Abstract
Background
Leptospira are the parasitic bacterial organisms associated with a broad range of mammalian hosts and are responsible for severe cases of human Leptospirosis. The epidemiology of leptospirosis is complex and dynamic. Multiple serovars have been identified, each adapted to one or more animal hosts. Adaptation is a dynamic process that changes the spatial and temporal distribution of serovars and clinical manifestations in different hosts. Serotyping based on repertoire of surface antigens is an ambiguous and artificial system of classification of leptospiral agents. Molecular typing methods for the identification of pathogenic leptospires up to individual genome species level have been highly sought after since the decipherment of whole genome sequences. Only a few resources exist for microbial genotypic data based on individual techniques such as Multiple Locus Sequence Typing (MLST), but unfortunately no such databases are existent for leptospires.
Results
We for the first time report development of a robust MLST method for genotyping of Leptospira. Genotyping based on DNA sequence identity of 4 housekeeping genes and 2 candidate genes was analyzed in a set of 120 strains including 41 reference strains representing different geographical areas and from different sources. Of the six selected genes, adk, icdA and secY were significantly more variable whereas the LipL32 and LipL41 coding genes and the rrs2 gene were moderately variable. The phylogenetic tree clustered the isolates according to the genome-based species.
Conclusion
The main advantages of MLST over other typing methods for leptospires include reproducibility, robustness, consistency and portability. The genetic relatedness of the leptospires can be better studied by the MLST approach and can be used for molecular epidemiological and evolutionary studies and population genetics.
Background
Leptospirosis is a zoonotic and an emerging infectious disease caused by the pathogenic Leptospira species and is identified in the recent years as a global public health problem because of its increased mortality and morbidity in different countries. Leptospirosis is frequently misdiagnosed as a result of its protean and non-specific presentation resembling many other febrile diseases, notably viral haemorrhagic fevers such as dengue [1]. There is, for certain, an underestimation of the leptospirosis problem due to lack of awareness and under-recognition through a lack of proper use of diagnostic tools.
The common mode of transmission of the infection in humans is either by direct or indirect contact with the urine of infected animals and may lead to potential lethal disease. A unique feature of this organism is to parasitize in a wide variety of wild and domestic animals [2]. Traditionally, two species have been identified, i.e. Leptospira interrogans and L. biflexa for pathogenic and non-pathogenic leptospires, respectively. The serovar is the basic identifier, characterized on the basis of serological criteria. To date nearly 300 serovars have been identified under the species L. interrogans alone that have been distributed among 25 different serogroups of antigenically similar serovars [3].
Previously a classification system based on DNA-DNA hybridization studies has been introduced, which now comprises 17 Leptospira species [4-7]. Among these, 7 species: L. interrogans, L. borgpetersenii, L. santarosai, L. noguchii, L. weilli, L. kirschneri and L. alexanderi are considered as the main agents of leptospirosis [5,6]. The enormous inventory of serovars, based mainly on an ever-changing surface antigen repertoire, throws an artificial and unreliable scenario of strain diversity. It is therefore difficult to track strains whose molecular identity keeps changing according to the host and the environmental niches they inhabit and cross through.
Other than the serological methods, molecular tools that have been employed so far for sub-classification and cataloguing of leptospiral agents include restriction endonuclease assay (REA) [8,9], pulsed field gel electrophoresis (PFGE) [10,11], restriction fragment length polymorphism (RFLP) [12], arbitrarily primed PCR [13], Variable Number of Tandem Repeats (VNTR) analysis [14] and fluorescent amplified fragment length polymorphism (FAFLP) [15]. All these techniques however, suffer from certain disadvantages that include requirement of large quantity of pure and high quality DNA, low discriminatory power, low reproducibility, ambiguous interpretation of data and problems associated with transfer of data between different laboratories [14].
MLST is a simple PCR based technique, which makes use of automated DNA sequencers to assign and characterize the alleles present in different target genes. The method allows one to generate sequence data in a low to high-throughput scale, which is unambiguous and suitable for epidemiological and population studies. The selected loci are generally the housekeeping genes, which evolve very slowly over an evolutionary time-scale [16] and hence qualify as highly robust markers of ancient and modern ancestry. The sequencing of multiple loci provides a balance between technical feasibility and resolution. MLST has been applied to the study of many other bacterial species like Neisseria meningitides [17], Streptococcus pneumoniae [18], Yersinia species [19], Campylobacter jejuni [20] and Helicobacter pylori [21].
Our present study is the first attempt to use the MLST, which currently differentiates the species and examines the intra and interspecies relationships of Leptospira. This method in future could be developed as a highly sophisticated genotyping system based on integrated genome analysis approaches to correctly identify and track leptospiral strains and is expected to greatly facilitate epidemiology of leptospirosis apart from deciphering the origins and evolution of leptospires in a global sense.
Methods
Bacterial strains
Bacterial strains (Table 1) were cultured by the WHO reference laboratory at the KIT Biomedical Research Centre at The Royal Tropical Institute, Amsterdam, The Netherlands (all isolates and reference strains labelled RK3) and at the Veterinary Sciences Division (VSD), The Queen's University of Belfast, United Kingdom (reference strains labelled RB3) and the WHO reference centre at Port Blair India (labelled isol 15). A total of 120 strains consisting of 79 isolates and 41 reference strains from different sources and geographical regions were analyzed by MLST. The 41 reference strains included in the study belonged to six Leptospira species (L. interrogans; L. kirschneri; L. noguchii; L. borgpetersenii; L. santarosai and L. alexanderi).
