Abstract
Innate immunity is crucial in the early stages of resistance to novel viral infection. The family of cytokines known as the interferons (IFNs) forms an essential component of this system: they are responsible for signalling that an infection is underway and for promoting an antiviral response in susceptible cells. We construct a spatial stochastic model, parameterized by experimental data and informed by analytic approximation, to capture the dynamics of virus–IFN interaction during in vitro infection of Madin–Darby bovine kidney cell monolayers by Herpes simplex virus 1. The dose dependence of infection progression, subsequent monolayer destruction and IFN-β production are investigated. Implications for in vivo infections, in particular the priming of susceptible cells by IFN-β during infection, are considered.
Keywords: innate immunity, viral infection, cytokines, interferon, stochastic model, dose dependence
1. Introduction
Viral infections in hosts provoke a response from the immune system, which works in higher vertebrates both to suppress acute stages of infection and establish a pathogen-specific immune state to deal with current and future infections. The specific response is developed and mounted by the adaptive immune system; in particular, the proliferation of T cells and B cells and the release of immunoglobins.
While the adaptive immune system is both fast and effective at targeting invasions by previously encountered pathogens, its role in host defence in the first days of novel infection is secondary to that of the innate immune system (Perelson et al. 1996; Medzhitov & Janeway 1997a,b). The latter is non-specific and immediate, requiring neither prior experience of the invading virus, nor time for lymphocytes to undergo proliferation before mounting a response. Cells and proteins of the innate immune system are ever present in a healthy host, and can respond to invading pathogens within the first minutes and hours of infection (Medzhitov & Janeway 2000). This system is a key process in preventing the establishment of new infections.
Here, we integrate data and models to explore the dynamic interaction between an invading virus and the non-adaptive immune response, mediated by cytokines. Cytokines are an essential component of the immune system. They are a family of small soluble proteins secreted by a multitude of different cells (though largely those of the immune system itself). They can be loosely classified into one of four families: the haematopoietins, which include many of the interleukins; the immunoglobin superfamily; the tumour necrosis factor family; the interferons (IFN; Janeway et al. 2005). Cytokines have many autocrine, paracrine and endocrine effects, such as inducing inflammation (Alheim et al. 1997; Svanborg et al. 1999), increasing body temperature, (Hober et al. 1993; Chai et al. 1996), or stimulating the adaptive response (Goodbourn et al. 2000; Luster 2002).
The IFNs are perhaps the most important cytokines in the initial innate response to viral infection. They are classified into two types: IFN-α (a family of related proteins) and the single protein IFN-β together form type I; IFN-γ is the sole and unrelated type II IFN (Young 1996). IFN-α and IFN-β are secreted by cells in response to viral infection and promote an antiviral response in otherwise susceptible cells (Kalvakolanu & Borden 1996). IFN-α secretion is restricted predominantly to leukocytes, but in contrast, IFN-β is produced by a wide variety of cells. For this reason, the dynamics of production and action of IFN-β can be studied in vitro in experimentally simple situations.
It is believed that the presence of double-stranded RNA (dsRNA) in the cytoplasm, originating from either the viral genome itself or from viral replication, induces IFN-β expression (Jacobs & Langland 1996), though other mechanisms may play a role. This pathway is complex and not fully understood (Goodbourn et al. 2000). IFN-β is produced and secreted from the infected cell and then binds to the surfaces of neighbouring cells, in turn inducing the production of antiviral proteins, such as Mx, PKRkinase, and oligoadenylate synthetase through the JaK/STAT pathway (Kalvakolanu & Borden 1996; Stark et al. 1998). These proteins interfere with and degrade virus-associated proteins and structures, thus rendering the cells effectively antiviral (Staeheli et al. 1986; Pavlovic et al. 1990; Samuel 1991; Landis et al. 1998; Lee & Vidal 2002).
The experimental system we studied is the in vitro infection of epithelial cell monolayers with Herpes simplex virus 1 (HSV-1). HSV-1 is a cytolytic DNA virus; it is stable and widely researched. Productive infection occurs over the course of 12–24 h (Barreca & O'Hare 2004), involving cytolysis and the release of a new generation of virions, typically of around 100 pfu per cell (pfu, plaque forming units; i.e. 100 infection-competent virions; Rampling et al. 2000). HSV-1 is generally resistant to IFN-mediated immune responses (Härle et al. 2002) through proteins, such as ICP0 (Mossman et al. 2000). However, the virus is sensitive to the IFN response it provokes in certain cell lines, such as MDBK (Madin–Darby bovine kidney) cells. This response is known to inhibit the spread of HSV-1 infection in monolayers in vitro (Barreca & O'Hare 2004).
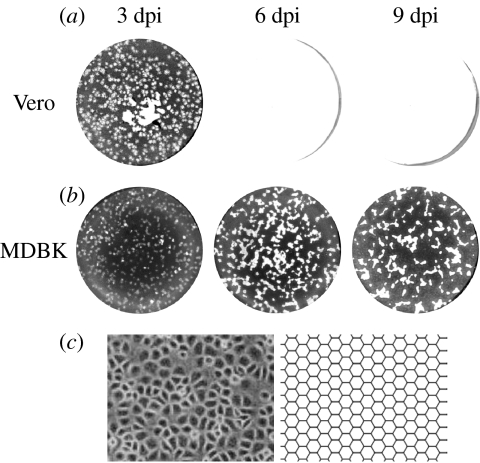
The spread of infection leaves a plaque of dead (lysed) cells (figure 1). In the experiments from which our data are taken, a viscous overlay containing neutralizing antibody was used to inhibit long-range diffusion of infectious virions (which would in any case be limited, owing to the comparatively large size of HSV virions and their attachment to immediately adjacent cells). This allows infection to progress cell-to-cell only, so that plaques of lysed cells grow smoothly outward from the initial site of infection. In cell lines without an IFN response to infection (in this case, Vero; Desmyter et al. 1968; Rhim et al. 1969; Emeny & Morgan 1979), infection spreads unchecked until the entire monolayer has been destroyed (figure 1a). In contrast, in MDBK cells, the production of IFN-β in response to the intracellular presence of virus acts to promote an antiviral response, which, when established, blocks further spread, and, in fact, allows the population to recover and undergo regrowth, leading to a reduction in plaque size (figure 1b).
