Abstract
The huge ecological and economic impact of biological invasions creates an urgent need for knowledge of traits that make invading species successful and factors helping indigenous populations to resist displacement by invading species or genotypes. High genetic diversity is generally considered to be advantageous in both processes. Combined with sex, it allows rapid evolution and adaptation to changing environments.
We combined paleogenetic analysis with continent-wide survey of genetic diversity at nuclear and mitochondrial loci to reconstruct the invasion history of a single asexual American water flea clone (hybrid Daphnia pulex×Daphnia pulicaria) in Africa. Within 60 years of the original introduction of this invader, it displaced the genetically diverse, sexual population of native D. pulex in Lake Naivasha (Kenya), despite a formidable numerical advantage of the local population and continuous replenishment from a large dormant egg bank. Currently, the invading clone has spread throughout the range of native African D. pulex, where it appears to be the only occurring genotype.
The absence of genetic variation did not hamper either the continent-wide establishment of this exotic lineage or the effective displacement of an indigenous and genetically diverse sibling species.
Keywords: ancient DNA, Daphnia pulex, extinction, paleogenetics, parthenogenesis, sex
1. Introduction
Massive international transport of people and goods lead to the inadvertent creation of a new Pangaea, a world where previously isolated species and gene pools are mixed and homogenized in the absence of significant geographic barriers. Biological invasions pose an increasing threat to local ecosystems and global biodiversity (Mooney & Cleland 2001; Ricciardi 2004; Clavero & Garcia-Berthou 2005). Knowledge of the ecological and genetic processes behind biological invasions is essential to understand why and under which conditions some species tend to be more invasive than others. This information is needed to develop measures and tools to avoid or control future invasions, along with the ecological and economic damage caused by them. High genetic diversity is generally thought to promote invasion (Jain & Martins 1979; Kolbe et al. 2004; Frankham 2005a) because genetically diverse colonizers have greater evolutionary potential and hence a higher probability of successful establishment and spread in the newly colonized range. At the same time, high genetic diversity reduces the extinction risk of local populations (Saccheri et al. 1998; Spielman et al. 2004; Frankham 2005b) and thus contributes to resistance against invasions (Tagg et al. 2005). Therefore, it can be considered important for both the invader and the invaded.
Genetic techniques have been successfully used to trace the geographic origins of invasive species and populations (e.g. Havel et al. 2000; Saltonstall 2002; Bonizzoni et al. 2004). Similarly, we found in a previous study that all Kenyan populations of the water flea Daphnia pulex consist solely of a single asexual clone of American origin (Mergeay et al. 2005). Most of these studies, however, lack the historical perspective on the genetic aspects of colonization, establishment and biotic interactions that are needed for proper understanding of invasion dynamics (Sakai et al. 2001). For the present study, we used a paleogenetic approach involving application of genetic techniques to the fossil record (DNA sequencing and genotyping of dormant eggs) to reconstruct the historical dynamics of this invading water flea clone in Lake Naivasha, a 135-km2 shallow freshwater lake in Kenya that has been a centre of regional economic development, from early colonial times. The sedimentary archive of subfossil ephippia (protective chitinous capsules that surround dormant eggs of water fleas, analogous to plant seeds) of this lake provides a continuous 1800-year record of the Daphnia population in the past and community dynamics in relation to climate-driven aquatic ecosystem change, and more recently to human impact (Verschuren 2001; Mergeay et al. 2004; Mergeay 2005). Although old dormant eggs may no longer be viable, they often still contain sufficient amounts of unfragmented DNA for genetic analysis. Using polymorphic genetic markers on eggs recovered from different dated sediment depths, we reconstructed genetic changes through time in the resident population of D. pulex since the early colonial settlement around the lake.
2. Material and methods
(a) Sampling: sediment coring and field surveys
The material studied comprises two short sediment cores (NC91-1S: 90 cm, ca 1870–1991 AD; NC01-1S: 158 cm, ca 1810–2001 AD) and a long core (NC01-D: 88–764 cm), retrieved near the deepest point of Lake Naivasha in July–August of 1991 and 2001, at a depth of 14.50 m. Coring procedures and dating details are provided in the electronic supplementary material. Subfossil Daphnia ephippia were extracted from contiguous 2 cm core increments by washing 15–70 ml of untreated sediment through a 150 μm mesh sieve and scanning the retained residue at 10–90× magnification under a stereo-dissecting microscope.