Table 1.
Details of leptospiral strains and isolates used for MLST based
| Labels | Genome Species | Serogroup | Serovar | Strain | Geographical area | Source |
| INT 01 | L. interrogans | Canicola | Sumneri | Sumner | Malaysia | RB3 |
| INT 02 | L. interrogans | Canicola | Portlandvere | MY 1039 | Jamaica | RB3 |
| INT 03 | L. interrogans | Pomona | Pomona | Pomona | Australia | RB3 |
| INT 04 | L. interrogans | Pomona | Proechimys | 1161 U | Panama | RB3 |
| INT 05 | L. interrogans | Pomona | Kenniwicki | LT 1026 | USA | RB3 |
| INT 06 | L. interrogans | Grippotyphosa | Grippotyphosa | Moskva V | Unknown | RB4 |
| INT 07 | L. interrogans | Grippotyphosa | Muelleri | RM 2 | Malaysia | RB3 |
| INT 08 | L. interrogans | Sejroe | Roumanica | LM 294 | Roumania | RB3 |
| INT 09 | L. interrogans | Sejroe | Saxkoebing | Mus 24 | Denmark | RB3 |
| INT 10 | L. interrogans | Sejroe | Hardjoprajitno | Hardjoprajitno | Indonesia | RB3 |
| INT 11 | L. interrogans | Icterohaemorrhagiae | Lai | Lai | China | RB3 |
| INT 12 | L. interrogans | Icterohaemorrhagiae | Copenhageni | M 20 | Denmark | RB3 |
| INT 13 | L. interrogans | Grippotyphosa | Valbuzzi | Valbuzzi | Australia | RB3 |
| INT 14 | L. interrogans | Pyrogenes | Manilae | LT 398 | Phillipins | RB3 |
| INT 15 | L. interrogans | Australis | Australis | Ballico | Ballico | RK3 |
| INT 16 | L. interrogans | Icterohaemorrhagiae | Icterohaemorrhagiae | RGA | Germany | RK3 |
| INT 17 | L. interrogans | Grippotyphosa | Ratnapura | Field Isolate 1 | South Andaman | Isol 15 |
| INT 18 | L. interrogans | Icterohaemorrhagiae | Copenhageni | Field Isolate 2 | South Andaman | Isol 15 |
| INT 19 | L. interrogans | Grippotyphosa | Ratnapura | Field Isolate 3 | South Andaman | Isol 15 |
| INT 20 | L. interrogans | Grippotyphosa | Ratnapura | Field Isolate 4 | South Andaman | Isol 15 |
| INT 21 | L. interrogans | Grippotyphosa | Valbuzzi | Field Isolate 5 | South Andaman | Isol 15 |
| INT 22 | L. interrogans | Icterohaemorrhagiae | Copenhageni | Field Isolate 6 | South Andaman | Isol 15 |
| INT 23 | L. interrogans | Grippotyphosa | Valbuzzi | Field Isolate 7 | North Andaman | Isol 15 |
| INT 24 | L. interrogans | Grippotyphosa | Valbuzzi | Field Isolate 8 | North Andaman | Isol 15 |
| INT 25 | L. interrogans | Grippotyphosa | Ratnapura | Field Isolate 9 | South Andaman | Isol 15 |
| INT 26 | L. interrogans | Grippotyphosa | Ratnapura | Field Isolate 10 | South Andaman | Isol 15 |
| INT 27 | L. interrogans | Grippotyphosa | Ratnapura | Field Isolate 11 | South Andaman | Isol 15 |
| INT 28 | L. interrogans | Grippotyphosa | Unknown | Field Isolate 12 | South Andaman | Isol 15 |
| INT 29 | L. interrogans | Grippotyphosa | Unknown | Field Isolate 13 | South Andaman | Isol 15 |
| INT 30 | L. interrogans | Sejroe | Sejroe | Field Isolate 14 | South Andaman | Isol 15 |
| INT 31 | L. interrogans | Pomona | Unknown | Field Isolate 15 | South Andaman | Isol 15 |
| INT 32 | L. interrogans | Grippotyphosa | Ratnapura | Field Isolate 16 | South Andaman | Isol 15 |
| INT 33 | L. interrogans | Australis | Ramisi | Field Isolate 17 | South Andaman | Isol 15 |
| INT 34 | L. interrogans | Grippotyphosa | Unknown | Field Isolate 18 | South Andaman | Isol 15 |
| INT 35 | L. interrogans | Grippotyphosa | Valbuzzi | Field Isolate 19 | South Andaman | Isol 15 |
| INT 36 | L. interrogans | Grippotyphosa | Valbuzzi | Field Isolate 20 | South Andaman | Isol 15 |
| INT 37 | L. interrogans | Hebdomadis | Goiano | Bovino 131 | Brazil | RB3 |
| INT 38 | L. interrogans | Canicola* | Canicola* | M12/90 | Brazil | Isol |
| INT 39 | L. interrogans | Icterohaemorrhagiae* | Copenhageni* | M9/99 | Brazil | Isol |
| INT 40 | L. interrogans | Australis* | Rushan* | L01 | Brazil | Isol |
| INT 41 | L. interrogans | Canicola* | Canicola* | L02 | Brazil | Isol |
