Abstract
The luxS gene, present in many bacterial genera, encodes the autoinducer 2 (AI-2) synthase. AI-2 has been implicated in bacterial signaling, and this study investigated its role in biofilm formation by Streptococcus gordonii, an organism that colonizes human tooth enamel within the first few hours after professional cleaning. Northern blotting and primer extension analyses revealed that S. gordonii luxS is monocistronic. AI-2 production was dependent on nutritional conditions, and maximum AI-2 induction was detected when S. gordonii was grown in the presence of serum and carbonate. In planktonic cultures, AI-2 production rose sharply during the transition from exponential to stationary phase, and the AI-2 concentration peaked approximately 4 h into stationary phase. An S. gordonii luxS mutant that did not produce AI-2 was constructed by homologous recombination. Complementation of the mutant by insertion of an intact luxS gene into the chromosome in tandem with the disrupted gene restored AI-2 production to a level similar to that of the wild-type strain. In planktonic culture, no growth differences were observed between the mutant and wild-type strains when five different media were used. However, when grown for 4 h as biofilms in 25% human saliva under flow, the luxS mutant formed tall microcolonies that differed from those formed by the wild-type and complemented mutant strains. Biofilms of the luxS mutant exhibited finger-like projections of cells that extended into the flow cell lumen. Thus, the inability to produce AI-2 is associated with altered microcolony architecture within S. gordonii biofilms formed in saliva during a time frame consistent with initial colonization of freshly cleaned enamel surfaces.
Oral bacteria adhere to one another by adhesin-receptor interactions between genetically distinct partner organisms (46). These interactions, referred to as coaggregation, may serve as a driving force behind the developmental architecture of dental plaque (20), a complex biofilm community that consists of more than 500 bacterial species in its mature state (30, 34). The initial colonizers of freshly cleaned enamel surfaces are predominantly streptococci (31), which form mixed-species microcolonies during early plaque development (32). In addition to intimate physical interactions, diffusible signals may also be important in oral bacterial interactions. For example, competence pathways described in oral streptococci in vitro are regulated by short cationic oligopeptides called competence-stimulating peptides (11, 18). Another diffusible signaling molecule, designated autoinducer 2 (AI-2) and synthesized by the LuxS enzyme, was recently identified in several genera of oral bacteria, including Actinobacillus, Fusobacterium, Porphyromonas, Prevotella, and Streptococcus (8, 10, 14-16, 27, 28).
The AI-2 signaling pathway was first described in the bioluminescent marine bacterium Vibrio harveyi (5). In V. harveyi, AI-2 works in conjunction with AI-1, an acyl-homoserine lactone signal, to regulate bioluminescence in a density-dependent manner (5, 6). Light production occurs when AI-1 and AI-2 each bind their respective sensor proteins, LuxN and LuxPQ, thereby initiating a complex phosphorylation cascade that culminates in both transcriptional derepression and activation of the luciferase operon (5, 6, 42). The AI-2 biosynthetic pathway is also well characterized in V. harveyi. The AI-2 precursor 4,5-dihydroxy-2,3-pentanedione (DPD) is produced from S-adenosylmethionine, a major methyl donor for cellular methyltransferase reactions, through three enzymatic steps (37). In the final step, LuxS cleaves S-ribosylhomocysteine to homocysteine and DPD. DPD is predicted to spontaneously cyclize and may undergo additional rearrangements, yielding a pool of molecules characterized as pro-AI-2 (9, 37, 47). V. harveyi LuxP binds pro-AI-2, and the crystal structure of this protein in complex with its bound ligand revealed that AI-2 is a furanosyl borate diester (9). Regulatory phosphorylation cascades through which AI-2 may be recognized as a signaling molecule in bacteria other than V. harveyi have not been characterized.
The AI-2 detection assay measures the induction of bioluminescence of a V. harveyi reporter strain in response to conditioned bacterial culture medium (40). The demonstration that conditioned medium from many gram-negative and gram-positive bacteria induces bioluminescence of the V. harveyi reporter led to the hypothesis that AI-2 may serve as a universal interspecies bacterial signal (4). In addition to regulation of the V. harveyi luciferase operon (37), AI-2-regulated genes have been reported within monospecies cultures of nonoral bacteria (e.g., see references 12, 24, 39, and 43), as well as among the oral isolates Porphyromonas gingivalis (8, 10), Actinobacillus actinomycetemcomitans (14, 15), and Streptococcus gordonii (27). It was reported that in P. gingivalis, genes involved in the acquisition of hemin, a growth factor essential for virulence, were differentially regulated in response to luxS inactivation (10). Additionally, a P. gingivalis luxS mutant exhibited reduced production of the virulence-associated Arg- and Lys-gingipain proteases (8). In A. actinomycetemcomitans, several genes involved in the acquisition and storage of iron were differentially regulated in a luxS mutant compared to the wild-type strain (15). It was also reported that a luxS mutant of the commensal oral bacterium S. gordonii exhibited altered expression of genes involved in carbohydrate metabolism (27). Thus, AI-2 may serve as a signaling molecule that regulates a variety of genes within planktonic cultures of oral bacteria.
In the present study, Northern blot and primer extension analyses demonstrated that S. gordonii DL1 luxS is monocistronic. The nutritional conditions under which S. gordonii produces AI-2 were characterized. Additionally, when grown as a biofilm in flow cells with saliva as the growth medium, an S. gordonii luxS mutant exhibited an altered spatial arrangement compared to the wild-type strain, which may reflect a loss of AI-2-mediated intercellular communication under conditions relevant to the organism's natural habitat.
MATERIALS AND METHODS
Culture conditions.