Field surveys in Ethiopia, Kenya and South Africa between 2001 and 2004, supplemented with limited samples from Botswana and Zimbabwe, yielded zooplankton collections from 177 African standing waters. Live zooplankton was collected using a conical tow net (25 cm diameter) of 150 μm mesh, washed in the net and fixed in 100% ethanol. In some waters, sediment samples containing ephippia were also taken (Mergeay et al. 2005) and treated as described earlier.
(b) DNA extraction, amplification and analyses
Ephippia of D. pulex were opened and single dormant eggs picked out and transferred to UV-sterilized 200 μl microfuge tubes for DNA extraction. Details of DNA extraction and PCR amplification are provided in the electronic supplementary material. Ten microsatellite loci (Dpu7, Dpu40, Dpu45, Dpu46, Dpu12/2, Dp183, Dp525alt, Dp464, Dp502 and Dp496; Colbourne et al. 2004) were used to assess genetic variation within and between modern populations of D. pulex. Three loci (Dpu7, Dpu40 and Dp183) that gave satisfactory yields in old dormant eggs from Lake Naivasha were retained for paleogenetic population analysis through time (12 successive time-averaged subpopulations, total n=517). Measures of allelic richness and 95% confidence intervals within subpopulations were calculated using the Fstat v.2.9.3 software package (Goudet 2001).
We used DNA sequencing for species identification of paleolimnological and modern samples, and for phylogeographic analyses of contemporary African samples, sequencing the 12S rRNA mitochondrial gene (12S) and subunit 5 of the NADH mitochondrial gene (ND5). The details are provided in the electronic supplementary material. Phenetic analyses were performed in Mega3 (Kumar et al. 2004) using the neighbour-joining algorithm with the K2P genetic distance method and 1000 bootstraps for branch support. African D. pulex haplotypes of ND5 were compared with 81 unique haplotypes of this species complex (Colbourne et al. 1998), of which a subset was retained for phenetic analysis (figure 3a). DNA sequences were deposited in GenBank, under accession numbers DQ235230–DQ235245 and AY745243.
Figure 3.
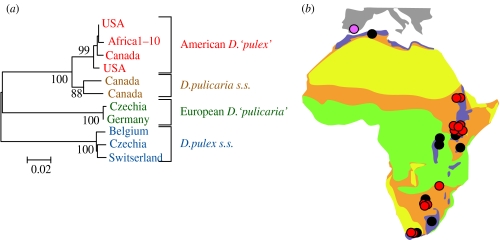
Geographical distribution of the invading American D. pulex clone in Africa. (a) Genetic tree obtained from an ND5 gene fragment showing the relationship of contemporary African D. pulex populations to other members of the D. pulex complex. Although morphologically indistinguishable, American and Old World D. pulex are genetically two distinct species (Colbourne et al. 1998). Scale bar indicates K2P genetic distance. Numbers at nodes indicate bootstrap support. (b) Comparison of the known distribution of the invading clone (red circles) with historical records of D. pulex in Africa (black circles). The pink circle shows the record of the same American 12S haplotype in Spain. Green: tropical forest; yellow: (semi)desert; lilac: temperate and mountain forest/grassland; orange: steppe and savannah.