| INT 42 | L. interrogans | Canicola* | Canicola* | L03 | Brazil | Isol |
| INT 43 | L. interrogans | Canicola* | Canicola* | L09 | Brazil | Isol |
| INT 44 | L. interrogans | Icterohaemorrhagiae* | Copenhageni* | L10 | Brazil | Isol |
| INT 45 | L. interrogans | Canicola* | Canicola* | L14 | Brazil | Isol |
| INT 46 | L. interrogans | Lyme* | Lyme* | K30B | UK | Isol |
| INT 47 | L. interrogans | Australis* | Australis* | K9H | UK | Isol |
| INT 48 | L. interrogans | Icterohaemorrhagiae* | Copenhageni* | Isolate 9 | Costa Rica | Isol |
| INT 49 | L. interrogans | Unknown* | Unknown* | Isolate 10 | Costa Rica | Isol |
| INT 50 | L. interrogans | Australis* | Lora* | 1992 | Tanzania | Isol |
| INT 51 | L. interrogans | Australis* | Lora* | 2324 | Tanzania | Isol |
| INT 52 | L. interrogans | Australis* | Lora* | 2364 | Tanzania | Isol |
| INT 53 | L. interrogans | Australis* | Lora* | 2366 | Tanzania | Isol |
| INT 54 | L. interrogans | Ballum* | Kenya* | 4885 | Tanzania | Isol |
| INT 55 | L. interrogans | Ballum* | Kenya* | 4883 | Tanzania | Isol |
| KIR 01 | L. kirschneri | Canicola | Kuwait | 136/2/2 | Kuwait | RB3 |
| KIR 02 | L. kirschneri | Canicola | Schueffneri | Vleermuis 90 C | Indonesia | RB3 |
| KIR 03 | L. kirschneri | Pomona | Mozdok | 5621 | Soviet Union (Russia) | RB3 |
| KIR 04 | L. kirschneri | Grippotyphosa | Vanderhoedeni | Kipod 179 | Israel | RB3 |
| KIR 05 | L. kirschneri | Pomona | Tsaratsovo | B 81/7 | Bulgaria | RB3 |
| KIR 06 | L. kirschneri | Grippotyphosa | Grippotyphosa | Moskva V | Russia | RK3 |
| KIR 07 | L. kirschneri | Grippotyphosa | Ratnapura | Wumalasena | Sri Lanka | RK3 |
| KIR 08 | L. kirschneri | Icterohaemorrhagiae* | Sokoine* | 745 | Tanzania | Isol |
| KIR 09 | L. kirschneri | Icterohaemorrhagiae* | Sokoine* | 771 | Tanzania | Isol |
| KIR 10 | L. kirschneri | Icterohaemorrhagiae* | Mwogolo* | 826 | Tanzania | Isol |
| KIR 11 | L. kirschneri | Icterohaemorrhagiae* | Mwogolo* | 845 | Tanzania | Isol |
| KIR 12 | L. kirschneri | Canicola* | Qunjian* | 2980 | Tanzania | Isol |
| KIR 13 | L. kirschneri | Icterohaemorrhagiae* | Sokoine* | 4602 | Tanzania | Isol |
| KIR 14 | L. kirschneri | Sejroe* | Ricardi/Saxkoebing* | 1499 | UK | Isol |
| KIR 15 | L. kirschneri | Sejroe* | Ricardi/Saxkoebing* | 1501 | UK | Isol |
| KIR 16 | L. kirschneri | Ballum* | Kenya | Njenga | Kenya | RK3 |
| NOG 01 | L. noguchii | Pyrogenes | Myocastoris | LSU 1551 | USA | RB3 |
| NOG 02 | L. noguchii | Louisiana | Louisiana | LSU 1945 | USA | RK3 |
| NOG 03 | L. noguchii | Panama | Panama | CZ214k | Panama | RK3 |
| NOG 04 | L. noguchii | Pyrogenes* | Guaratuba * | Isolate 4 | Costa Rica | Isol |
| SAN 01 | L. santarosai | Mini | Georgia | LT 117 | USA | RB3 |
| SAN 02 | L. santarosai | Sejroe | Recreo | 380 | Nicaragua | RB3 |
| SAN 03 | L. santarosai | Pyrogenes | Guaratuba | An 7705 | Brazil | RB3 |
| SAN 04 | L. santarosai | Pyrogenes | Varela | 1019 | Nicaragua | RB3 |
| SAN 05 | L. santarosai | Grippotyphosa | Canalzonae | CZ188 | Panama | RK3 |
| SAN 06 | L. santarosai | Bataviae* | Brasiliensis* | An 776 | Brazil | Isol |
| SAN 07 | L. santarosai | Sejroe* | Guaricura* | Bov.G | Brazil | Isol |
| SAN 08 | L. santarosai | Sejroe* | Guaricura* | M4/98 | Brazil | Isol |
| SAN 09 | L. santarosai | Grippotyphosa* | Bananal* | 2ACAP | Brazil | Isol |
| SAN 10 | L. santarosai | Grippotyphosa* | Bananal* | 16CAP | Brazil | Isol |
| SAN 11 | L. santarosai | Pyrogenes* | Alexi/Guaratuba/Princestown* | Isolate 1 | Costa Rica | Isol |
| SAN 12 | L. santarosai | Sarmin* | Weaveri/Rio* | Isolate 2 | Costa Rica | Isol |
| SAN 13 | L. santarosai | Tarassovi* | Rama* | Isolate 3 | Costa Rica | Isol |
| SAN 14 | L. santarosai | Tarassovi* | Rama* | Isolate 5 | Costa Rica | Isol |
| SAN 15 | L. santarosai | Bataviae* | Claytoni* | Isolate 6 | Costa Rica | Isol |