Figure 1.
Progression of HSV-1 infection in cell monolayers and comparison between epithelial cell monolayer and hexagonal lattice. (a) Vero cells, which do not possess an IFN-β response to infection, and so are unable to achieve an antiviral state, are destroyed by infection, leaving a plaque of dead cells (white). (b) MDBK cells, which begin to secrete IFN-β on infection, are able to slow, stop and reverse the course of infection. (c) Hexagons can approximate the spatial layout of epithelial cells. On the left is shown a population of MDBK cells; on the right, an example of a hexagonal lattice used to approximate such a population.
1.1 Structure of the paper
Despite their importance, the kinetics of the virus–IFN interaction have not been explored via dynamic models integrated with longitudinal viral data. By modelling the dynamics of plaque formation, it is possible to learn more about the time-scales involved in IFN-mediated antiviral dynamics. Questions relating to dose dependence can be explored. For example, how do the following key properties of the virus–cytokine interaction vary with initial infective dose: the time-scale of live infection; the kinetics of IFN production and duration of activity; and the severity of infection-induced pathology? In addition, we investigate how the priming of cells prior to infection affects plaque dynamics. We address these questions using a stochastic model of the in vitro infection, compared with experimental data and illuminated by analytic approximation.
2. Experimental data
The data consist of photographs of cell monolayers at increasing time post-infection (figure 1); it illustrates unrestricted growth of plaques in Vero cells, leading to total monolayer destruction (figure 1a), and the transition to an antiviral state and subsequent monolayer regrowth in MDBK cells (figure 1b).
In order to quantify the dynamics of plaque formation, it was necessary to calculate, for each plate, the percentage of the area of the monolayer that had formed plaque and normalize this against the number of plaques in the monolayer. The inoculum's ability to form plaques (its pfu) is a statistical average of the plaques that would form should the experiment be repeated many times over; the observed number of plaques will vary somewhat about this average. For large pfu, this variation is unimportant, but for smaller pfu (such as the 10 and 50 pfu experiments), the percentage plaque must be normalized against the actual number of plaques evident.
Calculating the percentage plaque by hand for a monolayer becomes more difficult as the inoculum pfu increases. Therefore, the photographs were instead passed through several image filters; after doing so, areas of plaque show up white and everything else is rendered black. The percentage plaque, now simply percentage of white pixels in the image, is then calculated from the image histogram.
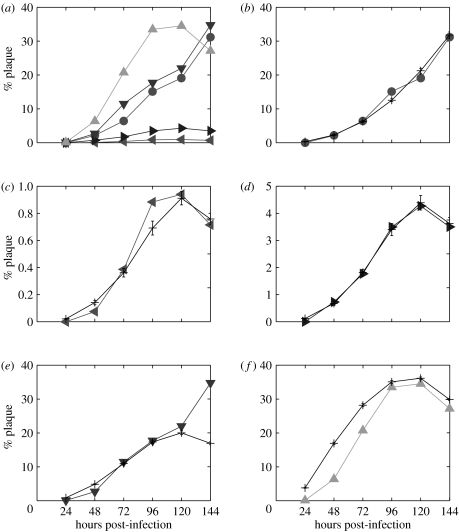
After transformation, the data take the form of a series of monolayer plaque percentages, for the Vero experiment at 50 pfu, and the MDBK experiments at 10, 50, 500 and 2500 pfu (figure 2a). These results illustrate the following observations.
Figure 2.
Experimental data compared with results of parameter fitting. (a) All data and model predictions for (b) Vero 50 pfu (filled circle), (c) MDBK 10 pfu (filled left triangle), (d) MDBK 50 pfu (filled right triangle), (e) MDBK 500 pfu (filled down triangle) and (f) MDBK 2500 pfu (filled up triangle). The last experimental data point in (e) diverges greatly from the model prediction. Model output was averaged over 20 realizations. Animations created from model output for infections in Vero and MDBK cells can be found in the electronic supplementary material.
2.1 Total monolayer destruction in Vero cells
The data for the Vero 50 pfu case support the qualitative observation that plaque growth in Vero cells is unrestricted. This leads to eventual total monolayer destruction, though this is not directly reflected in the graph, as the full dataset extends to only 6 days post-infection (dpi).
2.2 Regrowth in MDBK cells
The data for MDBK 10, 50 and 2500 pfu cases show quantitatively the antiviral effects of the IFN response to HSV infection that MDBK cells possess. Initially, the percentage plaque accelerates as the infection takes hold; subsequently, plaque growth slows as the increasing IFN concentration impedes new infection. It stops entirely once the antiviral state is reached at around 5 dpi; regrowth of the monolayer then takes over and the plaques shrink (observed as a decrease in percentage plaque).
The data for the MDBK 500 pfu infection appear anomalous, since although plaque growth appears to slow between the fourth and fifth days post-infection, the data point for the sixth day shows an unexpectedly high percentage plaque. This is perhaps due to a spurious experimental outcome. In light of this, the 6 dpi data point was not included during parameterization of the model. It may be, however, that this result is not anomalous, but rather indicative of another course that HSV infection may take in this system; we return to this point in the discussion.