S. gordonii DL1 was grown in either Todd Hewitt broth (THB; Difco Laboratories, Detroit, Mich.), brain heart infusion (BHI) medium (Becton Dickinson & Co., Sparks, Md.), Tryptone-yeast extract medium containing 0.5% glucose (TY-glucose) (19), Luria-Bertani (LB) medium, or 25% human saliva. Saliva was collected from at least six individuals, pooled, clarified by centrifugation, diluted with distilled water, and filter sterilized (33). S. gordonii overnight cultures were incubated anaerobically at 37°C with the GasPak System (Becton Dickinson & Co.). Escherichia coli was cultivated in LB broth at 37°C with shaking. Vibrio harveyi was grown in autoinducer bioassay medium (17) at 30°C with shaking. Horse serum (Quality Biological, Inc., Gaithersburg, Md.) was heat inactivated by being warmed to 60°C for 30 min. Horse serum (5%, vol/vol)-supplemented THB or BHI medium is designated THBS or BHIS, respectively. Where noted, antibiotics were added to growth media at the following concentrations: ampicillin, 100 μg/ml for E. coli; erythromycin, 10 μg/ml for S. gordonii and 300 μg/ml for E. coli; kanamycin, 1,000 μg/ml for S. gordonii and 50 μg/ml for E. coli; spectinomycin, 1,000 μg/ml for S. gordonii and 100 μg/ml for E. coli.
Nucleic acid isolation.
S. gordonii genomic DNA was purified as previously described (1). Plasmid DNA was isolated with the QIAprep Spin Miniprep Kit (Qiagen, Valencia, Calif.). Total cellular RNA was isolated from E. coli with the RNeasy Mini Kit (Qiagen) and from S. gordonii as previously described (13).
Northern blotting.
Denaturing electrophoresis and Northern transfer of RNA were performed as described by Sambrook et al. (36). Probe hybridization was done with Ultrahyb hybridization buffer (Ambion, Inc., Austin, Tex.) in accordance with the manufacturer's protocol, and images were generated with a Storm 860 imaging system (Molecular Dynamics, Sunnyvale, Calif.). A 399-bp internal fragment of S. gordonii luxS was synthesized and labeled for use as a probe by PCR with the primers 5′-GAT CAC ACC ATC GTT AAG G-3′ and 5′-GAT TAA CTT TGC CCA CTC C-3′. dATP concentrations were reduced 10-fold in PCR-labeling reaction mixtures to allow incorporation of [α-32P]dATP into the PCR product.
RNA primer extension.
Primer extension analysis was conducted with the avian myeloblastosis virus reverse transcriptase primer extension system (Promega, Madison, Wis.), in accordance with the manufacturer's protocols. RNA was isolated from mid-exponential-phase S. gordonii cultures grown in THBS or from mid-exponential-phase E. coli DH5α cultures grown in LB medium. cDNA synthesis was initiated by hybridizing 5 μg of total cellular RNA with 1 ng of [γ-33P]ATP end-labeled primer (5′-CAG GTG TTC AAT GGT ATG G-3′). Sequencing reactions for inclusion as markers on primer extension gels were generated in accordance with the manufacturer's instructions with the fmol DNA Cycle Sequencing System (Promega) with pSF151-luxS plasmid DNA (see section describing genetic complementation of the luxS mutant) as the template. Reaction products were run on a 6% polyacrylamide-1× Tris-borate-EDTA-7 M urea gel, and images were generated with the Storm 860 imaging system.
Analysis of AI-2 production.
Streptococcal starter cultures (10 ml), grown anaerobically overnight, were spun for 10 min at 867 × g and resuspended in 2 ml of the same medium. Concentrated starter cultures were used to inoculate fresh cultures at 1% of the total culture volume. Analysis of the relationship between AI-2 production and Na2CO3 concentration was conducted with BHIS containing 12, 24, or 48 mM Na2CO3. To analyze the relationship between AI-2 production and carbonate ion or buffering, 24 mM Na2CO3, NaHCO3, or morpholinepropanesulfonic acid (MOPS) was added to BHI medium. Na2CO3-, NaHCO3-, and MOPS-supplemented media were prepared by addition of the appropriate salt, followed by adjustment of the medium pH to 7.4 and filter sterilization. Na2CO3-, NaHCO3-, and MOPS-containing cultures were incubated in sealed tubes. Culture densities, expressed in Klett units, were determined with a Klett-Summerson photoelectric colorimeter (Arthur H. Thomas Co., Philadelphia, Pa.) with a red (660-nm) filter, and AI-2 production was assayed as follows. Cell-free culture supernatants were prepared by centrifugation (5 min at 16,000 × g) of 500-μl aliquots, followed by filtration through a 0.2-μm-pore-size membrane. Filtered supernatants were stored at −20°C until assayed for AI-2 activity as previously described (40). Ten microliters of each supernatant was added to triplicate wells of a white 96-well microtiter plate containing 90 μl of the overnight-grown V. harveyi BB170 reporter strain diluted 1:5,000 in autoinducer bioassay medium. The assay plate was then placed in a Wallac Victor2 1420 multilabel counter (Perkin-Elmer Life Sciences, Wellesley, Mass.) with the counting chamber temperature set at 30°C. The counter was programmed to measure the luminescence of each sample every 30 min, with each series of readings preceded by 20 min of stationary incubation, followed by 10 min of agitation (amplitude = 0.1 mm). Luminescence fold induction values were determined from the measurement taken 30 min prior to the time at which the negative control (90 μl of 1:5,000-diluted strain BB170 plus 10 μl of uninoculated sterile medium) underwent autoinduction of bioluminescence as a result of endogenous AI-2 production. V. harveyi BB152 was used as the positive control. To facilitate comparisons of AI-2 induction values from independent assays, all of the AI-2 production data reported herein were normalized as fold induction over a sterile growth medium control conducted in parallel with the samples analyzed.
luxS mutant construction.
The open reading frame (ORF) downstream of S. gordonii luxS was identified during an unrelated investigation, and the sequence of the ORF and flanking DNA, including that of the complete luxS gene, was obtained with the Universal Genome Walker kit (Clontech, Palo Alto, Calif.). Subsequently, The Institute for Genomic Research (http://tigrblast.tigr.org/ufmg/) made the incomplete S. gordonii genome sequence publicly available. To obtain the sequence of additional DNA flanking luxS, fragments from the database were assembled with SeqMan II DNA analysis software (DNA*, Madison, Wis.), yielding the 6.4-kb contiguous sequence shown in Fig. 1A. Nucleotides 10 to 5323 of S. gordonii contig 2411 were assembled with nucleotides 1 to 966 of S. gordonii contig 2273 with 120 nucleotides of homologous Streptococcus pneumoniae DNA (accession no. AE_008526) to span the gap within ORF3 between the two S. gordonii sequences. The percentage of nucleotide sequence identity between the homologous S. pneumoniae and S. gordonii sequences within ORF3 is 79%.
