Abstract
The genes encoding the enzymes of pyrimidine nucleotide biosynthesis (pyr genes) are regulated in Bacillus subtilis and many other bacterial species by transcriptional attenuation. When UMP or UTP is bound to the PyrR regulatory protein, it binds to pyr mRNA at specific sequences and secondary structures in the RNA. Binding to this site prevents formation of an antiterminator stem-loop in the RNA and permits formation of a downstream terminator, leading to reduced expression of the pyr genes lying downstream from the terminator. The functioning of several other transcriptional attenuation systems has been shown to involve transcriptional pausing; this observation stimulated us to use single-round transcription of pyr genes to test for formation of paused transcripts in vitro. Using templates with each of the three known B. subtilis pyr attenuation sites, we identified one major pause site in each in which the half-life of the paused transcript was increased four- to sixfold by NusA. In each case pausing at the NusA-stimulated site prevented formation of a complete antiterminator stem-loop, while it resulted in increased time for a PyrR binding loop to form and for PyrR to bind to this loop. Thus, the pausing detected in vitro is potentially capable of playing a role in establishing the correct timing for pyr attenuation in vivo. With two of three pyr templates the combination of NusA with PyrR markedly stimulated termination of transcription at the normal termination sites. This suggests that NusA, by stabilizing pausing, plays a role in termination of pyr transcription in vivo.
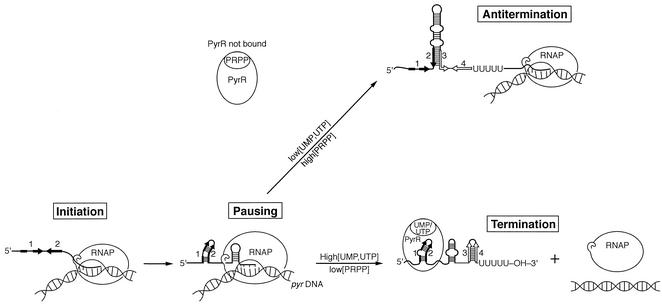
Transcription of the pyrimidine biosynthetic (pyr) operon in Bacillus subtilis and many other bacteria is regulated by a transcriptional attenuation mechanism involving the protein PyrR (for a review see reference 16). PyrR acts by binding in a uridine nucleotide-dependent fashion to pyr mRNA at three specific sites and altering the secondary structure of the downstream mRNA. In the absence of PyrR binding, the mRNA at each of the three sites folds into a large, stable stem-loop with internal bulges called the antiterminator (Fig. 1). Formation of the antiterminator stem-loop sequesters by base pairing sequences that are needed for the formation of a downstream factor-independent transcription terminator stem-loop. Under these conditions termination is prevented, and RNA polymerase continues to transcribe the coding sequences for the downstream pyr genes. Elevated levels of the nucleotides UMP and UTP promote the binding of PyrR to a specific sequence and secondary structure, called the binding loop, which overlaps the 5′ strand of the antiterminator (3). Binding of PyrR stabilizes the binding loop and prevents formation of the antiterminator, so that its 3′ strand is free to fold into the terminator stem-loop (Fig. 1). In the absence of PyrR binding, the antiterminator is the more stable secondary structure of the RNA. Thus, binding of PyrR promotes termination of transcription before the pyr gene coding sequences can be transcribed and provides a mechanism for repression of the pyr operon by exogenous pyrimidines. In Bacillus species this attenuation mechanism is cumulative for the downstream biosynthetic genes, because the binding loop-antiterminator-terminator RNA structures are found at three loci in the pyr operon, namely, the 5′ leader, between the first (pyrR) and second (pyrP) open reading frames, and between the second and third (pyrB) open reading frames. In other species only one or two attenuation regions are found in the pyr operon, or the pyr genes are scattered in multiple single genes and small operons, each containing a single attenuation region in its 5′ leader sequence (16).
FIG. 1.
Schematic representation of the roles of mRNA secondary structures and PyrR, as well as the possible role of pausing, in regulation of the B. subtilis pyr operon by transcriptional attenuation. Segments 1 and 2 of pyr mRNA base pair to form the PyrR binding loop; segments 3 and 4 base pair to form a terminator hairpin. Both structures are disrupted by base pairing of segments 2 and 3 as part of the antiterminator hairpin. See the text for details.