3. Model construction
Vero and MDBK are both epithelial cells, which grow and stick together to form membranes or boundaries. Epithelial cells will grow and divide in a region until it is completely covered; it has been shown that such growth results in an arrangement of cells best approximated by a hexagonal lattice (figure 1c; Nagai & Honda 2001). The discrete nature of the cells, the cell-to-cell spread of infection, and the relatively small number of infection events which take place (particularly at low pfu and during the early stages of infection) strongly suggest that we begin with an individual-based spatially explicit stochastic model, rather than, for example, an analytic spatial diffusion model (Cohen & Murray 1981; Murray 1989; Fearon & Locksley 1996; Gatenby & Gawlinski 1996).
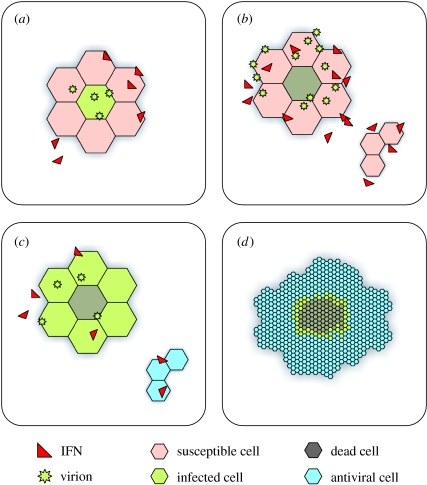
The monolayer is represented by a static array of cell objects, each of which is furnished both with parameters capturing biological states (infected, secreting IFN, antiviral, number of virions present in local area, etc.) and a list of other cell objects that are its neighbours. Transitions between the various cell states are governed probabilistically (figure 3; figure 1 in the electronic supplementary material).
Figure 3.
Progression of infection in the model: (a) virions successfully infect susceptible cells with probability ρ, which decreases monotonically with increasing IFN concentration (hence capturing the IFN-mediated suppression of infection). On infection, IFN production is begun; actual secretion starts after a delay of between 4 and 6 h. (b) The incubation period for infection is around 12 h, after which cytolysis occurs with a viable virion burst size normally distributed about 100; viable virions are scattered to neighbouring cells according to the previously determined neighbour weightings. IFN secreted by infected cells acts in an endocrine manner (i.e. globally) to reduce the probability of successful infection, ρ, and to initiate an antiviral state in uninfected cells. (c) Virions released at cytolysis go on to infect neighbouring susceptible cells. Away from infection, IFN brings about an antiviral state in susceptible cells after a time τ. (d) All susceptible cells have entered an antiviral state, leading to a cease in the spread of infection. Cell monolayer regrowth takes place as cells divide.
A cell's connections to its neighbours are weighted according to the physical distance between them on the lattice. Additional weight is given to the six adjacent cells, and cells more than two cell-widths distant cannot be neighbours (resulting in a maximum of 18 neighbours). Initially, to better approximate the heterogeneity in cell distribution in live monolayers, the addition of random variation to these weightings was also investigated: weightings were multiplied by random variables from several distributions on the interval [0,1]. However, these modifications were found to have no qualitative effect on model behaviour. Therefore, we use the regular lattice (without random weightings) in subsequent fits and simulations.
An overlay was used in the experiments to neutralize diffusion of infectious virions (therefore restricting the spread of infection from cell-to-cell). Accordingly, on cytolysis, new virions are distributed among the lysed cell's neighbours, according to the connection weightings, but do not diffuse or move in any other way in the model. Infectious particles will then enter the neighbouring cells during a time-step, with a probability ρ, infecting them and thus spreading the infection.
A time-step of 1 h is used in the model; this is sufficiently fine to capture the dynamics of cell infection, while being large enough to allow tractable model execution.
IFN molecules are many times smaller than virions (Janeway et al. 2005), and so diffuse much faster than them. For the purposes of this model, therefore, IFN diffusion is approximated as instantaneous; consequently, IFN is tracked at the global level, so that IFN concentration is the same at all points in the virtual monolayer.
The model was implemented in C++, and run on several different platforms. Neither special equipment nor computational arrangements were required. The computational capabilities and memory capacity of modern desktop computers were more than sufficient to simulate several days of infection in entire cell cultures (roughly 2×106 cells) in a few hours of runtime.
3.1 Parameter fitting
Many parameter values were taken from the literature as a first approximation for the purposes of modelling (Rampling et al. 2000; Barreca & O'Hare 2004): viral burst size is 100; incubation period is 12 h; cell division time is 24 h; and IFN secretion delay (the time between stimulation by dsRNA and first secretion of IFN) is 5 h. By choice of units of IFN, the IFN secretion rate can be scaled to 1.
Unknown parameters were estimated by least squares: the squared distance between the mean of an ensemble of model realizations and the experimental data was minimized. The three unknown parameters in the model are the virion infection probability ρ, the IFN suppression factor α, a summary parameter which encompasses the ways in which IFN impedes infection (and so effectively reduces ρ) and the antiviral time τ, the average time taken for an uninfected but IFN-exposed cell to reach an IFN-mediated antiviral state.
In the IFN-free (i.e. Vero) case, only ρ must be fit; this was estimated to be ρ=0.8, with a 95% confidence interval of [0.74,0.87]. All confidence intervals were calculated using the asymptotic properties of the likelihood ratio statistic (Fan et al. 2001).
Next, parameters controlling IFN-mediated behaviour were fit to the data from MDBK experiments; it was also necessary to re-fit the virion infection probability, ρ, since the MDBK cell line does not necessarily have the same natural susceptibility to HSV infection. Parameter values [and 95% confidence intervals] of ρ=0.09 [0.07,0.11], τ=95 [88,101] h and α=1.5 [1.2,1.8] were estimated. When used in the model, these parameter values lead to the model fits to experimental data shown in figure 2b–f.