3. Results
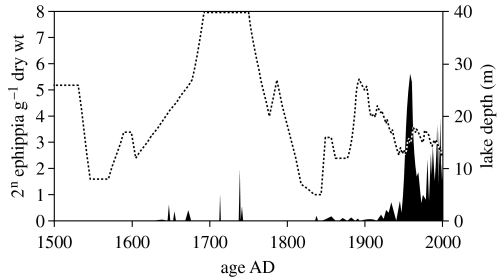
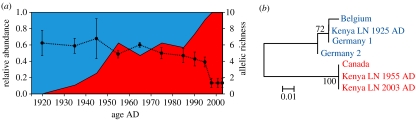
Daphnia pulex first colonized Lake Naivasha around 1630 AD (figure 1), but in the following century developed a sizeable population only intermittently and apparently disappeared around 1750. It colonized the lake a second time during a pronounced lowstand in the early nineteenth century, and from 1850 to ca 1930, it was continuously present at low densities. Since then, it enjoyed modest population expansion in the 1930s, a collapse during the 1940s lowstand episode, explosive population growth during lake-level rise in the late 1950s and early 1960s, and intermediate to high population densities prevailing since the 1970s (figure 1). Genetic analyses of three polymorphic microsatellite loci in dormant eggs of D. pulex (n=517) deposited from the 1920s to date revealed a striking change in the genetic structure of the Lake Naivasha population. Early in the twentieth century, it was characterized by a high genetic diversity as estimated by the allelic richness of the microsatellite loci (figure 2a). In the mid-1930s, a new multilocus genotype (MLG) appeared (Lake Naivasha genotype 1: LN1, a fixed heterozygote), which steadily increased in relative frequency until 1955, when it accounted for about 60% of the total population. After a ca 30-year period of relative genetic stability, this clone expanded further during the late 1980s and 1990s, until 1998, when it had completely displaced all other D. pulex genotypes. The overall genetic diversity of the Lake Naivasha population decreased gradually between the 1950s and the mid-1990s (figure 2a). Currently, LN1 is the only MLG detected in Lake Naivasha (n=300) and six other Kenyan waters studied (n=111) with the sporadic exception of derived genotypes: 2 out of 300 individuals from Lake Naivasha differed at one locus by a single microsatellite repeat unit (Mergeay et al. 2005). This transition in population genetic structure was confirmed by a parallel shift in mitochondrial haplotypes. Sequence data from the 12S gene revealed the presence of two highly distinct phylogenetic lineages in Lake Naivasha during the course of the past 100 years (figure 2b). In dormant eggs deposited before 1937 (n=6), we found only one haplotype belonging to the phylogenetic clade that groups European, probably most indigenous Asian and (as revealed here) African populations of the D. pulex species complex (the Old World clade; Colbourne et al. 1998). Among dormant eggs deposited after 1937, we found a second haplotype, which is nested in the North American clade of D. pulex (figure 2b). All analysed individuals with this American 12S haplotype (n=13) shared the nuclear microsatellite MLG LN1, while Old World haplotypes had different MLGs. Combining the data of microsatellite MLGs and mitochondrial DNA haplotypes, our data show the cryptic invasion of a single exotic genotype and subsequent gradual but steady displacement of a genetically diverse population of native D. pulex by that North American genotype within 60 years.
Figure 1.
Demographic changes in the Lake Naivasha Daphnia pulex population through time. Changes in ephippium density since 1500 AD. Stippled line shows climate-driven lake-level change through time (Verschuren 2001), highlighting the marked natural variability of the abiotic environment and aquatic habitat in Lake Naivasha.
Figure 2.
Genetic changes in the Lake Naivasha Daphnia pulex population through time. (a) Fraction of the invading clone (red area, left axis) versus other genotypes (blue area) and the changes in genetic diversity through time (dot plot with 95% confidence bars and stippled line, right axis). (b) Genetic tree obtained from a 12S fragment showing the relationship of Lake Naivasha D. pulex in 1925, 1955 and 2003 to D. pulex haplotypes from Belgium, Germany and Canada. Scale bar indicates K2P genetic distance. Numbers at nodes indicate bootstrap support. GenBank accession numbers: 12S Belgium, AY745245; 12S Germany 1, AY626356; 12S Germany 2, AY626357 12S Canada, AY626352.