Many features of the recognition of RNA by PyrR have been characterized previously (3, 5, 14). In these studies the attenuation system was treated as though it exists in a simple equilibrium between two states: the antitermination conformation without PyrR bound and the binding loop-PyrR complex promoting termination. However, it is clear from an examination of the scheme for PyrR action shown in Fig. 1 that kinetic aspects of this scheme must also be important. For example, the binding loop structure must fold for PyrR to bind to it before the much more stable antiterminator stem-loop is fully transcribed. Otherwise, termination would always be prevented. This consideration provoked us to consider the hypothesis that RNA polymerase might pause after transcription of sequences 1 and 2 (Fig. 1) during transcription of the antiterminator stem-loop. Such pausing would allow time for the binding loop to form and for PyrR to bind if uridine nucleotide levels are sufficiently high to activate the PyrR protein. Transcriptional pausing has been shown to be important in the proper functioning of coupled transcription-translation attenuation systems, such as the Escherichiacoli trp and his operons and the E. coli pyrBI operon (7). Recently, Yakhnin and Babitzke (18) reported that NusA-stimulated transcriptional pausing occurs in the B. subtilis trpEDCFBA operon at sites that are consistent with the hypothesis that pausing plays a role in attenuation in that operon.
In this paper we demonstrate that pausing also occurs during in vitro transcription of each of three B. subtilis pyr attenuation regions. Pausing at one site in each attenuation region is stimulated by B. subtilis NusA; these NusA-stimulated pause sites are located at positions consistent with the hypothesis that they function in PyrR binding and attenuation.
MATERIALS AND METHODS
Materials.
Histidine-tagged B. subtilis RNA polymerase was purified from B. subtilis MH5636, provided by F. Marion Hulett, University of Illinois, Chicago, by affinity chromatography on an Ni-nitrilotriacetic acid column (1 by 5 cm) as described by Qi and Hulett (12). Histidine-tagged B. subtilis NusA protein was purified from E. coli M15(pREP4) containing the pQE30-NusA plasmid (6), which was provided by Tina Henkin, Ohio State University. The same purification procedure that was used for isolation of RNA polymerase was used to purify NusA. Both purified proteins were dialyzed against 10 mM Tris-acetate (pH 7.9)-0.1 mM EDTA-50% glycerol and stored at −80°C. They were more than 95% pure as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis; their concentrations were determined by the Bradford assay by using reagents from Pierce.
PCR primers were chemically synthesized by the University of Illinois Biotechnology Center. Dharmacon Research, Inc., chemically synthesized the GCUG RNA transcription primer. Ribonucleotides were obtained from Promega, and the 3′-deoxyribonucleotides used for RNA sequencing were obtained from TriLink Biotechnologies, Inc. Vent DNA polymerase and the PCR buffer were obtained from Invitrogen, Inc. All other chemicals were obtained from Sigma Chemical Co.
DNA templates.
A 591-bp PvuII fragment isolated from pUC19/290 (17) was used as the template for PCR amplification of transcription template 1, which was a 290-bp DNA segment containing the pyr promoter and 5′ untranslated leader from nucleotide (nt) −60 to 230 (numbering from the start of pyr transcription) (13). Template 2, a 238-bp DNA fragment amplified from a 560-bp PvuII fragment of pLS622 (10), specified the pyr promoter from nt −51 to 8 fused to the pyrR-pyrP intercistronic attenuation sequence from nt 670 to 848. Template 3, a 308-bp DNA fragment amplified from a 622-bp PvuII fragment of pLS601 (10), specified the pyr promoter from nt −51 to 8 fused to the pyrP-pyrB intercistronic attenuation region from nt 2132 to 2381. The templates were purified by extraction of the PCR products with phenol-chloroform-isoamyl alcohol (25:24:1) and precipitation of the DNA from the aqueous layer with cold absolute ethanol. The precipitated DNA was washed with cold 70% ethanol, dried, and dissolved in RNase-free water, and the concentration was determined from the absorbance at 260 nm. In some cases a Qiagen kit was used to purify the PCR-amplified template DNA.
Single-round transcription in vitro.
Two-step, single-round transcription of B. subtilis pyr DNA templates in vitro was used to examine the kinetics of transcription. In the first step, stable halted transcripts were formed during incubation of a mixture containing 50 nM RNA polymerase, 50 nM template DNA, the oligoribonucleotide GCUG corresponding to nt 1 to 4 of the pyr transcripts at a concentration of 20 μM, and three ribonucleotide triphosphates (rNTPs) for 15 min at 37°C in 10 to 100 μl of transcription buffer (40 mM Tris-acetate [pH 7.9], 10 mM magnesium acetate, 100 mM potassium acetate, 14.3 mM 2-mercaptoethanol, 2% glycerol). For template 1, 12-nt halted complexes were formed by incubation as described above with 20 μM GCUG, 8 μM ATP, 8 μM UTP, and 1.25 μM [α-32P]GTP in the absence of CTP. For templates 2 and 3, 11- and 12-nt halted complexes, respectively, were formed under the same conditions except that GTP was omitted for template 2 and the transcripts were labeled with 1.25 μM [α-32P]ATP. Electrophoretic analysis of the transcription products after 15 min of incubation demonstrated that halted transcripts of the expected length (11 or 12 nt) were the predominant transcripts formed.