The lower value of ρ for MDBK cells suggests that they are less susceptible to infection by HSV-1 than Vero cells. It may also reflect the suppressive effect that the presence of an IFN response has on the early stages of infection. The time taken for the monolayer to reach an antiviral state (after which regrowth begins), τ=95 h, is relatively long; in an infection in vivo, the adaptive response would most likely be acting to suppress the infection by this time (Janeway et al. 2005). However, this value reflects the time at which plaque shrinking is witnessed in the MDBK data; the actual response time may therefore be faster. The IFN suppression factor α is a portmanteau parameter and dictates how IFN concentration reduces the infection probability. In the absence of IFN, a viable virion will, over the course of a time-step, successfully infect an adjacent susceptible cell with probability ρ. This probability is reduced by a factor for an IFN concentration I.
3.2 Visualizing the progression of infection
To better visualize plaque dynamics, animations of graphical model output were compiled using these parameter values (see the electronic supplementary material). The uncontrolled growth of plaques in Vero cell cultures and the slowing and reversal of plaque formation in MDBK cells are clearly illustrated.
4. Simulating virus–cell–IFN dynamics
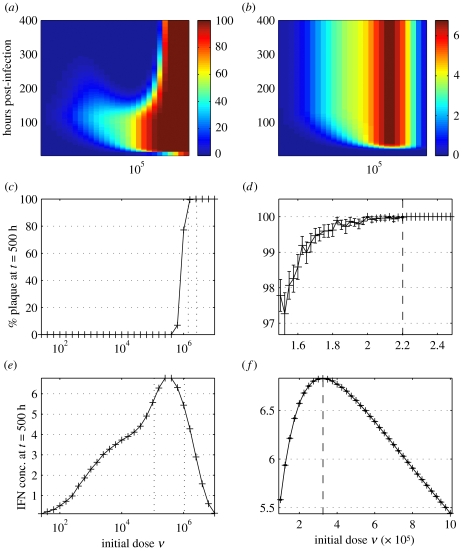
Now parameterized, the model can be used to simulate experiments. In particular, we focused on ‘virtual’ plates inoculated with varying doses ν (from 101 to 107 pfu). Infections were run through to 400 h, recording percentage plaque, IFN concentration and virion production every 2 h (figure 4). The results suggested that the qualitative dynamics of the infection change across the following three critical threshold initial doses.
Figure 4.
Model time-series profiles at initial doses ν from 101 to 107 pfu. Model output was averaged over 50 realizations. (a) Percentage plaque; (b) IFN concentration. (c, d) Determining the critical dosage for total monolayer destruction. Percentage plaque at t=500 h post-infection with (c) 101–107 pfu and (d) . The critical dosage is marked in (d). (e, f) Determining the critical dose for maximal IFN-β production. IFN concentration at t=500 h post-infection with (e) 101–107 pfu and (f) 105–106 pfu. The critical dosage is marked in (f).
4.1 Total monolayer destruction
The IFN-mediated antiviral state takes time to be established throughout the monolayer. Provided the infection has not destroyed all cells by this time, antiviral cells should remain; these can go on to divide and re-establish the monolayer. Hence, there exists a critical dosage, below which antiviral effects take hold before total destruction and above which destruction occurs before an antiviral state is reached. Running the model for sufficiently long (beyond any point at which an antiviral state might be achieved) allows us to determine the eventual state of the monolayer. If, by this time, plaque coverage is 100%, then recovery can never take place; if not, regrowth is underway and monolayer recovery will follow. Figure 4c,d shows the percentage plaque at t=500 h plotted against dose. From this it can be deduced that the critical dosage for total monolayer destruction is or roughly 0.9 pfu per cell.
4.2 Maximizing IFN-β production
From figure 4b, there is a second critical dosage, this time for maximal IFN-β production. As previously, IFN concentration at the end of infection was plotted against dose (figure 4e,f). We found that the critical dosage for maximal IFN-β production is or roughly 0.1 pfu per cell.
4.3 Minimizing recovery time
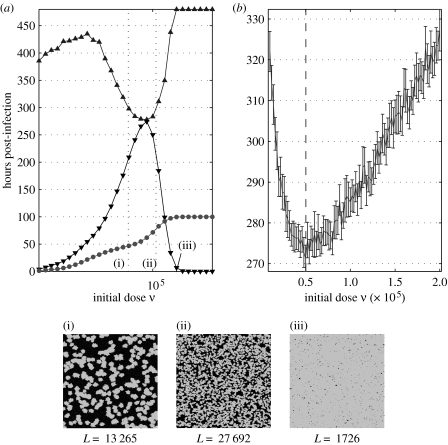
For the purposes of modelling, the recovery time is taken to mean the time at which total regrowth is completed (i.e. the time taken for all hexagons in the lattice to once again contain a live ‘cell’). The recovery time for doses from 101 to 107 pfu was calculated and plotted (figure 5), as well as the time and magnitude of percentage plaque peak (i.e. the highest value of the percentage of the monolayer covered with dead cells throughout infection). The non-monotinicity of recovery against dose suggested by the slightly upturned leftward protuberance of lighter colours into the darker region in figure 4a is clearly visible. In the cases where 100% plaque (i.e. total monolayer destruction) is achieved, regrowth is not possible (as there are no living cells to re-seed the plate), and so the algorithm used to find the recovery time terminates after t=480 h—this accounts for the plateau seen in the top right of figure 5a, which corresponds to peak plaque values of 100%. The critical dosage for minimal recovery time was found to be or 0.02 pfu per cell.
Figure 5.
Determining the dose for minimal recovery time. Time to recovery (filled up triangle) and time and magnitude of percentage plaque peak (filled down triangle) after infection with (a) 101–107 pfu and (b) 105–106 pfu. Model output was averaged over 50 realizations. The boundary length at peak (filled circle) is shown in (a) for comparison with the recovery time. The critical dosage is marked in (b). (i)–(iii) Visual representations of model output. Models were run until peak plaque percentages, at doses (i) , (ii) and (iii) . Below each are the boundary lengths L, calculated to be the total number of healthy cells in contact with at least one infected or dead cell.