Considering that all modern Kenyan D. pulex populations sampled thus far solely consist of this invading clone (Mergeay et al. 2005), we sampled D. pulex populations throughout Africa to establish its wider distribution. Historical and new ecological data (Sars 1916; Brehm-Eger 1912; Lowndes 1936; Brehm 1960; Green 1990, 1995; Hart 1992; Samraoui et al. 1998; Mergeay et al. 2006) suggest that this species is generally absent from the warm tropical lowlands and subtropical desert regions, being mainly restricted to standing waters in the cooler regions of coastal North and South Africa, and the mountainous regions of East Africa (figure 3b). In our collection of 177 modern zooplankton samples from a wide range of aquatic habitat types, we found D. pulex in 15 locations (see electronic supplementary material, table 1) geographically distributed in accordance with this known African distribution range (figure 3b). DNA sequencing of ND5 revealed only one haplotype in all 15 populations. This haplotype, in accordance with the paleogenetic results on 12S for the Lake Naivasha population, proved to be strongly embedded in the American D. pulex clade (figure 3a). A subsequent analysis of 10 polymorphic microsatellite loci in these populations again yielded no extant D. pulex populations with nuclear genotypes other than the American invader observed in Lake Naivasha. These results provide strong evidence that the invading clone is now present throughout the known geographic range of native African D. pulex (figure 3b). Although our samples did not include coastal North Africa, the presence of the Naivasha 12S haplotype in southern Spain (compare GenBank accession numbers AF277284 and AY745243) suggests that the invader is also present in the Mediterranean region.
As MLG LN1 is fixed heterozygous at three microsatellite loci (and in at least one allozyme locus), our results further established the asexual nature of the invading clone. This was confirmed in the laboratory by the observation that dormant eggs were produced in the absence of males, a process that normally requires sexual reproduction in cyclic parthenogenetic Daphnia. Allozyme analyses on individuals hatched from dormant eggs revealed that the invading genotype is heterozygous for the locus lactate dehydrogenase. This identifies the invader as a hybrid lineage of American D. pulex and American D. pulicaria (Hebert et al. 1989) common in North America.
4. Discussion
The absence of genetic variation in both the variable mitochondrial ND5 and the 10 hypervariable nuclear markers of all sampled modern D. pulex populations in Africa strongly suggests a single colonization of this lineage into Africa, followed by rapid continent-wide spread and complete displacement of indigenous African D. pulex throughout its native range. Most likely, the invading water flea clone was introduced accidentally to Lake Naivasha in 1927–1929, during stocking of largemouth bass (Micropterus salmoides) from the USA (Siddiqui 1979), following the suggestion by the then US President T. Roosevelt in 1910 that the sport fishing in British East Africa needed improvement (Robbins & MacCrimmon 1974). As this was the earliest introduction of American largemouth bass in Africa (Robbins & MacCrimmon 1974), Lake Naivasha is almost certainly the geographical origin of the American water flea lineage in Africa. Our paleogenetic analysis in Lake Naivasha thus traces the dynamics of the original introduction back in time, whereas our spatial survey across the continent reveals the extent of the subsequent spread of the invader. The present distribution of this American genotype in African lakes and reservoirs extends over more than 5500 km between Ethiopia and South Africa, testifying the high dispersal capacity of zooplankton dormant eggs. Although its spread may on occasion have been facilitated by human activities (e.g. translocation of fish), the presence of this genotype in remote regions with almost no history of fish stocking (e.g. northern Ethiopia) suggests a predominance of natural dispersal (e.g. by migrant waterfowl).
Similar to the seed banks of plants, dormant egg banks are supposed to buffer against local extinction through the storage effect (Chesson 1983), reduce establishment success of immigrant genotypes (De Meester et al. 2002) and increase local genetic diversity when old dormant eggs hatch in contemporary populations (Hedrick 1995). However, the extensive dormant egg bank of native D. pulex that existed in Lake Naivasha during the pre-invasion period (deposition rates of 102–103 eggs y−1 m−2, or an estimated total annual production of 1010–1011 dormant eggs) did not prevent the exotic clone from displacing the native population in the relatively short time of 60 years. The apparent competitive superiority of the invading clone is also reflected in the extent of its present distribution in Africa. Its large ecological amplitude and niche breadth are particularly striking: the 15 documented occurrences include all types of sampled freshwater habitat ranging from small temporary ponds, eutrophic sewage ponds, shallow turbid lakes and reservoirs, to clear lakes rich in aquatic macrophytes, both with and without fishes (electronic supplementary material, table 1).