| SAN 16 | L. santarosai | Shermani* | Shermani/Babudieri/Aguaruna* | Isolate 8 | Costa Rica | Isol |
| SAN 17 | L. santarosai | unknown* | (putative new serovar)# | Isolate 7 | Costa Rica | Isol |
| SAN 18 | L. santarosai | Icterohaemorrhagiae* | Copenhageni* | K13A | UK | Isol |
| ALE 01 | L. alexanderi | Manhao | Manhao | L60 | China | RK3 |
| BOR 01 | L. borgpetersenii | Sejroe | Istarica | Bratislava | Slovakia | RB3 |
| BOR 02 | L. borgpetersenii | Sejroe | Sejroe | M 84 | Denmark | RB3 |
| BOR 03 | L. borgpetersenii | Javanica | Dehong | De 10 | China | RB3 |
| BOR 04 | L. borgpetersenii | Javanica | Javanica | Veltrat Batavia | Indonesia | RB3 |
| BOR 05 | L. borgpetersenii | Javanica | Zhenkang | L 82 | China | RB3 |
| BOR 06 | L. borgpetersenii | Javanica | Poi | Poi | Italy | RK3 |
| BOR 07 | L. borgpetersenii | Mini | Mini | Sari | Italy | RK3 |
| BOR 08 | L. borgpetersenii | Ballum* | Kenya* | 153 | Tanzania | Isol |
| BOR 09 | L. borgpetersenii | Ballum * | Kenya* | 159 | Tanzania | Isol |
| BOR 10 | L. borgpetersenii | Ballum * | Kenya* | 723 | Tanzania | Isol |
| BOR 11 | L. borgpetersenii | Ballum * | Kenya* | 766 | Tanzania | Isol |
| BOR 12 | L. borgpetersenii | Ballum * | Kenya* | 1605 | Tanzania | Isol |
| BOR 13 | L. borgpetersenii | Ballum * | Kenya* | 1610 | Tanzania | Isol |
| BOR 14 | L. borgpetersenii | Ballum * | Kenya* | 2062 | Tanzania | Isol |
| BOR 15 | L. borgpetersenii | Ballum * | Kenya* | 2348 | Tanzania | Isol |
| BOR 16 | L. borgpetersenii | Ballum * | Kenya* | 2447 | Tanzania | Isol |
| BOR 17 | L. borgpetersenii | Ballum * | Kenya* | 4880 | Tanzania | Isol |
| BOR 18 | L. borgpetersenii | Ballum * | Kenya* | 4787 | Tanzania | Isol |
| BOR 19 | L. borgpetersenii | Hebdomadis* | Kremastos/Hebdomadis* | 873 | Ireland | Isol |
| BOR 20 | L. borgpetersenii | Hebdomadis* | Kremastos/Hebdomadis* | 871 | Ireland | Isol |
| BOR 21 | L. borgpetersenii | Sejroe* | Saxkoebing* | 1498 | Ireland | Isol |
| BOR 22 | L. borgpetersenii | Sejroe* | Ricardi/Saxkoebing* | 1522 | UK | Isol |
| BOR 23 | L. borgpetersenii | Sejroe* | Ricardi/Saxkoebing* | 1525 | UK | Isol |
| BOR 24 | L. borgpetersenii | Pomona* | Kunming* | RIM 139 | Portugal | Isol |
| BOR 25 | L. borgpetersenii | Pomona* | Kunming* | RIM 201 | Portugal | Isol |
| BOR 26 | L. borgpetersenii | Sejroe* | Ricardi/Saxkoebing* | RIM 156 | Portugal | Isol |
* – Unpublished presumptive classification, # – Unpublished putative new serovar, Isol – Isolates, RB – reference strains from Belfast lab, RK – reference strains from KIT. The numbers 3, 4 and 15 refer to the references describing strains or isolates.
Selection and validation of target genes for MLST
The candidate loci sequences were obtained from the strains L. interrogans Fiocruz L1-130 and L. interrogans Lai 56601 strains from the Leptolist server. Six genes, namely adk (Adenylate Kinase), icdA (Isocitrate dehydrogenase), LipL32 (outer membrane lipoprotein LipL32), rrs2 (16S rRNA), secY (pre-protein translocase SecY protein), and LipL41 (outer membrane Lipoprotein LipL41) (Table 2) were selected for MLST analysis. Many sequences of the rrs2, LipL32 and LipL41 are available in the GenBank [2]. PCR primers were designed for these genes based on GenBank records in the conserved regions flanking the variable internal fragments of the target regions. PCR primers for adk, icdA and secY were based on gene sequences of strains Fiocruz L1-130 and Lai 56601 [22,23] (Table 2). The Primer 3 software [24] was used to design the PCR primers for the amplification of the candidate loci. The PCR amplifications of the different MLST target genes were performed using 1.5 mM MgCl2, 200 μM of dNTP's (MBI Fermentas), 25–50 ng template DNA using Gene Amp 9700 (Applied Biosystems, Foster City, USA) PCR system.
Table 2.