FIG. 1.
(A) Linear map of 6.4 kb of genomic DNA flanking S. gordonii luxS. Predicted ORFs and their orientations are shown. A predicted stem-loop structure between ORF2 and luxS is indicated. (B) Schematic representations of the chromosomal regions encompassing the ermAM-disrupted gene of the S. gordonii luxS mutant and the tandem disrupted and intact gene copies of the complemented luxS mutant. The image is not drawn to scale.
An isogenic S. gordonii luxS mutant was constructed by gene replacement, inactivating the parental luxS gene by insertion of the erythromycin resistance gene ermAM (Fig. 1B). luxS and flanking DNA were PCR amplified from S. gordonii genomic DNA with the primers 5′-GGA ATT CCG AAA ATT CTT AAT TAT TTT ATC-3′ (EcoRI site shown in bold) and 5′-CCC AAG CTT CGG ATT CTC TAG ATT ATT AG-3′ (HindIII site shown in bold). The resultant PCR product contained the 483-nucleotide luxS ORF, 197 nucleotides of upstream DNA, and 80 nucleotides of downstream DNA, including a putative rho-independent transcriptional terminator. This PCR product was cloned into the E. coli-Streptococcus shuttle vector pDL278 (22), yielding pDL278-luxS. pDL278-luxS was transformed into E. coli XL10 Gold (Stratagene, La Jolla, Calif.) in accordance with the manufacturer's protocol, with selection for spectinomycin-resistant transformants, and the insert sequence was verified. ermAM was PCR amplified from pVA736 (25) with the following primers (27), which incorporate NdeI sites (shown in bold) at the ends of the resulting PCR product: 5′-GCA CAT ATG CTT AGA AGC AAA CTT AAG A-3′ and 5′-GCC CAT ATG CTT GGA AGC TGT CAG TAG T-3′. The ermAM PCR product was cloned into a unique NdeI site (27) at position 262 of the 483-nucleotide luxS ORF, yielding pDL278-luxS::ermAM, and transformed into E. coli XL10 Gold with selection for erythromycin-resistant transformants. The luxS::ermAM fragment was excised from pDL278-luxS::ermAM, gel purified, and transformed into S. gordonii to generate a luxS mutant. Transformants were plated on BHI medium containing erythromycin to select for isolates that had undergone double-crossover homologous recombination. Insertion of ermAM within luxS was verified by PCR; Southern blotting with ermAM as a probe confirmed the incorporation of a single ermAM cassette within the genome of the luxS mutant (data not shown).
Genetic complementation of the luxS mutant.
The S. gordonii luxS mutant was genetically complemented by chromosomal integration of an undisrupted copy of S. gordonii luxS and flanking DNA in tandem with the ermAM-disrupted luxS gene (Fig. 1B). luxS and flanking DNA were PCR amplified from S. gordonii genomic DNA with the EcoRI-incorporating primer described in the previous paragraph and the BamHI-incorporating primer 5′-CGG GAT CCC GGA TTC TCT AGA TTA TTA G-3′ (BamHI site shown in bold). The resultant PCR product was cloned into the streptococcal-integration shuttle vector pSF151 (44), yielding pSF151-luxS, and transformed into E. coli XL10 Gold with selection for kanamycin-resistant transformants, and the insert sequence was verified. pSF151-luxS was then transformed into the luxS mutant, and transformants that integrated pSF151-luxS into their chromosomes by single-crossover homologous recombination were identified with kanamycin and erythromycin resistance as selectable markers. Insertion of pSF151-luxS in tandem with the ermAM-disrupted chromosomal luxS gene was verified by PCR (data not shown). A Southern blot of genomic DNA from the luxS mutant and from the complemented mutant was hybridized to an ermAM probe. The probe hybridized to a single DNA fragment in the complemented mutant that was approximately 4.2 kb larger than the fragment to which the probe hybridized in the luxS mutant, consistent with the integration of a single copy of pSF151-luxS (4,270 bp; data not shown).
Cultivation and analysis of in vitro biofilms.
Mid-exponential-phase BHI medium-grown cultures of wild-type, luxS mutant, and complemented luxS mutant cells were washed three times in the medium used for growth in flow cells (either 25% sterile saliva, 10-fold-diluted BHI medium containing 2.4 mM Na2CO3, or 10-fold-diluted THBS; dilutions were made with distilled water). Washed cells were adjusted to an A600 of 0.05 and inoculated into flow cells conditioned with same medium in which the cells were washed. Cells were allowed to adhere under static conditions for 15 min, after which flow was initiated at 0.2 ml/min (33). Biofilms were stained with BacLight Live/Dead (Molecular Probes, Eugene, Oreg.) and examined by confocal microscopy with a 40× oil immersion lens with a numerical aperture of 1.25 attached to a TCS-SP2 microscope (Leica, Exton, Pa.). Confocal image stacks were collected at three random locations for each biofilm. In all cases, very few “dead” cells stained solely with propidium iodide were seen; therefore, maximum projections of green-red overlays, showing Syto-9- and propidium-iodide-stained cells together, were converted to grayscale for presentation. Volume and average colony height calculations were preformed on the three image stacks collected for each biofilm with algorithms described by Xavier et al. (48, 49).