The invasion of an asexual Daphnia clone we document here is reminiscent of invasions into Africa by the water fern Salvinia molesta and the water hyacinth Eichhornia crassipes: both are asexual pest species that invaded and spread throughout Africa and other tropical regions at an incredible pace (Thomas & Room 1986; Gopal 1987). Probably, the most striking difference is that these plants had no competition from resident congeneric species or other free-floating plants and could thus profit from an empty niche. In our survey of African lakes, we found 10 other Daphnia species that coexist and potentially compete with the invading clone (J. Mergeay, unpublished data). Currently, it is unclear whether the invasion of LN1 has also impacted other Daphnia species in Africa. Although Lake Naivasha's nine-species Daphnia community has changed considerably over the past century, species turnover (other than that between native and invading D. pulex genotypes) seems to have been fuelled primarily by anthropogenic influences on the ecosystem through the introduction of exotic fish species, changing land use and pollution (Mergeay et al. 2004), rather than by any direct competitive effects of the invader.
In plants, invasion success is often associated with the capacity to reproduce asexually during some stages of the life cycle (Sakai et al. 2001). Sexual Daphnia species have much in common with invasive plants, in that they alternate phases of asexual reproduction with regular bouts of sexual reproduction (so-called cyclic parthenogenesis). Doing so, they combine rapid population growth with the periodic generation of new genetic diversity. Moreover, they produce long-lived dormant stages that have high dispersal capacity. Upon colonization of new habitats, even by single individuals, rapid asexual population growth allows fast monopolization of available resources (De Meester et al. 2002). Furthermore, these cyclic parthenogens minimize the cost of male production, one of the principal costs of sex often used to explain the short-term evolutionary advantage of asexual species and lineages (Maynard Smith 1978; Lewis 1987; Doncaster et al. 2000). Moreover, sexual strains of D. pulex compensate for the cost of males by having a higher fecundity than asexual strains (Innes et al. 2000). As sexual recombination allows rapid spread of favourable genetic mutations through populations, maintaining recurrent investment in sexual reproduction promotes adaptation to new habitat conditions. This may be expected to be especially important in highly dynamic ecosystems such as Lake Naivasha (Crow 1992; Waxman & Peck 1999; Verschuren et al. 2000). Cyclic parthenogenetic Daphnia populations, similar to the indigenous African D. pulex, have been shown to genetically adapt to changing environmental conditions within a few years (Hairston et al. 1999; Cousyn et al. 2001). Although the resident population of Lake Naivasha had ample capacity to genetically adapt to environmental changes, they were outcompeted by a single asexual genotype with intrinsically lower evolutionary potential. How this obligate asexual exotic species could eradicate an indigenous sibling species that optimally combines the advantages of sexual and asexual reproduction is puzzling as well as alarming. Effects of heterosis, resulting from hybridisation, may contribute to the broad ecological tolerance of the invader (Kearney & Shine 2004), and hence to its success. Additionally, a reduced impact of specialized antagonists, such as parasites that are adapted specifically to local genotypes (Ebert 1994; Torchin et al. 2003) may have determined the prosperity of this invader.
This study documents the probable continent-wide displacement of a native species caused by the accidental introduction of a single exotic clone of a closely related species about 75 years ago. It shows that adequate genetic variation and a significant numerical advantage are no warranty against displacement and that invaders do not need sexual recombination to thrive in the diverse environments they colonize, even as they compete with genetically diverse native species.
Acknowledgments
This study was carried out with permission of the Permanent Secretary of the Ministry of Education, Science & Technology of Kenya and was funded by the KULeuven Research Fund project OT/04/23 (to L.D.M.) and the Research Foundation-Flanders project G0086.00 (to D.V.). We thank K. Mavuti, W. Shivoga and S. Higgins for logistic support, H. Eggermont and K. De Maeyer for assistance in the field, L. Brendonck, E. De Roeck, T. Dejenie, T. Nhiwatiwa and M. Barson for additional samples, and several anonymous reviewers for helpful comments. J.M. was supported by a PhD fellowship from IWT-Flanders and a KULeuven postdoctoral grant.