After the 15 min of incubation for the first step of transcription, elongation of the halted transcripts was initiated by addition of a solution in transcription buffer containing all four rNTPs and 100 μg heparin per ml to block initiation of new transcripts. The concentrations of the rNTPs in the elongation reaction mixtures were chosen to optimize detection of pausing intermediates and were as follows: for template 1, 10 μM GTP and each of the other rNTPs at a concentration of 160 μM; for template 2, 10 μM ATP, 20 μM GTP, and each of the other rNTPs at a concentration of 160 μM; and for template 3, 10 μM ATP and each of the other rNTPs at a concentration of 160 μM. In some cases, NusA and PyrR were included in the elongation reaction buffer at the concentrations indicated below. Elongation proceeded at 25°C, and 2-μl samples were removed for analysis of transcripts by electrophoresis at various times from 10 to 300 s. Elongation was stopped by addition of an equal volume of 98% formamide containing 10 mM EDTA, 0.1% bromophenol blue, and 0.1% xylene cyanole. To test for elongation of putative transcription intermediates into full-length transcripts, an equal volume of a chase solution containing each of the four rNTPs at a concentration of 1 mM in transcription buffer was added to a 2-μl sample of the elongation reaction mixture, and the reaction was allowed to continue for 5 min at 37°C before it was stopped.
To map the sequences of pausing intermediates, RNA sequence ladders were analyzed by electrophoresis together with the other transcription reaction mixtures. These ladders were generated by including one of the four 3′-deoxynucleotide triphosphates in each of four elongation reaction mixtures at the same concentration as the corresponding rNTP in that mixture. the other deoxynucleoside triphosphate. The sequencing reaction mixtures were incubated for 20 min at 25°C.
Stopped transcription reaction mixtures were denatured by heating them for 2 min at 90°C and were analyzed on denaturing 6% polyacrylamide gels polymerized with acrylamide-bisacrylamide (19:1). The dried gels were visualized by autoradiography; transcripts were analyzed quantitatively with a PhosphorImager by using ImageQuant software (Molecular Dynamics), and the PhosphorImager intensities were corrected for the number of radiolabeled nucleotides per transcript. The half-lives of paused transcripts were determined from semilog plots of the abundance of these transcripts relative to the abundance of the total transcripts as a function of time of elongation (at least five times from 15 to 300 s), as previously described (8).
RESULTS
NusA-stimulated pausing occurs during transcription in vitro of templates from all three B. subtilis pyr attenuation regions.
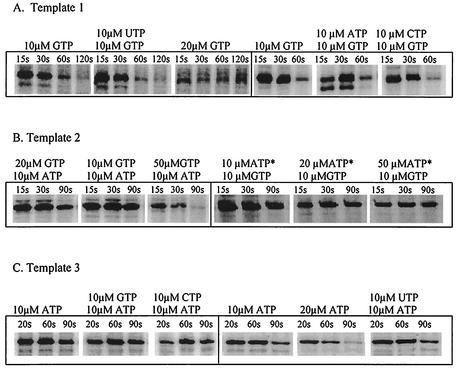
To test for pausing during transcription of pyr templates by B. subtilis RNA polymerase in vitro, we adapted the two-step, single-round transcription assay previously used for this purpose by workers in the laboratories of Landick (2, 8) and Babitzke (18). Transcription was initiated with the tetranucleotide GCUG and three rNTPs to allow formation of stable halted elongation complexes that were 11 or 12 nt long. For example, with the template specifying the pyr 5′ leader RNA, template 1, initiation with GCUG plus ATP, UTP, and GTP (GTP was radiolabeled in its α-phosphate with 32P) led to the stalled transcript 5′-GCUGAAUAGAUU-3′ because CTP was omitted from the initiation step (Fig. 2). Addition of heparin to block initiation of new transcripts plus all four rNTPs allowed synchronous elongation of the stalled transcripts. The concentrations of rNTPs added during the elongation phase were adjusted for each DNA template to give optimal detection of paused transcripts. Samples were taken at various times shortly after the elongation step was started, their lengths and sequences were analyzed by electrophoresis, and the amounts were quantified by PhosphorImager analysis. Pausing was defined by transient accumulation of transcripts of defined length during short elongation times (15 to 30 s) that disappeared with longer elongation times (i.e., they were chased into full-length terminated or runoff transcripts as predicted from the sequence of the template used).