Details of gene loci and the corresponding primer sequences used for MLST analysis
| Gene | Locus | Gene size (bp) | Co-ordinates | PCR product size (bp) | Size of polymorphic sequence (bp) | Function | Primer sequences |
| adk | LIC12852 | 564 | 3458298–3458861 | 531 | 430 | Adenylate Kinase | F-GGGCTGGAAAAGGTACACAA |
| R-ACGCAAGCTCCTTTTGAATC | |||||||
| icdA | LIC13244 | 1197 | 3979829–3981025 | 674 | 557 | Isocitarate Dehydrogenase | F-GGGACGAGATGACCAGGAT |
| R-TTTTTTGAGATCCGCAGCTTT | |||||||
| LipL41 | LIC12966 | 1068 | 3603575–3604642 | 520 | 518 | Outermenbrane Lipoprotein LipL41 | F-TAGGAAATTGCGCAGCTACA |
| R-GCATCGAGAGGAATTAACATCA | |||||||
| rrs2 | LIC11508 | 1512 | 1862433–1863944 | 541 | 452 | 16S ribosomal RNA | F-CATGCAAGTCAAGCGGAGTA |
| R-AGTTGAGCCCGCAGTTTTC | |||||||
| secY | LIC12853 | 1383 | 3458869–3460251 | 549 | 549 | Translocase pre-protein secY | F-ATGCCGATCATTTTTGCTTC |
| R-CCGTCCCTTAATTTTAGACTTCTTC | |||||||
| LipL32 | LIC11352 | 819 | 1666299–1667117 | 474 | 474 | Outermenbrane Lipoprotein LipL32 | F-ATCTCCGTTGCACTCTTTGC |
| R-ACCATCATCATCATCGTCCA |
Amplification parameters included an initial denaturation at 95°C for 5 min followed by 35 cycles of amplification comprising of denaturation (94°C for 30 sec), annealing (58°C for 30 sec) and primer extension (72°C for 1 min) steps and a final extension of 7 min at 72°C. All the amplified fragments were checked on 1.5% or 2% agarose gel with ethidium bromide staining and the amplicons were sequenced in both the directions using Big Dye Terminator cycle sequencing Kit (Applied Biosystems, Foster City, USA) on ABI 3100 DNA sequencers (Applied Biosystems, Foster City, USA).
MLST data analysis
The electropherograms were viewed by using Chromas Lite version 2.01 (Technelysium Pty Ltd, Australia) and the resulting DNA sequences corresponding to both the forward and reverse reads were aligned using the Seqscape software (Applied Biosystems, Foster City, USA). Low quality nucleotide sequences were trimmed from the ends while comparing with the reference sequence of the Fiocruz strain and all the processed sequences were subsequently aligned by Clustal X [25]. The Sequence Type Analysis and Recombinational Test (START) programme [26] was used to determine Guanine-Cytosine content, number of polymorphic sites and the ratio of non-synonymous to synonymous nucleotide substitutions (dN/dS). The phylogenetic analysis was performed using concatenated (2980bp) sequences in the order adk, icdA, LipL32, LipL41, rrs2 and secY for each strain using MEGA 3.1 [27] and the consensus tree was drawn based on 1000 bootstrap replicates with Kimura 2 parameter.
Results
Diversity among the candidate loci analyzed
The 5' parts of rrs2, LipL32, LipL41 and the 3' part of secY were considered for the analysis based on abundance of nucleotide substitution positions found in these regions. The sizes of the fragments analyzed for the selected housekeeping genes ranged between 430bp (adk) and 557bp (icdA). The positions of these MLST loci were scattered throughout the chromosome I of L. interrogans Fiocruz L1-130 (Table 2). Clustal X programme was used to align all the individual sequences separately and we observed that there were no large insertions and deletions in the selected region. According to our analysis the rrs2 gene was found to be highly conserved among all the isolates with the percentage of variable sites being 4.42. Other genes namely LipL32, LipL41, icdA, adk and secY, however, were significantly diverse with the percentages of variable sites being 11.3, 21.04, 22.8, 27.2 and 28.7 respectively. The locus with highest diversity was icdA with 51 different alleles found among the set of 120 different isolates studied. The ratio of non-synonymous (dN) to synonymous substitution (dS) was much less than 1.0 indicating that these genes are not under positive selection pressure (the selection is against the amino acid change), whereas the rrs2 gene showed dN/dS ratio as 1.369 suggesting a high flexibility for amino acid changes. The percentage of G + C content in these loci ranged from 39.16 (secY) to 51.92 (rrs2) (Table 3). The synonymous substitution which, plays a role in the divergence of strains was more frequent in icdA and secY with 126 different synonymous sites. When compared to synonymous substitutions, non-synonymous substitutions were more frequent in all the genes tested, but highest numbers of 429 and 423 were observed in case of icdA and secY respectively (Table 3).
Table 3.