Supplementary Material
References
- Bonizzoni M, Guglielmino C.R, Smallridge C.J, Gomulski M, Malacrida A.R, Gasperi G. On the origins of medfly invasion and expansion in Australia. Mol. Ecol. 2004;13:3845–3855. doi: 10.1111/j.1365-294X.2004.02371.x. doi: 10.1111/j.1365-294X.2004.02371.x [DOI] [PubMed] [Google Scholar]
- Brehm V. Crustacea: Cladocera. Mission zoologique de l' I.R. S.A.C. en Afrique orientale. Ann. Mus. Congo Tervuren, in 8°, Zool. 1960;81:27. (P. Basilewsky, N. Leleup, 1957) [Google Scholar]
- Brehm-Eger V. Wissenschaftliche Ergebnisse der Deutschen Zentral-Afrika-expedition 1907–1908. Verlag Klinkhardt & Biermann; Leipzig, Germany: 1912. Die Cladoceren. pp. 167–173. [Google Scholar]
- Chesson P.L. Coexistence of competitors in a stochastic environment: the storage effect. In: Friedman H.I, Strobeck C, editors. Population biology. Springer; New York, NY: 1983. pp. 188–198. [Google Scholar]
- Clavero M, Garcia-Berthou E. Invasive species are a leading cause of animal extinctions. Trends Ecol. Evol. 2005;20:110. doi: 10.1016/j.tree.2005.01.003. doi: 10.1016/j.tree.2005.01.003 [DOI] [PubMed] [Google Scholar]
- Colbourne J.K, Crease T.J, Weider L.J, Hebert P.D.N, Dufresne F, Hobaek A. Phylogenetics and evolution of a circumarctic species complex (Cladocera: Daphnia pulex) Biol. J. Linn. Soc. 1998;65:347–365. doi:10.1006/bijl.1998.0251 [Google Scholar]
- Colbourne J.K, Robison B, Bogart K, Lynch M. Five hundred and twenty-eight microsatellite markers for ecological genomic investigations using Daphnia. Mol. Ecol. Notes. 2004;4:485–490. doi 10.1111/j.1471-8286.2004.00721.x [Google Scholar]
- Cousyn C, De Meester L, Colbourne J.K, Brendonck L, Verschuren D, Volckaert F. Rapid local adaptation of zooplankton behavior to changes in predation pressure in the absence of neutral genetic changes. Proc. Natl Acad. Sci. USA. 2001;98:6256–6260. doi: 10.1073/pnas.111606798. doi: 10.1073/pnas.111606798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J.F. An advantage of sexual reproduction in a rapidly changing environment. J. Hered. 1992;83:169–173. doi: 10.1093/oxfordjournals.jhered.a111187. [DOI] [PubMed] [Google Scholar]
- De Meester L, Gomez A, Okamura B, Schwenk K. The monopolization hypothesis and the dispersal-gene flow paradox in aquatic organisms. Acta Oecol.-Int. J. Ecol. 2002;23:121–135. doi:10.1016/S1146-609X(02)01145-1 [Google Scholar]
- Doncaster C.P, Pound G.E, Cox S.J. The ecological cost of sex. Nature. 2000;404:281–285. doi: 10.1038/35005078. doi:10.1038/35005078 [DOI] [PubMed] [Google Scholar]
- Ebert D. Virulence and local adaptation of a horizontally transmitted parasite. Science. 1994;265:1084–1086. doi: 10.1126/science.265.5175.1084. [DOI] [PubMed] [Google Scholar]
- Frankham R. Resolving the genetic paradox in invasive species. Heredity. 2005a;94:385. doi: 10.1038/sj.hdy.6800634. doi: 10.1038/sj.hdy.6800634 [DOI] [PubMed] [Google Scholar]
- Frankham R. Genetics and extinction. Biol. Conserv. 2005b;126:131–140. doi: 10.1016/j.biocon.2005.05.002 [Google Scholar]
- Gopal B. Aquatic plant studies 1. Elsevier Science Publisher; Amsterdam, The Netherlands: 1987. Water hyacinth. pp. 113–157. [Google Scholar]
- Goudet, J. 2001 Fstat version 2.9.3. Lausanne, Switzerland, Institute of Ecology, Biology Building, UNIL, CH 1015.