FIG. 2.
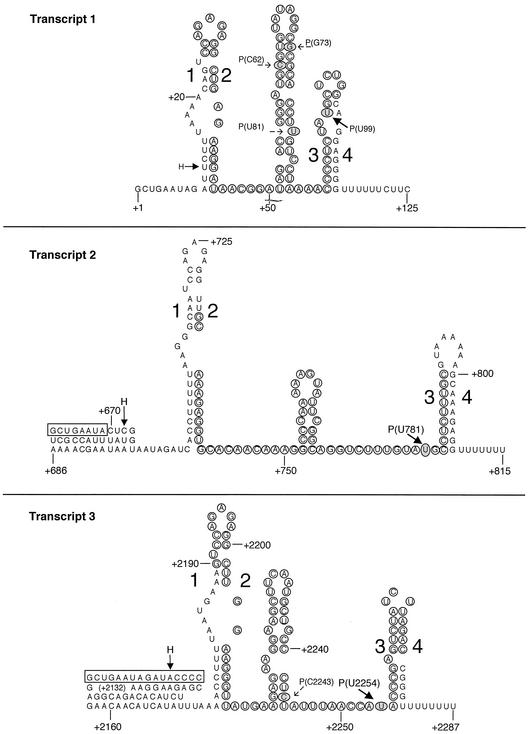
Sequences of the terminated transcripts from pyr templates 1, 2, and 3 in the binding loop-pause hairpin-terminator secondary structure. The nucleotide sequences are numbered from the start of transcription of the pyr operon (13), which is position 1. The sequences enclosed in boxes in transcripts 2 and 3 are alterations in the native transcripts that result from fusion of the B. subtilis pyr promoter plus nt 1 to 8 to intercistronic attenuation sequences. Circled nucleotides indicate sequences that form the stem-loop in the alternative antiterminator secondary structure for each transcript. Base-paired segments that form the binding loops are designated 1 and 2, and base-paired segments that form terminator stem-loops are designated 3 and 4, as shown in Fig. 1. The putative pause hairpins between the binding loops and the terminators were assigned as described in the Discussion. H indicates the 3′ nucleotides of the halted transcripts that were formed in the first step of the single-round transcription experiments described in the text. Major NusA-stimulated pause sites are indicated by P and a boldface arrow; minor pause sites are indicated by P and a dotted arrow.
Three DNA templates that were generated by PCR from previously described plasmids (10, 17) were used. RNAs transcribed from these templates are shown in Fig. 2. All three templates contained the wild-type B. subtilis pyr promoter at the 5′ end. Template 1 specified the pyr 5′ leader RNA from nt 1 to 230. Template 2 contained a fusion of the pyr promoter and nt 1 to 8 of the 5′ leader to nt 670 to 848 of the pyrR-pyrP intercistronic attenuation region. Template 3 contained a fusion of the pyr promoter and nt 1 to 8 of the 5′ leader to nt 2132 to 2381 of the pyrP-pyrB intercistronic attenuation region (with 8 nonnative nucleotides inserted as a consequence of the cloning methods used). Since the 5′ ends of the transcripts from all three templates were identical for the first 8 nt, the same initiation protocol with GCUC was used for all three, except that GTP was omitted from the initiation step for template 2 instead of CTP.
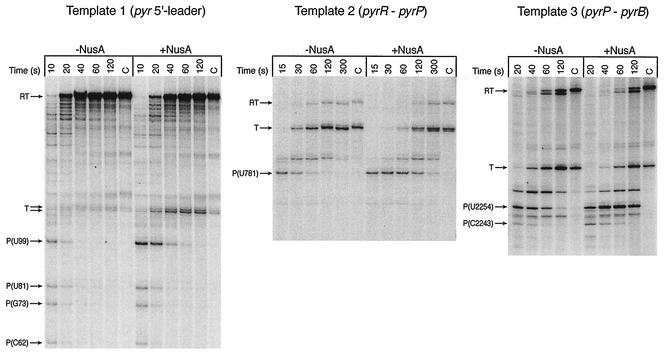
One or more paused transcripts were detected with each of the three pyr templates (Fig. 3). The lengths and sequences of these transcripts were determined by including RNA sequencing ladders in parallel lanes during electrophoretic analysis of the single-round transcripts. For template 1, the sites of pausing were at C62, G73, U81, and U99 (Fig. 3). The paused transcript at U99 was the most abundant, and its formation was much more sensitive to the addition of B. subtilis NusA than formation of the other transcipts (Fig. 4). With template 2, only one significant paused transcript, the 3′ terminus of which mapped to U781, was detected (Fig. 3), and its abundance and stability were strongly increased by NusA (Fig. 3 and 4). (A second, less abundant transient transcript with a 3′ terminus at C791, which accumulated much more slowly, was also observed; NusA did not alter its abundance.) Transcription of template 3 yielded two NusA-sensitive paused transcripts; one of these was at U2254 and was much more abundant and strongly affected by NusA than the other, which was at C2243 (Fig. 3). Two other possible paused transcripts were detected at A2249 and C2264, which were chased into readthrough and terminated transcripts, but their half-lives were longer than those of the other paused transcripts and were not affected much by NusA (Fig. 3).