Allelic diversity parameters observed for the six target genes used for MLST analysis of leptospires
| Gene | G+C% | No. of alleles | Polymorphic sites | Synonymous sites | Non-synonymous sites | % of variable nucleotide sites | dN/dSratio |
| adk | 41.55 | 40 | 117 | 100 | 329 | 27.2 | 0.039 |
| icd1 | 40.9 | 51 | 127 | 126 | 429 | 22.8 | 0.017 |
| LipL32 | 46.46 | 36 | 54 | 112 | 362 | 11.3 | 0.091 |
| LipL41 | 42.88 | 52 | 109 | 123 | 393 | 21.04 | 0.055 |
| rrs2 | 51.92 | 29 | 20 | 112 | 338 | 4.42 | 1.369 |
| secY | 39.16 | 49 | 158 | 126 | 423 | 28.7 | 0.019 |
Clustering analysis of Leptospires based on MLST
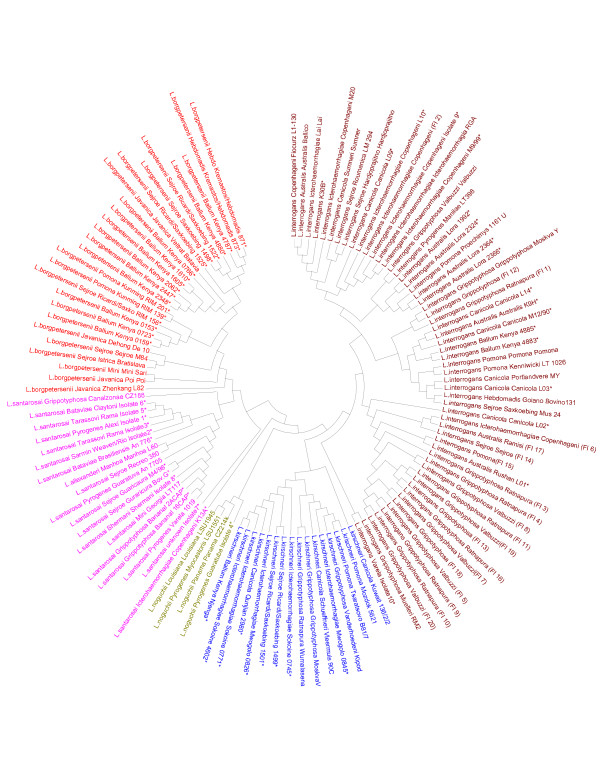
The neighbor-joining tree was constructed for representative isolates based on a 'super locus' of 2980bp comprising concatenated sequence of all the six loci. For this, the genes were fused in the order – adk, icdA, LipL32, LipL41, rrs2 and secY. The phylogenetic tree generated five different clusters where L. interrogans (56 samples), L. noguchii (4 samples), L. kirschneri (16 samples), L. santarosai (18 samples), L. alexanderi (1 sample), L. borgpetersenii (26 samples) separated according to their genome species (Figure 1).
Figure 1.
Genetic relatedness among Leptospira isolates based on the concatenated sequences of the six housekeeping and candidate gene loci analyzed (see table 1 for detailed information on isolates/strains). * Unpublished presumptive serological classification.
MLST analysis also clearly identified each of the field isolates up to the species level and in general, classification based on these observations corroborated with previous taxonomic status of these isolates determined either by serological criteria or by genomic methods such as FAFLP (data not shown). There are two isolates for which serological classification seemed to be in contrast to MLST identification, i.e. INT 46, L. interrogans serovar Lyme and SAN 18, L. santarosai serovar Copenhageni. It should be noted that in these cases serovar designation is based on preliminary serological analysis, which may be incorrect. L. alexanderi was found to be genomically highly similar to L. santarosai and clustered accordingly. This could therefore be a subspecies of L. santarosai.
L. interrogans isolate SAN 17 from Costa Rica, indicated as putative new serovar (Table 1) along with another L. interrogans member belonging to serovar Muelleri of the serogroup Grippotyphosa, formed an isolated branch under the L. interrogans cluster arguing for a separate taxonomic status, possibly another subspecies of L. interrogans.
Discussion
The present study was a first attempt in the development of MLST for Leptospira species; the main objective being the selection of the housekeeping and candidate genes that are species specific, stable and evolve slowly. The availability of the complete sequence of L. interrogans Lai 56601 and Fiocruz L1-130 helped us in selecting the candidate loci. Genetically diverse group of strains was used for the study to evaluate the sequence diversity among the tested housekeeping genes. The six genes selected and studied here appear to be distinctly resolving to reveal a wide variety of genotypes among the isolates analyzed. This indicates a significant heterogeneity and sequence variation at each locus (Table 3).
The six loci selected were found to be suitable for MLST typing as they can be amplified and sequenced in all the isolates irrespective of species as these loci are unlinked on the L. interrogans chromosome I and exhibit a modest degree of sequence diversity and resolution. A total of 585 polymorphic sites were observed in the 'super locus' of 2980bp. Non-synonymous sites were more abundant as compared to synonymous sites (Table 3) indicating that the amino acid sequence variability possibly represents acclimatization to the specific host and environmental restrictions [2].