4.4 Summary of qualitative plaque dynamics
These results highlight the existence of three distinct critical viral dosage levels, each an order of magnitude larger than the next (table 1). The first, , defines the boundary between total eventual recovery and total destruction of the monolayer. Since virion production is directly proportional (ignoring stochastic variation) to the number of infected cells, this critical dosage is also the one at which maximum virion production occurs (since all cells become infected). Increasing the dose above this point merely results in a shorter live infection—the end result (total destruction) remains the same, as does (on average) the number of virions produced by the infection. It is perhaps surprising that is so high: 0.9 pfu per cell indicates that, on average, 90% of the monolayer will be destroyed in the first round of infection, and yet, from these results, doses at or just below this will still result in eventual monolayer recovery. In an experimental setting, however, this theoretical recovery time may well exceed the time for which such cultures can successfully be kept alive in vitro under infection conditions.
Table 1.
Critical doses suggested by modelling results.
| ν | value (pfu per cell) | description |
|---|---|---|
| νD | 0.9 | destruction versus recovery |
| νI | 0.1 | maximum IFN-production |
| νR | 0.02 | minimum recovery time |
The second critical dosage, , is that at which maximum final IFN concentration is reached. This result is less surprising: a delicate balance is struck at an intermediate dosage, between too small an infection which stimulates only a small number of cells to begin IFN-β secretion, and too large an infection which results in the secreting cells being killed off too quickly.
4.4.1 Recovery times and boundary lengths
The third threshold dosage, , is perhaps the most interesting. As the dose ν increases, one would expect the peak percentage plaque to increase also—and, indeed, both experimental data and model output reflect this expectation (figure 5a). One might also expect that, as peak percentage plaque increases, so too would the recovery time. However, while the time to recovery does initially increase with dose (and hence peak plaque), this trend fails at around ; approaching , a sharp decrease in recovery time is witnessed (figure 5b). Increasing ν further results in a second increase in recovery time, which, at , is undefined (since recovery cannot occur after total destruction).
The reason for this non-monotonic dependence of recovery time on ν becomes apparent when one considers where regrowth occurs, namely, at the boundary between healthy cells and plaque (as healthy cells divide into unpopulated regions). Time to recovery is dependent not just on the amount of plaque that must be recovered, but also on the length of the boundary at which this regrowth can take place. Figure 5a shows visual representations of model output for doses , and ; shown below each frame (a 360×360 cell region of a complete plate) is the boundary length, calculated to be the total number of healthy cells in contact with at least one infected or dead cell.
As ν increases, the number of plaques increase, but, because of the infection-suppressing action of IFN-β, each plaque is smaller, so that the resulting boundary is longer. Further increasing ν leads to a corresponding further reduction in the size of each plaque, but now there are so many that overlapping of the plaques results in a greatly decreased boundary length—there are no longer enough healthy cells to promote a rapid recovery.
4.5 An analytic approximation
To investigate how expected plaque boundary length varies with plaque size and number, we can abstract the problem to one of overlapping discs in the plane. Consider scattering n discs of radius r into an area A, so that the coordinates of the centre of each are uniformly distributed in their respective ranges. Now consider a point x on the circumference of one of the discs. Ignoring interactions with the boundary of the area A, the probability that x is covered by another disc is equal to the probability that the centre of a second disc lies in a circle of radius r about x, i.e.
Hence, the probability that none of the n−1 other discs cover x is
This is the same for any point on the circumference, so that we can calculate the expected effective circumference (i.e. circumference of this disc not covered by any other disc) by multiplying this probability by the actual circumference, ; finally, multiplying this by n gives us the expected total effective circumference,
This analytic derivation can be compared against stochastically generated results (figure 6). The model was used to compute the total plaque boundary in Vero cells at regular time intervals and for various pfu up to 2500. The strong similarity between analytic and stochastic results supports the analysis undertaken, and justifies the assumption that boundary effects can be ignored.
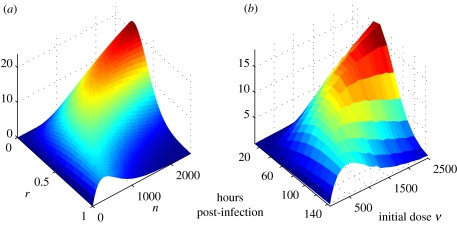
Figure 6.
Analytically derived effective circumference compared against stochastic simulations of plaque boundary length. In (a), the total effective circumference of n discs of radius r scattered in an area A has been plotted (scaled so that its form can be more easily compared with the stochastic results). In (b), the model was used to calculate the plaque boundary area (i.e. the number of cells multiplied by the area of a cell) of infection in Vero cells, in which infection progresses approximately linearly with time, at a range of plaque forming units. Model output was averaged over 50 realizations.
This allows us to calculate the critical values of r and n at which is maximal,
These equations give the critical values for r, given n, and n, given r, respectively; the maximum value for is unbounded as and .
We can equate the number of plaques n with the dose ν. In contrast with the analytic approximation, the plaque radius r does not vary freely, but grows in time at a rate sensitive to the IFN concentration, which is, in turn, affected by the dose ν. Though this additional dependence alters the quantitative behaviour, there exists still a critical dose, (cf. ), for which the boundary length is maximal.
Figure 5a includes a plot of hours to recovery and boundary length at peak percentage plaque (scaled by a positive constant factor, so that the two may be easily compared). As anticipated, the maximum boundary length at peak percentage plaque coincides with the minimum recovery time, so that .