FIG. 3.
NusA-stimulated transcriptional pausing from three pyr templates. Two-step, single round-transcription reactions were performed as described in Materials and Methods. The transcripts were analyzed after the times of elongation at 25°C shown in the presence and absence of 0.24 μM NusA. Lanes C contained the transcripts formed in a 5-min chase at 37°C in the presence of all four rNTPs at a concentration of 0.5 mM. RT and T indicate the positions of readthrough (runoff) and terminated transcripts, respectively. The 3′-terminal residues of the paused transcripts were mapped from sequencing ladders that are not shown.
FIG. 4.
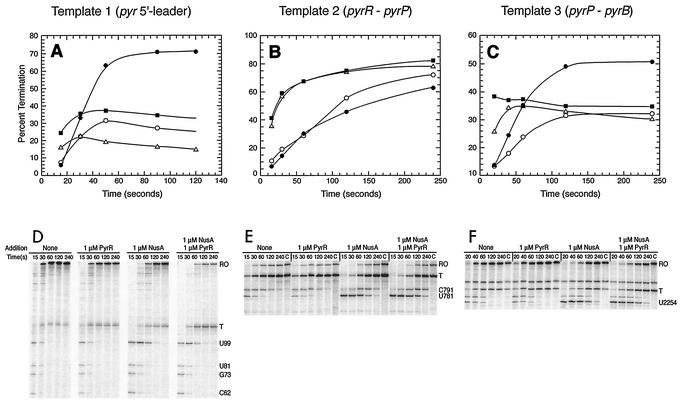
Effects of NusA and PyrR on transcriptional pausing and termination from three pyr templates. (A to C) Levels of termination calculated as described in the legend to Fig. 7 for different transcription elongation times with no addition (▵) and with 1 μM PyrR (▪), with 1 μM NusA (○), and with 1 μM PyrR plus 1 μM NusA (•) added. (D to F) Electrophoretic analysis of transcripts after various elongation times from which the data in panels A to C were calculated. The gels illustrate the patterns of paused transcripts formed under the conditions used. RO and T denote positions of runoff (read through) and terminated transcripts, respectively. C denotes a 5-min chase as in Fig. 3.
It was difficult on the basis of the sequencing ladders alone to assign the 3′ termini of the paused transcripts with a high degree of certainty. For example, the pause site at U99 appeared on some gels to be located instead at A98. To test our identification of the pause sites, we examined the effects of changing the concentration of the rNTP that should contribute the next nucleotide following a putative pause site. For example, increasing the concentration of GTP from 10 to 20 μM (while maintaining the specific radioactivity in [32P]GTP constant) in the elongation step with template 1 resulted in a two- to threefold decrease in the amount of NusA-stimulated pausing at U99, as predicted if the next nucleotide added after pausing was G100 (Fig. 5A). On the other hand, decreasing the concentration of UTP, CTP, or ATP from 160 to 10 μM had only relatively small effects on the amount of paused transcript (Fig. 5A). Similar specific effects of the GTP concentration on the amount of the paused transcript with template 2 confirmed our identification of U781 as the 3′ terminus of the paused transcript with that template (Fig. 5B). With template 3 increasing the concentration of ATP specifically decreased the amount of pausing, as expected for a paused transcript terminating at U2254 and followed by A2255 (Fig. 5C).
FIG. 5.
Effects of nucleotide concentration during the elongation step of transcription of pyr templates in vitro on the abundance of NusA-stimulated paused transcripts. The concentration of rNTPs during elongation was 160 μM, except as shown, and the NusA concentration was 0.24 μM. In each row the left three panels and the right three panels present separate experiments. (A) Template 1. The 3′ terminus of the paused transcript was identified as U99. Transcripts were labeled with [α-32P]GTP having a constant specific radioactivity. (B) Template 2. The 3′ terminus of the paused transcript was identified as U781. Transcripts were labeled with [α-32P]ATP having a constant specific radioactivity. An asterisk indicates that the concentration of RNA polymerase used was one-third the concentration used in the other experiments. (C) Template 3. The 3′ terminus of the paused transcript was identified as U2254. Transcripts were labeled with [α-32P]GTP having a constant specific radioactivity.