- Green J. Zooplankton associations in Zimbabwe. J. Zool. 1990;222:259–283. [Google Scholar]
- Green J. Altitudinal distribution of tropical planktonic Cladocera. Hydrobiologia. 1995;307:75–84. doi:10.1007/BF00031999 [Google Scholar]
- Hairston N.G, Jr, Lampert W, Cáceres C.E, Holtmeier C.L, Weider L.J, Gaedke U, Fischer J.M, Fox J.A, Post D.M. Rapid evolution revealed by dormant eggs. Nature. 1999;401:446. doi:10.1038/46731 [Google Scholar]
- Hart R.C. Experimental studies of food and suspended sediment effects on growth and reproduction of 6 planktonic cladocerans. J. Plankton Res. 1992;14:1425–1448. [Google Scholar]
- Havel J.E, Colbourne J.K, Hebert P.D.N. Reconstructing the history of intercontinental dispersal in Daphnia lumholtzi by use of genetic markers. Limnol. Oceanogr. 2000;45:1414–1419. [Google Scholar]
- Hebert P.D.N, Beaton M.J, Schwartz S.S, Stanton D.J. Polyphyletic origins of asexuality in Daphnia pulex. I. Breeding-system variation and levels of clonal diversity. Evolution. 1989;43:1004–1015. doi: 10.1111/j.1558-5646.1989.tb02546.x. doi:10.2307/2409581 [DOI] [PubMed] [Google Scholar]
- Hedrick P.W. Genetic polymorphism in a temporally varying environment: effects of delayed diapause. Heredity. 1995;75:164–170. [Google Scholar]
- Innes D.J, Fox C.J, Winsor G.L. Avoiding the cost of males in obligately asexual Daphnia pulex (Leydig) Proc. R. Soc. B. 2000;267:991–997. doi: 10.1098/rspb.2000.1101. doi:10.1098/rspb.2000.1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S.K, Martins P.S. Ecological genetics of the colonising ability of rose clover (Trifolium hirtum All.) Am. J. Bot. 1979;66:361–366. doi:10.2307/2442390 [Google Scholar]
- Kearney M, Shine R. Morphological and physiological correlates of hybrid parthenogenesis. Am. Nat. 2004;164:803–813. doi: 10.1086/425986. doi:10.1086/425986 [DOI] [PubMed] [Google Scholar]
- Kolbe J.J, Glor R.E, Schettino L.R.G, Lara A.C, Larson A, Losos J.B. Genetic variation increases during biological invasion by a Cuban lizard. Nature. 2004;431:177–181. doi: 10.1038/nature02807. doi: 10.1038/nature02807 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. Mega3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. doi:10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Lewis W.M., Jr . The cost of sex. In: Stearns S.C, editor. The evolution of sex and its consequences. Birkhaüser Verlag; Basel, MA: 1987. pp. 33–57. [Google Scholar]
- Lowndes M.A.F.L.S. Scientific results of the Cambridge Expedition to the East African Lakes 1930-1 No. 16. The smaller Crustacea. Linn. J. Zool. 1936;16:1–33. [Google Scholar]
- Maynard Smith J. Cambridge University Press; Cambridge, UK: 1978. The evolution of sex. [Google Scholar]
- Mergeay, J. 2005 Paleoecology and paleogenetics in the tropics: the Daphnia of Lake Naivasha, Kenya, PhD thesis. Katholieke Universiteit Leuven.