4.6 Priming cells
During infections in hosts, cells may be primed with IFN (produced in infected regions elsewhere in the body, or as a result of vaccination (Pacheco et al. 2005)) prior to exposure to the virus itself. In vitro experiments confirm that monolayers treated with IFN show increased resistance to viral infection (Barreca & O'Hare 2004). Naive MDBK cells treated for 12 h with medium (shown to contain IFN-β) harvested at 3 dpi from an initial infection (7000 pfu) lead to a approximately 96% reduction in plaque count on subsequent infection using a standard dose of 1000 pfu, compared with untreated cells similarly infected.
Using the model, the IFN concentration in the medium resulting from a 7000 pfu infection at 3 dpi was estimated (approx. 3.4 units per cell); this quantity was then used to pre-treat 25 virtual monolayers for 12 h, after which 1000 pfu infections were carried out. An average plaque count reduction to 2.4% (with confidence interval [1.3%,3.4%]) was observed, consistent with the experimental results of Barreca & O'Hare (2004).
The stark contrast between progression of infection in primed and unprimed MDBK cells can clearly be seen in the animations of model output included in the electronic supplementary material. The animations show simulated infections in (equally sized) cell cultures with the same initial dose; plaques form more slowly and in much smaller numbers in the primed monolayer.
5. Discussion
This work demonstrates that it is possible to capture the early dynamics of the IFN-β response to viral infection—at least in the case of HSV-1 infection of MDBK cells—even after many simplifications and assumptions. No within-cell modelling was undertaken, so that heterogeneities in cell behaviour, such as progression of infection, were not included. Instead, cells were treated as ‘black boxes’ onto which virions and IFN-β bind, from which IFN-β is secreted, and out of which new virions emerge on infection-mediated cytolysis. In addition, diffusion of IFN-β molecules was assumed to be instantaneous, whereas in reality they would diffuse at finite speed according to local concentration gradients.
Despite the simplicity of the model, the results highlight a number of interesting biological issues, which prompt further study.
5.1 Behaviour of epithelial cell monolayers in vivo
Our work tentatively suggests that, in the absence of the cellular components of immunity, epithelial cell monolayers recover fastest from multiplicities of infection of up to around 10−2 (i.e. an initial dose of 10−2 pfu per cell). At this dose, recovery time is shortest, though total IFN production has not yet peaked. From the perspective of host health, attempting to minimize recovery time and cytokine production simultaneously may be a primary consideration, as cytokines are responsible for many of the clinical symptoms of viral infections (Hober et al. 1993; Chai et al. 1996; Alheim et al. 1997; Janeway et al. 2005).
For intermediate virus inocula, high concentrations of IFN are produced as a result of infection, though recovery may eventually take place. In these situations in vivo, patches of epithelial cells suffering such widespread infection may act more as IFN producers than barriers to further infection, stimulating both the transition to antiviral states elsewhere and a response from the cellular innate (and, later, adaptive) immune response. For high doses, total destruction is inevitable, as all cells become infected before an antiviral state can be achieved. This depletion of susceptible cells itself may prove to be a major limitation for primary in vivo infections on epithelial monolayers, such as infection of mucosal surfaces by influenza (Hamilton 2002; J. Wood 2006, personal communication).
The anomalous result in the MDBK 500 pfu dataset (i.e. the apparent lack of antiviral state establishment and monolayer regrowth by 6 dpi) might not be an artefact of data analysis, but may instead suggest that infection can occasionally escape regulation by IFNs. To investigate this, the infection in a single cell monolayer must be quantified throughout its course. This would require experiments making use of non-destructive monitoring of plaques over time (rather than fixing and staining at daily intervals), such as green fluorescent protein (GFP) expression (Elliott & O'Hare 1999).
5.2 Priming cells in vivo
Experimental and model results highlight the importance of priming of cells (i.e. pre-exposure to IFN) in viral infections—greatly reduced plaque counts following IFN treatment exemplify its antiviral function (Barreca & O'Hare 2004). In a host, the presence of cellular immune system components would likely increase the efficacy of priming, through amplification of IFN production and the marshalling of leukocytes to combat virus-infected cells. For example, dendritic cells have been shown to be potent producers of IFN-β in response to viral infection (Ferbas et al. 1994; Cella et al. 1999); their presence would result in increased IFN concentrations throughout infection, and therefore increased suppression of infection.
5.3 Applications and future work
The existence of (theoretical) critical dosages may prove useful in establishing the optimal dose for maximum IFN production. This technique may also be used to study other phenomenon in viral infections, such as latent infections and recrudescence of HSV. The employment of non-destructive techniques for monitoring of plaques, such as the GFP expression technique mentioned above, would capture the dynamics of plaque formation in a single tissue culture over the entire course of infection. These datasets would, in turn, allow more extensive parameterization and testing of the model.
This work could easily be adapted to other viruses, such as influenza, and would also provide a mechanistic spatial component for within-host models (which have been predominantly non-spatial to date (Perelson 2002), though some spatial modelling has been undertaken, e.g. Kirschner & Marino 2005). Extensions to models of this kind would require the inclusion of elements to capture the cellular and perhaps even structural responses of innate immunity (e.g. the enhanced signalling and cytokine secretion capabilities of dendritic cells, or the ciliary escalator and mucosal secretions of the upper respiratory tract); the effects of non-local virion disperal and virion diffusion would have to be considered, and would likely have significant impact on model dynamics and the course of infection.
Within-host models are often used to inform treatment regimens (Perelson et al. 1997; Frost et al. 2001; Hamilton 2002; Culshaw et al. 2003). In viral infections, cytokines are both crucial to the innate immune response and responsible for many of the infection's symptoms. Therefore, minimization of cytokine production coupled with effective combating of infection would be the objective, and within-host models may highlight courses of treatment that would best fulfil this. In conclusion, this work illustrates the dynamic nature of the interaction between viruses and the innate immune system. There is considerable scope for more work in this area, integrating laboratory approaches with theory.