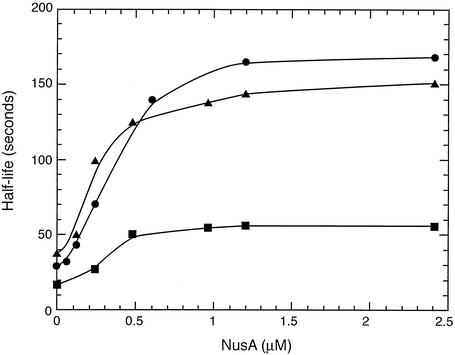
The dependence of the half-lives of the most abundant paused transcripts formed from each template on the concentration of NusA was similar for all three paused transcripts (Fig. 6). The concentration dependence curves were sigmoid and showed half-maximal effects at about 0.5 μM NusA. The half-lives of the paused transcripts were increased four- to sixfold by saturating NusA. In contrast to the three abundant paused transcripts, one from each template, the half-lives of other, minor putative paused transcripts observed with template 1 (C62, G73, and U81) and template 3 (C2243) were only increased twofold or less by saturating NusA.
FIG. 6.
Half-lives of the major paused transcripts as a function of NusA concentration in the elongation step. The 3′ termini of the paused transcripts were U99 from template 1(▪), U781 from template 2(•), and U2254 (▴) from template 3. Half-lives were determined from semilog plots of the abundance of the paused transcripts during 300 s of elongation as described in Materials and Methods.
Effects of NusA, PyrR, and PyrR plus NusA on pausing and termination.
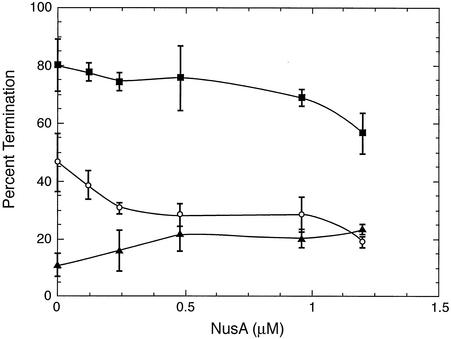
Because NusA has been shown to stimulate termination of transcription at certain factor-independent terminators (15), we examined its effects on termination during 5-min elongation reactions with the three pyr templates (Fig. 7). No consistent pattern emerged. Termination with template 1 was stimulated twofold by NusA, but termination with templates 2 and 3 was not affected and was decreased, respectively, by NusA. In all cases the site of termination was the same as that observed in previous multiple-round in vitro transcription studies without addition of NusA (9).
FIG. 7.
Effects of NusA concentration on the levels of termination of transcripts from pyr template 1 (▴), template 2 (▪), and template 3 (○). The level of termination was calculated from PhosphorImager analysis of the amounts of terminated transcripts divided by the sum of readthrough plus terminated transcripts after correction for the differences in the number of phosphate atoms per mole of transcript radiolabeled by [α-32P]GTP or [α-32P]ATP. Samples were analyzed after transcription elongation for 5 min at 25°C. The data are the means of three or more determinations; the error bars indicate the standard deviations of the means.
The effects of the PyrR regulatory protein with and without NusA on pausing and on termination were also studied with all three pyr templates (Fig. 4). PyrR and NusA were each added to the transcription elongation reaction mixtures at a concentration of 1 μM, at which they exerted about 90% of their maximal effects (Fig. 6 and 7; data not shown for PyrR). The concentration of UTP (160 μM) in the elongation reaction mixture was sufficient to fully activate PyrR binding to RNA (9). The level of termination as a function of elongation time is shown in Fig. 4; the effects on pausing were evaluated qualitatively from the autoradiograms of the transcription products shown in Fig. 4. Several generalizations emerged from an examination of the data in Fig. 4. Addition of PyrR had no effect on the pattern of transcriptional pausing in the presence or absence of NusA; PyrR altered neither the sites of pausing nor the half-lives of paused transcripts. For templates 1 and 3, PyrR increased termination, as expected from previous multiround in vitro transcription experiments (9), although the magnitude of the increase was smaller than that previously described. For templates 1 and 3 the combination of PyrR and NusA strongly increased termination. The behavior of transcription from template 2 in the experiments shown in Fig. 4 differed substantially from the behavior of transcription from the other two templates. Termination was quite high in all experiments and was not increased by PyrR or by PyrR plus NusA. As with the other templates, NusA appeared to suppress termination, although this effect disappeared with longer elongation times. We suggest that the differences may have resulted from the very high intrinsic level of termination seen with this template and from the strong effect of NusA on the formation and stability of the paused transcript from this template.