RESULTS
S. gordonii luxS and flanking DNA.
A contiguous 6.4-kb sequence of S. gordonii genomic DNA that contains five ORFs, including luxS, is represented in Fig. 1A. The amino acid sequence encoded by ORF1 (1,167 bp) is 82% identical and 91% similar to that of a conserved hypothetical protein of unknown function in S. pneumoniae (accession no. NP_357927). ORF2 (1,413 bp) encodes a putative protein with 36% identity and 47% similarity to a conserved hypothetical protein of unknown function in Streptococcus agalactiae (accession no. NP_687339). ORF3 (1,608 bp) encodes an amino acid sequence with 71% identity and 75% similarity to a hypothetical S. pneumoniae protein (accession no. NP_346175) containing a histidine-aspartate phosphohydrolase domain (2). The amino acid sequence encoded by ORF4 (627 bp) is 91% identical and 94% similar to a putative S. pneumoniae guanylate kinase (accession no. NP_359175). The S. gordonii luxS ORF (483 bp; accession no. AAL87464) is encoded on the DNA strand complementary to that encoding ORF1 to -4. S. gordonii LuxS is 81% identical and 91% similar to Streptococcus pyogenes LuxS (accession no. AAG28749) (24). The 3′ end of S. gordonii luxS is separated from the 3′ end of ORF2 by 60 nucleotides that contain features of a rho-independent transcriptional terminator (7).
luxS Northern blot and primer extension analyses.
Total RNA was isolated from mid-exponential-phase wild-type and luxS mutant S. gordonii cultures grown in THBS and examined for the presence of a luxS transcript by Northern blotting. The probe revealed a single band when hybridized to total RNA from the wild-type strain, consistent with the predicted size (483 bp) of the luxS ORF (data not shown). The probe revealed two bands of approximately 1,075 and 1,300 bp when hybridized to total RNA from the luxS mutant. The 1,300-bp band is consistent with the predicted length of a transcript consisting of the 5′ end of luxS fused to ermAM, and the 1,075-bp band likely represents a degradation product of this transcript.
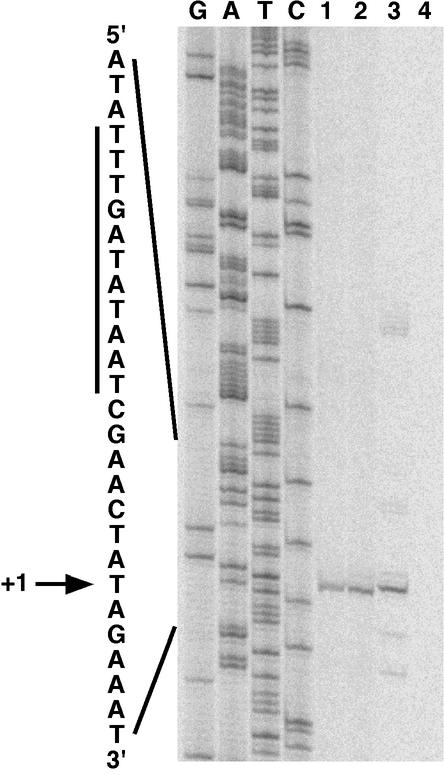
RNA primer extension analysis was performed to map the transcription initiation site of luxS in wild-type S. gordonii, in the complemented luxS mutant, and in E. coli DH5α(pSF151-luxS), a strain that harbors S. gordonii luxS on a plasmid (Fig. 2). In all of the strains tested, the transcription start site was a thymine located 27 nucleotides upstream from the luxS translation initiation codon. A potential extended −10 promoter region (TTTGATATAAT) that varied by only one nucleotide from the gram-positive consensus sequence (TRTGNTATAAT, where R is purine and N is any base) (35, 45) was spaced seven nucleotides upstream of the transcription start site; no −35 hexamer was apparent. Taken together, the identification of a transcription start directly upstream of the luxS start codon, the luxS transcript size, the opposite orientations of the ORFs flanking luxS (Fig. 1A), and the presence of a potential rho-independent transcriptional terminator downstream of the gene demonstrate that S. gordonii luxS is monocistronic.
FIG. 2.
Primer extension mapping of the S. gordonii luxS transcription start site. Labeled cDNA from reverse transcription reactions was run next to luxS sequencing reaction products (lanes G, A, T, and C) generated with the same primer. Reverse transcription products from reaction mixtures containing total RNA isolated from wild-type S. gordonii (lane 1), the complemented luxS mutant (lane 2), E. coli DH5α(pSF151-luxS) (lane 3), and E. coli DH5α without a plasmid (lane 4) are shown. An expanded view of the complementary nucleotide sequence surrounding the transcription start site (+1) is shown, and the putative extended −10 hexamer is highlighted with a vertical bar.
Planktonic growth comparisons.
When grown planktonically in THBS (Fig. 3A), the luxS mutant, wild-type, and complemented mutant strains exhibited similar doubling times of 47 ± 2, 50 ± 2, and 52 ± 3 min, respectively. Cultures of the wild-type strain also exhibited similar doubling times in THB and TY-glucose medium (Fig. 3A). It was previously shown that when grown planktonically in 25% saliva, S. gordonii attained a maximum cell density of approximately 107 CFU/ml (33). Therefore, to facilitate monitoring of S. gordonii growth in saliva, cultures were inoculated at a cell density of approximately 104 CFU/ml. After three consecutive subcultures in 25% saliva, the mutant, wild-type, and complemented mutant strains exhibited similar doubling times and attained final densities of 4.3 × 106, 5.9 × 106, and 5.6 × 106 CFU/ml, respectively. The final cell densities attained in saliva corresponded to less than 10 Klett units, which is below the inoculation density of cultures grown in laboratory media (Fig. 3A).
FIG. 3.