Acknowledgements
We thank John McCauley for helpful discussions during this work. T.J.H. is supported by the MRC; C.B. and P.O. are supported by Marie Curie Cancer Care; J.R.G. is supported by the Royal Society; B.T.G. is supported by the BBSRC.
Supplementary Material
Animation of graphical model output showing the progression of infection in part of a ‘virtual’ tissue culture from t=0 to t=6 days post infection; the time between frames is 6 hours.
Animation of graphical model output showing the progression of infection in part of a ‘virtual’ tissue culture from t=0 to t=6 days post infection; the time between frames is 6 hours.
Animation of graphical model output showing the progression of infection in part of a ‘virtual’ tissue culture from t=0 to t =6 days post infection; the time between frames is 6 hours.
References
- Alheim K, Chai Z, Fantuzzi G, Hasanvan H, Malinowsky D, Santo E.D, Ghezzi P, Dinarello C.A, Bartfai T. Hyperresponsive febrile reactions to interleukin (IL) 1α and IL-1β, and altered brain cytokine mRNA and serum cytokine levels, in IL-1β-deficient mice. Proc. Natl Acad. Sci. USA. 1997;94:2681–2686. doi: 10.1073/pnas.94.6.2681. doi:10.1073/pnas.94.6.2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreca C, O'Hare P. Suppression of herpes simplex virus 1 in MDBK cells via the interferon pathway. J. Virol. 2004;78:8641–8653. doi: 10.1128/JVI.78.16.8641-8653.2004. doi:10.1128/JVI.78.16.8641-8653.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Salio M, Sakakibara Y, Langen H, Julkunen I, Lanzavecchia A. Maturation, activation, and protection of dendritic cells induced by double-stranded RNA. J. Exp. Med. 1999;189:821–829. doi: 10.1084/jem.189.5.821. doi:10.1084/jem.189.5.821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Z, Gatti S, Toniatti C, Poli V, Bartfai T. Interleukin (IL)-6 gene expression in the central nervous system is necessary for fever response to lipopolysaccharide or IL-1 beta: a study on IL-6-deficient mice. J. Exp. Med. 1996;183:311–316. doi: 10.1084/jem.183.1.311. doi:10.1084/jem.183.1.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D.S, Murray J.D. A generalized diffusion model for growth and dispersal in a population. J. Math. Biol. 1981;12:237–249. [Google Scholar]
- Culshaw R.V, Ruan S, Spiteri R.J. Optimal HIV treatment by maximising immune response. J. Math. Biol. 2003;48:545–562. doi: 10.1007/s00285-003-0245-3. doi:10.1007/s00285-003-0245-3 [DOI] [PubMed] [Google Scholar]
- Desmyter J, Melnick J.L, Rawls W.E. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero) J. Virol. 1968;2:955–961. doi: 10.1128/jvi.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott G, O'Hare P. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 1999;73:4110–4119. doi: 10.1128/jvi.73.5.4110-4119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emeny J.M, Morgan M.J. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 1979;43:247–252. doi: 10.1099/0022-1317-43-1-247. [DOI] [PubMed] [Google Scholar]
- Fan J, Zhang C, Zhang J. Generalized likelihood ratio statistics and Wilks phenomenon. Ann. Stat. 2001;29:153–193. [Google Scholar]
- Fearon D.T, Locksley R.M. The instructive role of innate immunity in the acquired immune response. Science. 1996;272:50–53. doi: 10.1126/science.272.5258.50. [DOI] [PubMed] [Google Scholar]
- Ferbas J.J, Toso J.F, Logar A.J, Navratil J.S, Rinaldo C.R., Jr CD4+ blood dendritic cells are potent producers of IFN-alpha in response to in vitro HIV-1 infection. J. Immunol. 1994;152:4649–4662. [PubMed] [Google Scholar]
- Frost S.D.W, Dumaurier M.-J, Wain-Hobson S, Brown A.J.L. Genetic drift and within-host metapopulation dynamics of HIV-1 infection. Proc. Natl Acad. Sci. USA. 2001;98:6975–6980. doi: 10.1073/pnas.131056998. doi:10.1073/pnas.131056998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby R.A, Gawlinski E.T. A reaction–diffusion model of cancer invasion. Cancer Res. 1996;56:5745–5753. [PubMed] [Google Scholar]
- Goodbourn S, Didcock L, Randall R.E. Interferons: cell signalling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 2000;81:2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- Hamilton, R. B. 2002 From horse to herd: linking within-host and population level epidemic dynamics. Ph.D. thesis, University of Cambridge.