Several molecular tools that have been so far described for the characterization of Leptospira are associated with several drawbacks. Methods like PFGE, RFLP, and REA need large quantity of purified DNA, present tedious methodology, have low discriminatory levels, are hard to interpret the data, suffer from lack of reproducibility, require specialized equipment such as counter clamped homogenous electric field electrophoresis systems and give poor data transfer. The VNTR or MLVA technique described by Majed et al [14] and Slack et al [28] are more specific to L. interrogans. MLST overcomes all these disadvantages as this technique is simple, and easy to standardize on an automated DNA sequencer that is more widely available in most of the laboratories and above all the sequence data generated are unambiguous, specific and explicit. The main advantage of MLST is the transfer of data that can be shared and compared between different laboratories easily through the Internet. To date, a large number of organisms have been typed by MLST, which proved to be a highly discriminatory technique [29]. MLST analysis on Leptospira strains showed that the similar serovars and the serogroups of different species are not clustered together (Figure 1). This method is more suitable in identifying the species of leptospires as indicated by the clustering patterns up to species level (Figure 1). The tree generated gives an idea on the phylogenetic organization of the Leptospira. The L. interrogans seems to be like a clonal branch as the isolates are more closely related and emerge from L. kirschneri indicating that they have evolved from this species. The L. interrogans and the L. kirschneri emerge from L. noguchii branch indicating it as a monophyletic group [2]. Due to the greater sequence diversity observed in all the six genes except rrs2, the dendrogram generated could differentiate effectively the L. interrogans, L. kirschneri, L. noguchii, L. santarosai and L. borgpetersenii.
Conclusion
With this new technique of MLST, we believe the issues related to ever-increasing serotype diversity would be effectively addressed via high throughput genome profiling. This will help establish population genetic structure of this pathogen with diverse host range and under different ecological conditions and will provide a scope for genotype-phenotype correlation to be established. Analyses based on the allelic profiles generated by our method may be successfully used to gain insights into the evolution and phylogeographic affinities of leptospires as it has been done for many other organisms. Large-scale, global genotyping, therefore, largely constitutes the essential mandate of studying leptospirosis in different hosts at the population level. Such approaches always generate extremely valuable information that can be translated into a wealth of databases to search for strain specific markers for epidemiology or to construct evolutionary history of the strains for a particular epidemiological catchment area. This task becomes greatly simplified if the genotypic data are categorized, stacked, archived and made electronically portable to facilitate easy access, extensive comparisons, remote access and retrieval in sets.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
NA and SMD carried out all the experiments related to primer designing, DNA sequencing and phylogenetic analyses and wrote the manuscript. NA and RAH designed the study and edited the manuscript. MDLAV, RSM, PV and WAE performed isolations of Leptospira. WAE and RAH performed serological and (other) molecular characterizations of the isolates, extracted DNA from isolates and reference strains and provided geographic and epidemiological data.
Acknowledgments
Acknowledgements
We thank Prof. Seyed E. Hasnain, University of Hyderabad, India for discussions and helpful suggestions. We thank three anonymous experts who served as referees for this work and their constructive suggestions have helped the manuscript a great deal to become worth publication. We also thank S. A. Vasconcello from the Univesidada de São Paulo, Brazil for providing some of the isolates and staff of the WHO/FAO/OIE Leptospirosis Reference Centre, KIT Biomedical Research for technical and material support in the (provisional) typing of Leptospira isolates. NA would like to thank Dept. of Biotechnology, Govt. of India for the financial support in terms of core grants to CDFD. Authors also acknowledge the financial support of the European Union (Lepto and dengue Project, INCO-Dev ICA4-CT-2001-10086 and RATZOOMAN Project, INCO-Dev ICA4-CT-2002-10056).
Contributor Information
Niyaz Ahmed, Email: niyaz.cdfd@gmail.com.
S Manjulata Devi, Email: manju@cdfd.org.in.
M de los Á Valverde, Email: mvalverde@inciensa.sa.cr.
P Vijayachari, Email: vijayacharip@yahoo.com.
Robert S Machang'u, Email: machangu2001@yahoo.com.
William A Ellis, Email: bill.ellis@dardni.gov.uk.
Rudy A Hartskeerl, Email: r.hartskeerl@kit.nl.
References
- Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Suchard MA, Kelley MM, Dundoo M, Alt DP, Zuerner RL. Molecular Evolution and Mosaicism of Leptospiral Outer Membrane Proteins Involves Horizontal DNA Transfer. J Bacteriol. 2004;186:2818–2828. doi: 10.1128/JB.186.9.2818-2828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmety E, Dikken H. Classification of the species Leptospira interrogans and history of its serovars. University Press Groningen The Netherlands; 1993. [Google Scholar]
- Brenner DJ, Kaufmann AF, Sulzer KR, Steigerwalt AG, Rogers FC, Weyant RS. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol. 1999;49:839–858. doi: 10.1099/00207713-49-2-839. [DOI] [PubMed] [Google Scholar]