DISCUSSION
The sites of NusA-stimulated transcriptional pausing in vitro in the pyr operon are consistent with a role in attenuation in vivo.
With each of the pyr templates a major NusA-stimulated pause site was identified by our experiments. In each case the location of the pause site yielded a transcript that should permit formation of the upstream PyrR binding loop and the top of the antiterminator stem-loop, which presumably serves as part of the pausing signal. However, the paused transcripts do not contain the 3′ nucleotides that base pair with binding loop sequences and prevent formation of the binding loop. Likewise, the paused transcripts are not capable of forming the terminator stem-loop. Thus, we speculate that pausing could serve as a kinetic checkpoint in the attenuation mechanism. Paused transcripts should allow the binding loop RNA secondary structure to form and allow time for the PyrR-UMP/UTP complex to bind to it, if uridine nucleotides are present at a sufficiently high concentration. If PyrR does not bind to the paused transcripts within their relative short lifetimes, transcriptional elongation should continue, the complete antiterminator sequence should be transcribed, and the resulting antitermination should lead to elevated expression of the downstream pyr genes. While this is an attractive model, we emphasize that direct evidence for its functioning in vivo has not been obtained. Workers in our laboratory are currently attempting to test whether pausing plays a role in regulation of pyr gene expression in vivo.
Comparisons to transcriptional pausing in the B. subtilis trp operon.
Yakhnin and Babitzke (18) recently used an in vitro transcription system similar to that used in our experiments to demonstrate the occurrence of NusA-stimulated transcriptional pausing in the B. subtilis trp operon, which in its fundamental properties was quite similar to pausing with the three pyr templates. Specifically, the site of pausing was at a position that should allow binding of the terminator protein TRAP to the paused transcript but which should not permit antitermination or termination. NusA increased the half-lives of paused trp transcripts four- to fivefold, which was similar to the four- to sixfold stabilization of pyr pausing intermediates. However, the concentration dependence of stabilization by NusA differed for the two systems. Approximately fivefold-higher concentrations of NusA were required for half-maximal stabilization of pyr pause transcripts than for half-maximal stabilization of trp transcripts, and the NusA concentration dependence was sigmoid in the pyr system and hyperbolic for trp. A sigmoid concentration dependence of the effects of NusA on termination of transcription in the trp system was observed, however. TRAP interfered with pausing in the trp system, but PyrR did not affect pyr pausing. This difference is probably accounted for by differences in the binding sites for the two regulatory proteins on their respective RNA targets. The TRAP binding site overlaps much of the antiterminator stem-loop and probably interferes with formation of the hairpin at the top of the antiterminator that is part of the pausing signal. The binding sequences for PyrR (3) do not overlap the corresponding secondary structure at the top of the pyr antiterminators. Thus, PyrR binding interferes with antiterminator folding but should not affect stability of the putative pause hairpin. NusA alone also stimulated termination of trp transcription in vitro, but it did so with only one of the three pyr templates. However, with both systems the combination of NusA with the regulatory protein resulted in increased termination. It is possible that NusA participates not only in transcriptional pausing but also in termination in vivo with both of these attenuation systems.
What defines a pause site in B. subtilis?
The RNA structural elements that are important for transcriptional pausing in vitro have been well defined for E. coli RNA polymerase and coupled transcription-translation attenuation systems (7, 11). For such systems pausing at class I (NusA-stimulated) sites (1) occurs after an RNA hairpin consisting of at least 5 or 6 bp. The stem of this pause hairpin is separated by 10 or 11 nt from the 3′-terminal nucleotide of the paused RNA. Pausing is strongly favored when U or C lies at the 3′ terminus of the paused transcript. The available data on pause sites in B. subtilis are more limited, and no systematic study of the effects of structural variants on pausing, such as the study performed with the E. coli his pause site (4), has been conducted. The trp pause sites identified by Yakhnin and Babitzke (18) were at U residues located 11 nt downstream from the base of a well-defined RNA hairpin and were thus in accordance with the previously derived generalizations for E. coli pause sites. The NusA-stimulated pause sites identified in our work also fit reasonably well. However, the putative pause hairpins contain unpaired bases and are less readily defined than generally is the case for E. coli pause sites (Fig. 2). Pausing occurred at a U residue 12 nt downstream from a reasonable hairpin with pyr template 1, at a U residue 12 nt downstream from a rather unstable hairpin with pyr template 2, and at a U residue 10 nt downstream from one of three possible hairpins with pyr template 3. In summary, it appears from our limited observations that class I (1) pause sites for B. subtilis RNA polymerase are determined by the same general RNA structural features that characterize such pause sites in E. coli.