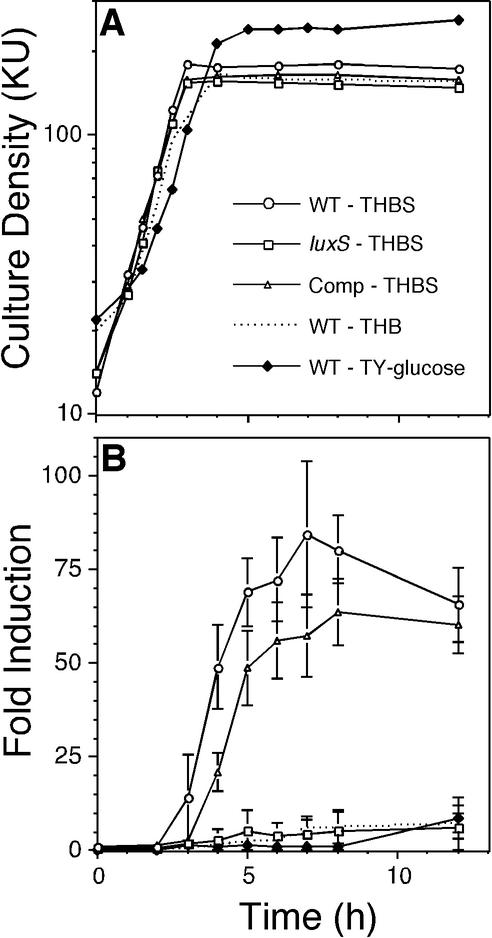
S. gordonii exhibits maximum induction of AI-2 production in THBS. Time courses of culture densities (A) and AI-2 induction levels (B) of wild-type (WT; open circles), luxS mutant (open squares), and complemented (Comp) luxS mutant (open triangles) S. gordonii strains grown in THBS are shown. Culture densities and AI-2 induction levels of the wild-type strain grown in THB lacking serum (dotted lines, no symbols) and in TY-glucose medium (filled diamonds) are also shown. All time course experiments were repeated at least six times, and the averages of three representative independent experiments with standard deviations are shown (B). KU, Klett units.
Characterization of S. gordonii AI-2 production.
Maximum AI-2 production (84-fold induction over an uninoculated medium control) was observed when wild-type S. gordonii was cultivated in THBS (Fig. 3B). AI-2 production was initially observed as the culture entered stationary phase, and AI-2 induction levels peaked approximately 4 h into stationary phase (Fig. 3B). Under these same growth conditions, background levels of AI-2 production (sixfold induction) were observed in culture supernatants of the luxS mutant, but the complemented mutant, harboring tandem ermAM-disrupted and intact chromosomal copies of luxS, produced amounts of AI-2 (64-fold induction) similar to those of the wild-type strain (Fig. 3B). S. gordonii exhibited background levels of AI-2 production (sevenfold induction; Fig. 3B) when cultivated in THB lacking serum. Thus, during cultivation in THB, induction of S. gordonii AI-2 production was serum dependent.
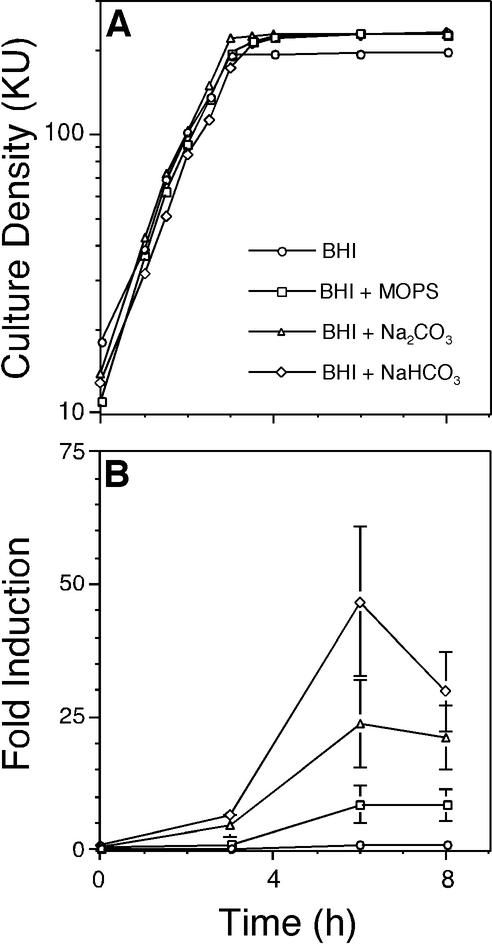
To further evaluate the effects of serum on AI-2 production in liquid culture, maximum AI-2 induction levels in BHI medium (no induction) and in BHIS (threefold induction) were compared. Thus, during cultivation in BHI medium, components in addition to serum are required for induction of AI-2 production by S. gordonii. THB contains 24 mM Na2CO3, while BHI medium lacks Na2CO3. To evaluate the effects of Na2CO3 on AI-2 production, BHIS was supplemented with various amounts of Na2CO3 and growth (Fig. 4A) and AI-2 production (Fig. 4B) were evaluated. Analysis of AI-2 production in BHIS supplemented with 12, 24, or 48 mM Na2CO3 revealed maximum AI-2 induction levels of 21-, 49-, and 59-fold, respectively (Fig. 4B). Supplementation of BHIS cultures with Na2CO3 did not affect doubling times (Fig. 4A). To determine whether Na2CO3, carbonate ion, or buffering stimulates AI-2 production, 24 mM Na2CO3, NaHCO3, or MOPS was added to BHI medium cultures and growth (Fig. 5A) and AI-2 production (Fig. 5B) were evaluated. Wild-type cultures grown under these conditions yielded maximum AI-2 induction levels of 24-fold (Na2CO3), 47-fold (NaHCO3), and ninefold (MOPS), whereas cells grown in unsupplemented BHI medium did not produce AI-2. The demonstration that two different carbonate salts, but not MOPS, stimulated AI-2 production suggested that carbonate ion is required to induce AI-2 production in S. gordonii. Further, comparison of the AI-2 induction levels of cells grown in BHI medium containing 24 mM Na2CO3 (24-fold; Fig. 5B) to the higher AI-2 induction levels observed in BHIS cultures supplemented with the same amount of Na2CO3 (49-fold; Fig. 4B) demonstrated that during S. gordonii cultivation in BHI medium, carbonate and serum both contributed to AI-2 production.
FIG. 4.
Induction of S. gordonii AI-2 production by Na2CO3 in a concentration-dependent manner. Time courses of culture densities (A) and AI-2 induction levels (B) of wild-type S. gordonii grown in BHIS supplemented with 0 mM (circles), 12 mM (squares), 24 mM (triangles), and 48 mM (diamonds) Na2CO3. All time course experiments were repeated at least six times, and the averages of three representative independent experiments with standard deviations are shown (B). KU, Klett units.
FIG. 5.
Contribution of carbonate ion to S. gordonii AI-2 production. Time courses of culture densities (A) and AI-2 induction levels (B) of wild-type S. gordonii grown in BHI medium (circles) or BHI medium supplemented with 24 mM MOPS (squares), 24 mM Na2CO3 (triangles), and 24 mM NaHCO3 (diamonds). All time course experiments were repeated at least six times, and the averages of three representative independent experiments with standard deviations are shown (B). KU, Klett units.