- Härle P, Sainz B, Jr, Carr D.J.J, Halford W.P. The immediate-early protein, ICP0, is essential for the resistance of herpes simplex virus to interferon-α/β. Virology. 2002;293:295–304. doi: 10.1006/viro.2001.1280. doi:10.1006/viro.2001.1280 [DOI] [PubMed] [Google Scholar]
- Hober D, Poli L, Roblin B, Gestas P, Chungue E, Granic G, Pecarere P.I.J, Vergez-Pascal R, Wattre P. Serum levels of tumor necrosis factor-alpha (TNF-alpha), interleukin-6 (IL-6), and interleukin-1 beta (IL-1 beta) in dengue-infected patients. Am. J. Trop. Med. Hyg. 1993;48:324–331. doi: 10.4269/ajtmh.1993.48.324. [DOI] [PubMed] [Google Scholar]
- Jacobs B.L, Langland J.O. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. doi:10.1006/viro.1996.0259 [DOI] [PubMed] [Google Scholar]
- Janeway C.A, Travers P, Walport M, Shlomchik M. 6th edn. Garland Publishing; New York, NY: 2005. Immunobiology: the immune system in health and disease. [Google Scholar]
- Kalvakolanu D.V, Borden E.C. An overview of the interferon system: signal transduction and mechanisms of action. Cancer Invest. 1996;14:25–53. doi: 10.3109/07357909609018435. [DOI] [PubMed] [Google Scholar]
- Kirschner D, Marino S. Mycobacterium tuberculosis as viewed through a computer. Trends Microbiol. 2005;13:206–211. doi: 10.1016/j.tim.2005.03.005. doi:10.1016/j.tim.2005.03.005 [DOI] [PubMed] [Google Scholar]
- Landis H, Simon-Jödicke A, Klöti A, Paolo C.D, Schnorr J.-J, Schneider-Schaulies S, Hefti H.P, Pavlovic J. Human MxA protein confers resistance to Semliki forest virus and inhibits the amplification of a Semliki forest virus-based replicon in the absence of viral structural proteins. J. Virol. 1998;72:1516–1522. doi: 10.1128/jvi.72.2.1516-1522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H, Vidal S.M. Functional diversity of Mx proteins: variations on a theme of host resistance to infection. Genome Res. 2002;12:527–530. doi: 10.1101/gr.20102. doi:10.1101/gr.20102 [DOI] [PubMed] [Google Scholar]
- Luster A.D. The role of chemokines in linking innate and adaptive immunity. Curr. Opin. Immunol. 2002;14:129–135. doi: 10.1016/s0952-7915(01)00308-9. doi:10.1016/S0952-7915(01)00308-9 [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway C.A. Innate immunity: impact on the adaptive immune response. Curr. Opin. Immunol. 1997a;9:4–9. doi: 10.1016/s0952-7915(97)80152-5. doi:10.1016/S0952-7915(97)80152-5 [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway C.A. Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997b;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. doi:10.1016/S0092-8674(00)80412-2 [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway C.A. Innate immunity. N. Engl. J. Med. 2000;343:338–344. doi: 10.1056/NEJM200008033430506. doi:10.1056/NEJM200008033430506 [DOI] [PubMed] [Google Scholar]
- Mossman K.L, Saffran H.A, Smiley J.R. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 2000;74:2052–2056. doi: 10.1128/jvi.74.4.2052-2056.2000. doi:10.1128/JVI.74.4.2052-2056.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.D. Springer; New York, NY: 1989. Mathematical biology. [Google Scholar]
- Nagai T, Honda H. A dynamic cell model for the formation of epithelial tissues. Phil. Mag. B. 2001;81:699–719. [Google Scholar]
- Pacheco J.M, Brum M.C.S, Moraes M.P, Golde W.T, Grubman M.J. Rapid protection of cattle from direct challenge with foot-and-mouth disease virus (FMDV) by a single inoculation with an adenovirus-vectored FMDV subunit vaccine. Virology. 2005;337:205–209. doi: 10.1016/j.virol.2005.04.014. doi:10.1016/j.virol.2005.04.014 [DOI] [PubMed] [Google Scholar]
- Pavlovic J, Zürcher T, Haller O, Staeheli P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol. 1990;64:3370–3375. doi: 10.1128/jvi.64.7.3370-3375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelson A.S. Modelling viral and immune system dynamics. Nat. Rev. Immunol. 2002;2:28–36. doi: 10.1038/nri700. doi:10.1038/nri700 [DOI] [PubMed] [Google Scholar]
- Perelson A.S, Neumann A.U, Markowitz M, Leonard J.M, Ho D.D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- Perelson A.S, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D.D. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. doi:10.1038/387188a0 [DOI] [PubMed] [Google Scholar]
- Rampling R, et al. Toxicity evaluation of replication-competent herpes simplex virus (ICP 34.5 null mutant 1716) in patients with recurrent malignant glioma. Gene Ther. 2000;7:859–866. doi: 10.1038/sj.gt.3301184. doi:10.1038/sj.gt.3301184 [DOI] [PubMed] [Google Scholar]
- Rhim J.S, Schell K, Creasy B, Case W. Biological characteristics and viral susceptibility of an African green monkey kidney cell line (Vero) Proc. Soc. Exp. Biol. Med. 1969;132:670–678. doi: 10.3181/00379727-132-34285. [DOI] [PubMed] [Google Scholar]
- Samuel C.E. Antiviral actions of interferon. Interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. doi:10.1016/0042-6822(91)90112-O [DOI] [PubMed] [Google Scholar]
- Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell. 1986;44:147–158. doi: 10.1016/0092-8674(86)90493-9. doi:10.1016/0092-8674(86)90493-9 [DOI] [PubMed] [Google Scholar]
- Stark G.R, Kerr I.M, Williams B.R.G, Silverman R.H, Schreiber R.D. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. doi:10.1146/annurev.biochem.67.1.227 [DOI] [PubMed] [Google Scholar]
- Svanborg C, Godaly G, Hedlund M. Cytokine responses during mucosal infections: role in disease pathogenesis and host defence. Curr. Opin. Microbiol. 1999;2:99–103. doi: 10.1016/s1369-5274(99)80017-4. doi:10.1016/S1369-5274(99)80017-4 [DOI] [PubMed] [Google Scholar]
- Young H.A. Regulation of interferon-gamma gene expression. J. Interferon Cytokine Res. 1996;16:563–568. doi: 10.1089/jir.1996.16.563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Animation of graphical model output showing the progression of infection in part of a ‘virtual’ tissue culture from t=0 to t=6 days post infection; the time between frames is 6 hours.
Animation of graphical model output showing the progression of infection in part of a ‘virtual’ tissue culture from t=0 to t=6 days post infection; the time between frames is 6 hours.
Animation of graphical model output showing the progression of infection in part of a ‘virtual’ tissue culture from t=0 to t =6 days post infection; the time between frames is 6 hours.