Acknowledgments
We are especially indebted to Alex Yakhnin and Paul Babitzke for communicating the results of their studies prior to publication, for invaluable assistance with the design of the two-step, single-round in vitro transcription system used in our studies, and for helpful comments on the manuscript. We also gratefully acknowledge Murali Palangat and Robert Landick for important assistance with experimental methodology for studies of transcription in vitro and for helpful comments on the manuscript. Charles Yanofsky is acknowledged for helpful comments on the manuscript.
This research was supported by Public Health Service grant GM47112 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Artsimovitch, I., and R. Landick. 2000. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. USA 97:7090-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artsimovitch, I., V. Svetlov, L. Anthony, R. R. Burgess, and R. Landick. 2000. RNA polymerases from Bacilus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro. J. Bacteriol. 182:6027-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonner, E. R., J. N. D'Elia, B. K. Billips, and R. L. Switzer. 2001. Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR. Nucleic Acids Res. 29:4851-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, C. L., and R. Landick. 1993. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J. Mol. Biol. 233:25-42. [DOI] [PubMed] [Google Scholar]
- 5.Ghim, S.-Y., and R. L. Switzer. 1996. Characterization of cis-acting mutations in the first attenuator region of the Bacillussubtilis pyr operon that are defective in regulation of expression by pyrimidines. J. Bacteriol. 178:2351-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundy, F. J., T. R. Moir, M. T. Haldeman, and T. M. Henkin. 2002. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: disparity between conservation and functional requirements. Nucleic Acids Res. 30:1645-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landick, R., C. L. Turnbough, and C. Yanofsky. 1996. Transcription attenuation, p. 1263-1286. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D. C.
- 8.Landick, R., D. Wang, and C. L. Chan. 1996. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: his leader pause site as paradigm. Methods Enzymol. 274:334-353. [DOI] [PubMed] [Google Scholar]
- 9.Lu, Y., and R. L. Switzer. 1996. Transcriptional attenuation of the Bacillus subtilis pyr operon by the PyrR regulatory protein and uridine nucleotides in vitro. J. Bacteriol. 178:7206-7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu, Y., R. J. Turner, and R. L. Switzer. 1995. Roles of the three transcriptional attenuators of the Bacillus subtilis pyrimidine biosynthetic operon in the regulation of its expression. J. Bacteriol. 177:1315-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mooney, R. A., I. Artsimovitch, and R. Landick. 1998. Information processing by RNA polymerase: recognition of regulatory signals during RNA chain elongation. J. Bacteriol. 180:3265-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi, Y., and F. M. Hulett. 1998. PhoP∼P and RNA polymerase σA holoenzyme are sufficient for transcription of Pho regulation promoters in Bacillus subtilis: PhoP∼P activator sites within the coding region stimulate transcription in vitro. Mol. Microbiol. 28:1187-1197. [DOI] [PubMed] [Google Scholar]
- 13.Quinn, C. L., B. T. Stephenson, and R. L. Switzer. 1991. Functional organization and nucleotide sequence of the Bacillus subtilis pyrimidine biosynthetic operon. J. Biol. Chem. 266:9113-9127. [PubMed] [Google Scholar]
- 14.Savacool, H. K., and R. L. Switzer. 2002. Characterization of the interaction of Bacillus subtilis PyrR with pyr mRNA by site-directed mutagenesis of the protein. J. Bacteriol. 184:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt, M. C., and M. J. Chamberlin. 1987. nusA protein of Escherichia coli is an efficient transcription termination factor for certain termination sites. J. Mol. Biol. 195:809-818. [DOI] [PubMed] [Google Scholar]
- 16.Switzer, R. L., R. J. Turner, and Y. Lu. 1999. Regulation of the Bacillus subtilis pyrimidine biosynthetic operon by transcriptional attenuation: control of gene expression by an mRNA-binding protein. Prog. Nucleic Acids Res. Mol. Biol. 62:329-367. [DOI] [PubMed] [Google Scholar]
- 17.Turner, R. J., Y. Lu, and R. L. Switzer. 1994. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J. Bacteriol. 176:3708-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yakhnin, A. V., and P. Babitzke. 2002. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism in vitro. Proc. Natl. Acad. Sci. USA 99:11067-11072. [DOI] [PMC free article] [PubMed] [Google Scholar]