016/S0264-410X(00)00129-8. [DOI] [PubMed] [Google Scholar]
- Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, Vinetz JM. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3:757–771. doi: 10.1016/S1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- McBride AJ, Athanazio DA, Reis MG, Ko AI. Leptospirosis. Curr Opin Infect Dis. 2005;18:376–386. doi: 10.1097/01.qco.0000178824.05715.2c. [DOI] [PubMed] [Google Scholar]
- Koizumi N, Watanabe H. Leptospirosis vaccines: past, present, and future. J Postgrad Med. 2005;51:210–214. [PubMed] [Google Scholar]
- Rappuoli R, Covacci A. Reverse vaccinology and genomics. Science. 2003;302:602. doi: 10.1126/science.1092329. [DOI] [PubMed] [Google Scholar]
- Wack A, Rappuoli R. Vaccinology at the beginning of the 21st century. Curr Opin Immunol. 2005;17:411–418. doi: 10.1016/j.coi.2005.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruto D, Adu-Bobie J, Capecchi B, Rappuoli R, Pizza M, Masignani V. Biotechnology and vaccines: application of functional genomics to Neisseria meningitidis and other bacterial pathogens. J Biotechnol. 2004;113:15–32. doi: 10.1016/j.jbiotec.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Grandi G. Genomics and proteomics in reverse vaccines. Methods Biochem Anal. 2006;49:379–393. doi: 10.1002/0471973165.ch20. [DOI] [PubMed] [Google Scholar]
- Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, Zhang YX, Xiong H, Lu G, Lu LF, Jiang HQ, Jia J, Tu YF, Jiang JX, Gu WY, Zhang YQ, Cai Z, Sheng HH, Yin HF, Zhang Y, Zhu GF, Wan M, Huang HL, Qian Z, Wang SY, Ma W, Yao ZJ, Shen Y, Qiang BQ, Xia QC, Guo XK, Danchin A, Saint Girons I, Somerville RL, Wen YM, Shi MH, Chen Z, Xu JG, Zhao GP. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422:888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- Nascimento AL, Verjovski-Almeida S, Van Sluys MA, Monteiro-Vitorello CB, Camargo LE, Digiampietri LA, Harstkeerl RA, Ho PL, Marques MV, Oliveira MC, Setubal JC, Haake DA, Martins EA. Genome features of Leptospira interrogans serovar Copenhageni. Braz J Med Biol Res. 2004;37:459–477. doi: 10.1590/S0100-879X2004000400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento AL, Ko AI, Martins EA, Monteiro-Vitorello CB, Ho PL, Haake DA, Verjovski-Almeida S, Hartskeerl RA, Marques MV, Oliveira MC, Menck CF, Leite LC, Carrer H, Coutinho LL, Degrave WM, Dellagostin OA, El-Dorry H, Ferro ES, Ferro MI, Furlan LR, Gamberini M, Giglioti EA, Goes-Neto A, Goldman GH, Goldman MH, Harakava R, Jeronimo SM, Junqueira-de-Azevedo IL, Kimura ET, Kuramae EE, Lemos EG, Lemos MV, Marino CL, Nunes LR, de Oliveira RC, Pereira GG, Reis MS, Schriefer A, Siqueira WJ, Sommer P, Tsai SM, Simpson AJ, Ferro JA, Camargo LE, Kitajima JP, Setubal JC, Van Sluys MA. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J Bacteriol. 2004;186:2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamberini M, Gomez RM, Atzingen MV, Martins EA, Vasconcellos SA, Romero EC, Leite LC, Ho PL, Nascimento AL. Whole-genome analysis of Leptospira interrogans to identify potential vaccine candidates against leptospirosis. FEMS Microbiol Lett. 2005;244:305–313. doi: 10.1016/j.femsle.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Cullen PA, Xu X, Matsunaga J, Sanchez Y, Ko AI, Haake DA, Adler B. Surfaceome of Leptospira spp. Infect Immun. 2005;73:4853–4863. doi: 10.1128/IAI.73.8.4853-4863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nally JE, Whitelegge JP, Aguilera R, Pereira MM, Blanco DR, Lovett MA. Purification and proteomic analysis of outer membrane vesicles from a clinical isolate of Leptospira interrogans serovar Copenhageni. Proteomics. 2005;5:144–152. doi: 10.1002/pmic.200400880. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R, Perez A, Quinones Mdel C, Cruz R, Alvarez A, Armesto M, Fernandez C, Menendez J, Rodriguez I, Baro M, Diaz M, Rodriguez J, Sierra G, Obregon AM, Toledo ME, Fernandez N. [Efficacy and safety of a vaccine against human leptospirosis in Cuba] Rev Panam Salud Publica. 2004;15:249–255. doi: 10.1590/s1020-49892004000400005. [DOI] [PubMed] [Google Scholar]
- Yan Y, Chen Y, Liou W, Ding J, Chen J, Zhang J, Zhang A, Zhou W, Gao Z, Ye X, Xiao Y. An evaluation of the serological and epidemiological effects of the outer envelope vaccine to leptospira. J Chin Med Assoc. 2003;66:224–230. [PubMed] [Google Scholar]
- Martinez Sanchez R, Obregon Fuentes AM, Perez Sierra A, Baly Gil A, Diaz Gonzalez M, Baro Suarez M, Menendez Capote R, Ruiz Perez A, Sierra Gonzalez G, Lopez Chavez AU. [The reactogenicity and immunogenicity of the first Cuban vaccine against human leptospirosis] Rev Cubana Med Trop. 1998;50:159–166. [PubMed] [Google Scholar]
- Ikoev VN, Gorbunov MA, Vachaev BF, Iagovkin EA, Kondratenko VF, Anan'ina Iu V, Ansimova TI, Kostina NI, Iur'eva IL, Nikitin MG. [The evaluation of the reactogenicity and immunogenic activity of a new concentrated inactivated leptospirosis vaccine] Zh Mikrobiol Epidemiol Immunobiol. 1999:39–43. [PubMed] [Google Scholar]