- Ramadass P, Jarvis BDW, Corner RJ, Penny D, Marshall RB. Genetic characterization of pathogenic Leptospira species by DNA hybridization. Int J Syst Bacteriol. 1992;42:215–219. doi: 10.1099/00207713-42-2-215. [DOI] [PubMed] [Google Scholar]
- Yasuda BH, Steigerwalt AG, Sulzer LR, Kauhnann AF, Rogers F, Brenner DJ. Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int J Syst Bacteriol. 1987;37:407–415. [Google Scholar]
- Levett PN, Morey RE, Galloway RL, Steigerwalt AG. Leptospira broomii sp. nov., isolated from humans with Leptospirosis. Int J Syst Evol Microbiol. 2006;56:671–673. doi: 10.1099/ijs.0.63783-0. [DOI] [PubMed] [Google Scholar]
- Savio ML, Rossi C, Fusi P, Tagliabue S, Pacciarini ML. Detection and identification of Leptospira interrogans serovars by PCR coupled with restriction endonucleas eanalysis of amplified DNA. J Clin Microbiol. 1994;32:935–941. doi: 10.1128/jcm.32.4.935-941.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PD, Levett PN. Differentiation of Leptospira species and serovars by PCR-restriction Endonuclease analysis, arbitrarily primed PCR and low-stringency PCR. J Med Microbiol. 1997;46:173–181. doi: 10.1099/00222615-46-2-173. [DOI] [PubMed] [Google Scholar]
- Herrmann JL, Baril C, Belienger E, Perolat P, Baranton G, Girons IS. Genome conservation in isolates of Leptospira interrogans. J Bacteriol. 1991;173:7582–7588. doi: 10.1128/jb.173.23.7582-7588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JL, Bellenger E, Perolat P, Baranton G, Girons IS. Pulsed-field gel electrophoresis of NotI digests of leptospiral DNA: a new rapid method of serovars identification. J Clin Microbiol. 1992;30:1696–1702. doi: 10.1128/jcm.30.7.1696-1702.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuerner RL, Herrmann JL, Girons IS. Comparison of geneticmaps for two Leptospira interrogans serovars provides evidence for two chromosomes and intra species heterogeneity. J Bacteriol. 1993;175:5445–5451. doi: 10.1128/jb.175.17.5445-5451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perolat P, Merien F, Ellis WA, Baranton G. Characterization of Leptospira isolates from serovars hardjo by Ribotyping, arbitrarily primed PCR, and mapped restriction site polymorphisms. J Clin Microbiol. 1994;32:1949–1957. doi: 10.1128/jcm.32.8.1949-1957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majed Z, Bellenger E, Postic D, Pourcel C, Baranton G, Picardeau M. Identification of variable-number tandem-repeat loci in Leptospira interrogans sensu stricto. J Clin Microbiol. 2005;43:539–545. doi: 10.1128/JCM.43.2.539-545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayachari P, Ahmed N, Sugunan AP, Ghousunnisa S, Rao KR, Hasnain SE, Sehgal SC. Use of Fluorescent Amplified Fragment Length Polymorphism for Molecular Epidemiology of Leptospirosis In India. J Clin Microbiol. 2004;42:3575–3580. doi: 10.1128/JCM.42.8.3575-3580.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Spratt BG. Multilocus sequence typing. Trends Microbiol. 1999;7:482–487. doi: 10.1016/S0966-842X(99)01609-1. [DOI] [PubMed] [Google Scholar]
- Maiden MCJ, Bygraves JA, Spratt BG. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae : identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- Kotetishvili M, Kreger A, Wauters G, Morris JG, Sulakvelidze A, Stine OC. Multilocus Sequence Typing for Studying Genetic Relationships among Yersinia Species. J Clin Microbiol. 2005;43:2674–2684. doi: 10.1128/JCM.43.6.2674-2684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle KE, Colles FM, Wareing DRA, Ure R, Fox AJ, Bolton FE, Bootsma HJ, Willems RJL, Urwin R, Maiden MCJ. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi SM, Ahmed I, Khan AA, Rahman SA, Alvi A, Sechi LA, Ahmed N. Genomes of Helicobacter pylori from native Peruvians suggest admixture of ancestral and modern lineages and reveal a western type cag-pathogenicity island. BMC Genomics. 2006;7:191. doi: 10.1186/1471-2164-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko AI, Reis MG, Dourado CMR, Johnson WD, Jr, Riley LW. Urban epidemic of severe Leptospirosis in Brazil. Lancet. 1999;354:820–825. doi: 10.1016/s0140-6736(99)80012-9. [DOI] [PubMed] [Google Scholar]
- Nascimento AL, Ko AI, Martins EA, Monteiro-Vitorello CB, Ho PL, Haake DA, Verjovski-Almeida S, Hartskeerl RA, Marques MV, Oliveira MC, Menck CF, Leite LC, Carrer H, Coutinho LL, Degrave WM, Dellagostin OA, El-Dorry H, Ferro ES, Ferro MI, Furlan LR, Gamberini M, Giglioti EA, Goes-Neto A, Goldman GH, Goldman MH, Harakava R, Jeronimo SM, Junqueira-de-Azevedo IL, Kimura ET, Kuramae EE, Lemos EG, Lemos MV, Marino CL, Nunes LR, de Oliveira RC, Pereira GG, Reis MS, Schriefer A, Siqueira WJ, Sommer P, Tsai SM, Simpson AJ, Ferro JA, Camargo LE, Kitajima JP, Setubal JC, Van Sluys MA. Comparative genomics of two Leptospira interrogans pathogenesis. J Bacteriol. 2004;186:2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://frodo.wi.mit.edu/
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/S0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Jolley KA, Feil EJ, Chan MS, Maiden MC. Sequence type analysis and recombinational tests (START) Bioinformatics. 2001;17:1230–1231. doi: 10.1093/bioinformatics/17.12.1230. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and Sequence Alignment. Brief Bioinfor. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Slack AT, Dohnt MF, Symonds ML, Smythe LD. Development of a Multiple-Locus Variable number of tandem repeat Analysis (MLVA) for Leptospira interrogans and its application to Leptospira interrogans serovars Australis isolates from Far North Queensland, Australia. Annals Clin Microbiol and Antimicrobiol. 2005;4:10. doi: 10.1186/1476-0711-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden MCJ. High-throughput sequencing in the population analysis of bacterial pathogens of humans. Int J Med Microbiol. 2000;290:183–190. doi: 10.1016/S1438-4221(00)80089-2. [DOI] [PubMed] [Google Scholar]