- Mergeay J, Declerck S, Verschuren D, De Meester L. Daphnia community analysis in shallow Kenyan lakes and ponds using sediment archives. Freshwat. Biol. 2006;51:399–411. doi: 10.1111/j.1365-2427.2005.01494.x [Google Scholar]
- Mergeay J, Verschuren D, De Meester L. Cryptic invasion and dispersal of an American Daphnia in East Africa. Limnol. Oceanogr. 2005;50:1278–1283. [Google Scholar]
- Mergeay J, Verschuren D, Van Kerckhoven L, De Meester L. Two hundred years of a diverse Daphnia community in Lake Naivasha (Kenya): effects of natural and human-induced environmental changes. Freshwat. Biol. 2004;49:998–1013. doi: 10.1111/j.1365-2427.2004.01244.x [Google Scholar]
- Mooney H.A, Cleland E.E. The evolutionary impact of invasive species. Proc. Natl Acad. Sci. USA. 2001;98:5446–5451. doi: 10.1073/pnas.091093398. doi: 10.1073/pnas.091093398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardi A. Assessing species invasions as a cause of extinction. Trends Ecol. Evol. 2004;19:619. doi: 10.1016/j.tree.2004.09.02 [Google Scholar]
- Robbins W.H, MacCrimmon H.R. Biomanagement and Research Enterprises; Sault Ste. Marie, Canada: 1974. The black bass in America and overseas. [Google Scholar]
- Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. Inbreeding and extinction in a butterfly metapopulation. Nature. 1998;392:491–494. doi:10.1038/33136 [Google Scholar]
- Sakai A.K, et al. The population biology of invasive species. Annu. Rev. Ecol. Syst. 2001;32:305–332. doi:10.1146/annurev.ecolsys.32.081501.114037 [Google Scholar]
- Saltonstall K. Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc. Natl Acad. Sci. USA. 2002;99:2445–2449. doi: 10.1073/pnas.032477999. doi: 10.1073/pnas.032477999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samraoui B, Segers H, Maas S, Baribwegure D, Dumont H.J. Rotifera, Cladocera, Copepoda, and Ostracoda from coastal wetlands in northeast Algeria. Hydrobiologia. 1998;386:183–193. doi:10.1023/A:1003538730152 [Google Scholar]
- Sars G.O. The freshwater Entomostraca of Cape Province (Union of South Africa). Part I: Cladocera. Ann. S. Afr. Mus. 1916;XV:303–351. [Google Scholar]
- Siddiqui A.Q. Changes in fish species composition in Lake Naivasha, Kenya. Hydrobiologia. 1979;64:131–138. doi:10.1007/BF00023188 [Google Scholar]
- Spielman D, Brook B.W, Frankham R. Most species are not driven to extinction before genetic factors impact them. Proc. Natl Acad. Sci. USA. 2004;101:15 261–15 264. doi: 10.1073/pnas.0403809101. doi: 10.1073/pnas.0403809101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagg N, Innes D.J, Doncaster C.P. Outcomes of reciprocal invasions between genetically diverse and genetically uniform populations of Daphnia obtusa (Kurz) Oecologia. 2005;143:527–536. doi: 10.1007/s00442-005-0016-5. doi: 10.1007/s00442-005-0016-5 [DOI] [PubMed] [Google Scholar]
- Thomas P.A, Room P.M. Taxonomy and control of Salvinia molesta. Nature. 1986;320:581–584. doi:10.1038/320581a0 [Google Scholar]
- Torchin M.E, Lafferty K.D, Dobson A.P, McKenzie V.J, Kuris A.M. Introduced species and their missing parasites. Nature. 2003;421:628–630. doi: 10.1038/nature01346. doi: 10.1038/nature01346 [DOI] [PubMed] [Google Scholar]
- Verschuren D. Reconstructing fluctuations of a shallow East African lake during the past 1800 yrs from sediment stratigraphy in a submerged crater basin. J. Paleolimnol. 2001;25:297–311. doi:10.1023/A:1011150300252 [Google Scholar]
- Verschuren D, Laird K.R, Cumming B.F. Rainfall and drought in equatorial east Africa during the past 1100 years. Nature. 2000;403:410–414. doi: 10.1038/35000179. doi:10.1038/35000179 [DOI] [PubMed] [Google Scholar]
- Waxman D, Peck J.R. Sex and adaptation in a changing environment. Genetics. 1999;153:1041–1053. doi: 10.1093/genetics/153.2.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.