S. gordonii AI-2 production was also evaluated in several other media. Maximum induction levels observed in culture supernatants from the lag, exponential, and stationary phases of growth from cells grown in 25% saliva (twofold induction), LB medium supplemented with 0.5% glucose (no induction), and TY-glucose (ninefold induction; Fig. 3B) were similar to the background AI-2 induction levels (sixfold induction; Fig. 3B) detected in the luxS mutant.
Biofilm growth.
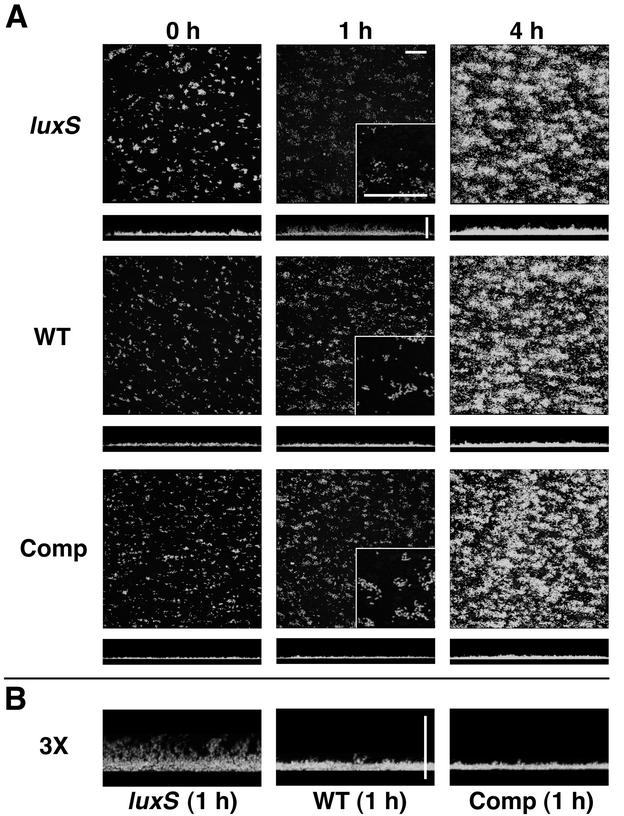
S. gordonii biofilms were grown and evaluated in flow cells with 10-fold dilutions of the same media that stimulated AI-2 production in planktonic culture; full-strength media did not support biofilm formation. In 10-fold-diluted THBS, attachment of cells to the substratum occurred but biomass decreased after 1 h and was nearly undetectable after 4 h (data not shown). When they were grown in flow cells with 10-fold-diluted BHI medium containing 2.4 mM Na2CO3, no differences were noted between luxS mutant and wild-type biofilms (Fig. 6) or complemented mutant biofilms (data not shown). At 0 h, these biofilms consisted of short chains of cells that were evenly distributed over the flow cell surface. After 1 h, the chains had elongated but cells had not formed microcolonies. After 4 h, chains continued to elongate and greater surface coverage was achieved but microcolonies still had not formed (Fig. 6).
FIG. 6.
S. gordonii luxS mutant biofilm phenotype is not apparent in BHI medium. (A) Time course (left to right) of biofilm development in medium-conditioned flow cells (10-fold-diluted BHI medium containing 2.4 mM Na2CO3) by luxS mutant (luxS) and wild-type (WT) S. gordonii strains. x-z reconstructions of each biofilm are shown below each x-y image. Digital zooms (magnification, 3×) of the lower left corner of each 1-h x-y image are shown as insets. (B) Digital zooms (magnification, 3×) of the center of each 1-h x-z reconstruction. Scale bars, 50 μm.
In contrast, when grown as biofilms with 25% human saliva as the sole nutrient source, luxS mutant, wild-type, and complemented mutant cells aggregated and formed microcolonies visible at 0 h (Fig. 7A). After 1 h of growth, microcolony organization was still apparent but only a minor increase in surface coverage occurred. When viewed in the x-z plane, microcolonies of the luxS mutant exhibited finger-like projections of cells that extended into the lumen of the flow cell and were taller than microcolonies of the wild-type and complemented mutant strains (Fig. 7B). After 4 h of growth, the biofilms all looked similar in the x-y plane and were characterized by denser microcolonies and greater surface coverage than at previous time points. However, when viewed in the x-z plane, taller microcolonies again were evident in luxS mutant biofilms compared to the wild-type and complemented mutant strains. AI-2 was not detected in the bulk liquid phases of saliva- or laboratory medium-grown biofilms.
FIG. 7.
S. gordonii luxS mutant biofilm phenotype in saliva. (A) Time courses (left to right) of biofilm development in saliva-conditioned flow cells by luxS mutant (luxS), wild-type (WT), and complemented luxS mutant (Comp) S. gordonii strains. x-z reconstructions of each biofilm are shown below each x-y image. Digital zooms (magnification, 3×) of the lower left corner of each 1-h x-y image are shown as insets. (B) Digital zooms (magnification, 3×) of the center of each 1-h x-z reconstruction. Scale bars, 50 μm.
Mathematical algorithms (48, 49) were used to determine the average microcolony heights and biovolumes of 4-h biofilms formed in saliva. This analysis underscored visual observations (Fig. 7A, 4-h x-z panels) that the microcolonies within luxS mutant biofilms (average height of 2.88 μm) were taller than microcolonies within wild-type (average height of 1.86 μm) and complemented mutant (average height of 2.06 μm) biofilms. A volumetric analysis of 4-h biofilms revealed that the luxS mutant, wild-type, and complemented mutant strains occupied similar biovolumes of 6.37 × 104, 7.25 × 104, and 7.70 × 104 μm3, respectively, consistent with the observation that the strains exhibited the same growth rates in planktonic culture. Thus, similar biovolume values for saliva-grown biofilms formed by the three strains suggest that the biofilms all contained similar amounts of biomass while quantitative analysis of average microcolony heights showed that the luxS mutant formed taller microcolonies than the wild-type and complemented mutant strains. Together, visual and mathematical analyses of saliva-grown biofilms revealed that the presence of an intact luxS gene did not affect biomass accumulation but rather exerted effects on biomass architecture.