- Zuerner R, Haake D, Adler B, Segers R. Technological advances in the molecular biology of Leptospira. J Mol Microbiol Biotechnol. 2000;2:455–462. [PubMed] [Google Scholar]
- Branger C, Sonrier C, Chatrenet B, Klonjkowski B, Ruvoen-Clouet N, Aubert A, Andre-Fontaine G, Eloit M. Identification of the hemolysis-associated protein 1 as a cross-protective immunogen of Leptospira interrogans by adenovirus-mediated vaccination. Infect Immun. 2001;69:6831–6838. doi: 10.1128/IAI.69.11.6831-6838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Chao G, Zuerner RL, Barnett JK, Barnett D, Mazel M, Matsunaga J, Levett PN, Bolin CA. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect Immun. 2000;68:2276–2285. doi: 10.1128/IAI.68.4.2276-2285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Mazel MK, McCoy AM, Milward F, Chao G, Matsunaga J, Wagar EA. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect Immun. 1999;67:6572–6582. doi: 10.1128/iai.67.12.6572-6582.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N, Watanabe H. Leptospiral immunoglobulin-like proteins elicit protective immunity. Vaccine. 2004;22:1545–1552. doi: 10.1016/j.vaccine.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Palaniappan RU, Chang YF, Hassan F, McDonough SP, Pough M, Barr SC, Simpson KW, Mohammed HO, Shin S, McDonough P, Zuerner RL, Qu J, Roe B. Expression of leptospiral immunoglobulin-like protein by Leptospira interrogans and evaluation of its diagnostic potential in a kinetic ELISA. J Med Microbiol. 2004;53:975–984. doi: 10.1099/jmm.0.45568-0. [DOI] [PubMed] [Google Scholar]
- Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, Galeotti CL, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood DW, Jeffries AC, Saunders NJ, Granoff DM, Venter JC, Moxon ER, Grandi G, Rappuoli R. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FS. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- Yu CS, Lin CJ, Hwang JK. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004;13:1402–1406. doi: 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sung WK, Krishnan A, Li KB. Protein subcellular localization prediction for Gram-negative bacteria using amino acid subalphabets and a combination of multiple support vector machines. BMC Bioinformatics. 2005;6:174. doi: 10.1186/1471-2105-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi G. Genomics, Proteomics and Vaccines. West Sussex , John Viley and Sons, Ltd; 2004. [Google Scholar]
- Allan E, Wren BW. Genes to genetic immunization: identification of bacterial vaccine candidates. Methods. 2003;31:193–198. doi: 10.1016/S1046-2023(03)00133-6. [DOI] [PubMed] [Google Scholar]
- Yu ES. Leptospira and Leptospirosis. Beijing: People's Medical Publishing House; 1992. [Google Scholar]
- Lee JW, Lee SY, Song H, Yoo JS. The proteome of Mannheimia succiniciproducens, a capnophilic rumen bacterium. Proteomics. 2006;6:3550–3566. doi: 10.1002/pmic.200500837. [DOI] [PubMed] [Google Scholar]
- Chakravarti DN, Fiske MJ, Fletcher LD, Zagursky RJ. Application of genomics and proteomics for identification of bacterial gene products as potential vaccine candidates. Vaccine. 2000;19:601–612. doi: 10.1016/S0264-410X(00)00256-5. [DOI] [PubMed] [Google Scholar]
- Rey S, Gardy JL, Brinkman FS. Assessing the precision of high-throughput computational and laboratory approaches for the genome-wide identification of protein subcellular localization in bacteria. BMC Genomics. 2005;6:162. doi: 10.1186/1471-2164-6-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro H, Croda J, Flannery B, Mazel M, Matsunaga J, Galvao Reis M, Levett PN, Ko AI, Haake DA. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect Immun. 2001;69:4958–4968. doi: 10.1128/IAI.69.8.4958-4968.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Matsunaga J. Characterization of the leptospiral outer membrane and description of three novel leptospiral membrane proteins. Infect Immun. 2002;70:4936–4945. doi: 10.1128/IAI.70.9.4936-4945.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PA, Haake DA, Bulach DM, Zuerner RL, Adler B. LipL21 is a novel surface-exposed lipoprotein of pathogenic Leptospira species. Infect Immun. 2003;71:2414–2421. doi: 10.1128/IAI.71.5.2414-2421.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Berven FS, Flikka K, Jensen HB, Eidhammer I. BOMP: a program to predict integral beta-barrel outer membrane proteins encoded within genomes of Gram-negative bacteria. Nucleic Acids Res. 2004;32:W394–9. doi: 10.1093/nar/gkh351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setubal JC, Reis M, Matsunaga J, Haake DA. Lipoprotein computational prediction in spirochaetal genomes. Microbiology. 2006;152:113–121. doi: 10.1099/mic.0.28317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin JH, Sheng YY, Zhang ZM, Shi YZ, He P, Hu BY, Yang Y, Liu SG, Zhao GP, Guo XK. Genome-wide transcriptional analysis of temperature shift in L. interrogans serovar lai strain 56601. BMC Microbiol. 2006;6:51. doi: 10.1186/1471-2180-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]