DISCUSSION
The work described herein demonstrated that S. gordonii luxS is monocistronic, and it defined the nutritional conditions under which the organism produces AI-2. Additionally, the effects of luxS on the development of monospecies biofilms in flowing saliva by S. gordonii, an initial colonizer of human enamel surfaces, are described.
AI-2 production.
S. gordonii AI-2 production was evaluated in a variety of laboratory media, and maximum AI-2 induction was observed when the organism was cultivated in medium containing serum and carbonate. AI-2 was not detected in the supernatants of planktonic saliva-grown S. gordonii cultures or in the bulk liquid phase from flow cells. The inability to detect AI-2 in planktonic saliva-grown cultures may have been a result of the low maximum cell densities (106 to 107 CFU/ml) attained under these conditions. During planktonic cultivation in laboratory media, AI-2 did not reach detectable levels until cultures attained densities of 108 to 109 CFU/ml. Thus, if saliva-grown cultures produced the same amount of AI-2 per cell as laboratory medium-grown cultures, AI-2 concentrations in saliva culture supernatants would be below the detection limit of the AI-2 assay. Furthermore, in flow cells, AI-2 would not accumulate to a detectable concentration in the bulk liquid phase unless AI-2 production exceeded dilution by flow.
Surette and Bassler reported that AI-2 production by Salmonella enterica serovar Typhimurium responds to conditions such as the availability of a rich carbon source, high osmolarity, and low pH (41). Our results obtained by planktonic culture are consistent with these findings, and serum and carbonate, which are both naturally present in the human mouth, established nutritional conditions that contributed to S. gordonii AI-2 production. Both saliva (3) and serum (38) contain bicarbonate as the primary buffering agent. Furthermore, the physiological bicarbonate concentration in serum and saliva (3) is consistent with the concentration used in our experiments. In solution, carbonate ion (CO32−), bicarbonate ion (HCO31−), carbonic acid (H2CO3), and carbon dioxide (CO2) all exist in equilibrium, with their amounts determined by pH. As the pH of a carbonate-containing culture acidifies, carbonate dissociates to yield carbon dioxide. Thus, carbon dioxide may serve as an environmental signal that induces AI-2 production in S. gordonii. Acid production within dental plaque causes dissociation of salivary bicarbonate into carbonic acid, which is then converted into carbon dioxide and water by carbonic anhydrase (23). Metabolically, streptococci are capable of fixing the carbon from carbon dioxide by carboxylation of phosphoenolpyruvate to yield oxaloacetate (21, 26). Oxaloacetate is then transaminated to yield aspartate, which can be converted to methionine through several additional enzymatic steps (29). AI-2 is a side product of the bacterial activated-methyl cycle, a methionine-recycling pathway (47). Thus, stimulation of AI-2 production by carbonate may result from the increased levels of intracellular methionine produced through carbon dioxide fixation, which increases flux through the activated-methyl cycle, a precursor to AI-2 production. While growing as a biofilm that is bound to the acquired pellicle, S. gordonii would be well situated to fix soluble carbon dioxide liberated by pellicle-bound carbonic anhydrase (23), triggering the cells to produce AI-2. On a tooth surface, elevated carbon dioxide concentrations would most likely occur following acid production by dental plaque.
Biofilm phenotype.
Under conditions that resulted in high AI-2 concentrations in planktonic cultures, biofilms either did not form (THBS; data not shown) or did not exhibit phenotypic differences (BHI medium plus carbonate; Fig. 6). Furthermore, when biofilms were cultivated in BHI medium (Fig. 6A; see elongated chains of cells), growth resembled that of planktonic BHI medium-grown cultures, where streptococci exhibit characteristic chains of cells. All saliva-grown biofilms differed significantly in cellular organization from those grown in BHI medium (compare Fig. 6A and 7A; clumps of cells or microcolonies in Fig. 7A). In addition, a phenotypic difference between the luxS mutant and wild-type strains was apparent in saliva-grown biofilms (finger-like projections of cells extending from luxS mutant biofilms in Fig. 7B). These results suggest that the S. gordonii luxS mutant phenotype depends on an intrinsic property of biofilm growth in saliva.
S. gordonii binds to components within the salivary pellicle, including mucins, agglutinins, proline-rich proteins, statherin, and α-amylase (20). Saliva-grown biofilms formed by the luxS mutant, wild-type, and complemented mutant strains all exhibited a microcolony architecture (Fig. 7A) that was not apparent in BHI medium-grown biofilms (Fig. 6A). Thus, binding of S. gordonii to a salivary pellicle may facilitate initial microcolony formation within saliva-grown biofilms. Saliva-grown biofilms of the luxS mutant, however, consisted of taller microcolonies that exhibited finger-like projections of cells with morphological similarities to the chains of cells observed in BHI medium-grown biofilms (compare Fig. 6B and 7B). Within wild-type microcolonies formed in saliva, a localized concentration of AI-2 higher than that in the surrounding liquid could arise, resulting in the development of a biofilm architecture different from that observed in the luxS mutant. In contrast, localized regions with elevated AI-2 levels may not exist within BHI medium-grown biofilms that lack microcolony organization. Thus, while all three strains formed microcolonies when cultivated as biofilms in saliva, disruption of luxS altered the biofilm architecture, resulting in the development of taller microcolonies that exhibited finger-like projections of cells extending into the flow cell lumen, similar to the chains of cells observed in BHI medium-grown biofilms.
As a member of the human oral flora and an initial colonizer of enamel surfaces, S. gordonii evolved under conditions of continuous salivary flow; if the cells fail to adhere to a surface and form microcolonies, they are removed from the ecosystem. Thus, it is intriguing that the phenotype of an S. gordonii luxS mutant becomes apparent only when the organism is grown under conditions representative of the human oral environment, namely, as a biofilm in flowing saliva.
Acknowledgments
We thank P. Egland (National Institute of Dental and Craniofacial Research, National Institutes of Health), A. Firoved (National Institute of Allergy and Infectious Diseases, National Institutes of Health), and J. Foster (National Institute of Dental and Craniofacial Research, National Institutes of Health) for critical evaluation of the manuscript, R. McNab for sharing information regarding the S. gordonii luxS disruption strategy used in this study, and B. Bassler for supplying V. harveyi strains BB152 and BB170 and for advice regarding the AI-2 bioassay.
REFERENCES
- 1.Andersen, R. N., N. Ganeshkumar, and P. E. Kolenbrander. 1993. Cloning of the Streptococcus gordonii PK488 gene, encoding an adhesin which mediates coaggregation with Actinomyces naeslundii PK606. Infect. Immun. 61:981-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23:469-472. [DOI] [PubMed] [Google Scholar]
- 3.Bardow, A., B. Madsen, and B. Nauntofte. 2000. The bicarbonate concentration in human saliva does not exceed the plasma level under normal physiological conditions. Clin. Oral Investig. 4:245-253. [DOI] [PubMed] [Google Scholar]
- 4.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 6.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 7.Brendel, V., G. H. Hamm, and E. N. Trifonov. 1986. Terminators of transcription with RNA polymerase from Escherichia coli: what they look like and how to find them. J. Biomol. Struct. Dyn. 3:705-723. [DOI] [PubMed] [Google Scholar]
- 8.Burgess, N. A., D. F. Kirke, P. Williams, K. Winzer, K. R. Hardie, N. L. Meyers, J. Aduse-Opoku, M. A. Curtis, and M. Camara. 2002. LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiology 148:763-772. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 10.Chung, W. O., Y. Park, R. J. Lamont, R. McNab, B. Barbieri, and D. R. Demuth. 2001. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J. Bacteriol. 183:3903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cvitkovitch, D. G. 2001. Genetic competence and transformation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:217-243. [DOI] [PubMed] [Google Scholar]
- 12.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dû, L. D., and P. E. Kolenbrander. 2000. Identification of saliva-regulated genes of Streptococcus gordonii DL1 by differential display using random arbitrarily primed PCR. Infect. Immun. 68:4834-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Demuth. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69:7625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong, K. P., L. Gao, and D. R. Demuth. 2003. luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect. Immun. 71:298-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frias, J., E. Olle, and M. Alsina. 2001. Periodontal pathogens produce quorum sensing signal molecules. Infect. Immun. 69:3431-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg, E. P., J. W. Hastings, and S. Ulitzur. 1979. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch. Microbiol. 120:87-91. [Google Scholar]
- 18.Håvarstein, L., and D. Morrison. 1999. Quorum sensing and peptide pheromones in streptococcal competence for genetic transformation, p. 9-19. In G. Dunny and S. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, DC.
- 19.Jenkinson, H. F., S. D. Terry, R. McNab, and G. W. Tannock. 1993. Inactivation of the gene encoding surface protein SspA in Streptococcus gordonii DL1 affects cell interactions with human salivary agglutinin and oral actinomyces. Infect. Immun. 61:3199-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lachica, V. F., and P. A. Hartman. 1969. Inhibition of CO2 fixation in group D streptococci. Can. J. Microbiol. 15:57-60. [DOI] [PubMed] [Google Scholar]
- 22.LeBlanc, D. J., L. N. Lee, and A. Abu-Al-Jaibat. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130-145. [DOI] [PubMed] [Google Scholar]
- 23.Leinonen, J., J. Kivels, S. Parkkila, A.-K. Parkkila, and H. Rajaniemi. 1999. Salivary carbonic anhydrase isoenzyme VI is located in the human enamel pellicle. Caries Res. 33:185-190. [DOI] [PubMed] [Google Scholar]
- 24.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 25.Macrina, F. L., K. R. Jones, and P. H. Wood. 1980. Chimeric streptococcal plasmids and their use as molecular cloning vehicles in Streptococcus sanguis (Challis). J. Bacteriol. 143:1425-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, W. R., and C. F. Niven, Jr. 1960. Mode of CO2 fixation by the minute streptococci. J. Bacteriol. 79:295-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moat, A. G., and J. W. Foster. 1995. Microbial physiology, 3rd ed. Wiley-Liss, Inc., New York, N.Y.
- 30.Moore, W. E., and L. V. H. Moore. 1994. The bacteria of periodontal diseases. Periodontol. 2000 5:66-77. [DOI] [PubMed] [Google Scholar]
- 31.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369-380. [DOI] [PubMed] [Google Scholar]
- 32.Palmer, R. J., Jr., S. M. Gordon, J. O. Cisar, and P. E. Kolenbrander. 2003. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 185:3400-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer, R. J., Jr., K. Kazmerzak, M. C. Hansen, and P. E. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabelnikov, A. G., B. Greenberg, and S. A. Lacks. 1995. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J. Mol. Biol. 250:144-155. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 38.Sherwood, L. 1997. Human physiology: from cells to systems, 3rd ed. Wadsworth Publishing Company, Belmont, Calif.
- 39.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 42.Swartzman, E., M. R. Silverman, and E. A. Meighen. 1992. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J. Bacteriol. 174:7490-7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 44.Tao, L., D. J. LeBlanc, and J. J. Ferretti. 1992. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120:105-110. [DOI] [PubMed] [Google Scholar]
- 45.Voskuil, M. I., and G. H. Chambliss. 1998. The −16 region of Bacillus subtilis and other gram-positive bacterial promoters. Nucleic Acids Res. 26:3584-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whittaker, C. J., C. M. Klier, and P. E. Kolenbrander. 1996. Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 50:513-552. [DOI] [PubMed] [Google Scholar]
- 47.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 48.Xavier, J. B., A. Schnell, S. Wuertz, R. Palmer, D. C. White, and J. S. Almeida. 2001. Objective threshold selection procedure (OTS) for segmentation of scanning laser confocal microscope images. J. Microbiol. Methods 47:169-180. [DOI] [PubMed] [Google Scholar]
- 49.Xavier, J. B., D. C. White, and J. S. Almeida. 2003. Automated biofilm morphology quantification from confocal laser scanning microscopy imaging. Water Sci. Technol. 47:31-37. [PubMed] [Google Scholar]