Abstract
NADP is essential for biosynthetic pathways, energy, and signal transduction. In living organisms, NADP biosynthesis proceeds through the phosphorylation of NAD with a reaction catalyzed by NAD kinase. We expressed, purified, and characterized Bacillus subtilis NAD kinase. This enzyme represents a new member of the inorganic polyphosphate [poly(P)]/ATP NAD kinase subfamily, as it can use poly(P), ATP, or other nucleoside triphosphates as phosphoryl donors. NAD kinase showed marked positive cooperativity for the substrates ATP and poly(P) and was inhibited by its product, NADP, suggesting that the enzyme plays a major regulatory role in NADP biosynthesis. We discovered that quinolinic acid, a central metabolite in NAD(P) biosynthesis, behaved like a strong allosteric activator for the enzyme. Therefore, we propose that NAD kinase is a key enzyme for both NADP metabolism and quinolinic acid metabolism.
The importance of NAD(P) in energy metabolism has long been known; NAD is used almost exclusively in oxidative degradation, and NADP is confined to reductive biosynthesis reactions. In recent years it has become evident that NAD(P) not only is a key molecule in energy transduction but also is involved in a plethora of different biochemical processes, including DNA repair and recombination, protein ADP ribosylation, and calcium-mediated signaling (25).
The multiple functions of NAD(P) in energy metabolism, transcription, signaling pathways, and detoxification reactions make it obvious that nicotinamide dinucleotide is a key molecule for cell viability, which implies that the cellular concentration of this compound must be tightly regulated. Whereas the biosynthetic pathways resulting in the synthesis of NAD(P) have been described in detail (2, 17), the mechanisms that regulate the NAD(P) metabolic flux are still poorly understood. In Escherichia coli and Salmonella enterica serovar Typhimurium, the multifunctional protein NadR has been shown to be an NAD-dependent repressor of transcription of genes involved in NAD biosynthesis (22, 19), suggesting that the pathway is entirely regulated at the transcriptional level in these organisms. On the other hand, no observations have been reported concerning the control of enzymatic activity along the biosynthetic pathway, either through allosteric effectors or through covalent modifications. To date, the mechanisms that regulate the rate of NAD(P) flux through its metabolic pathway are far from fully elucidated in both prokaryotes and eukaryotes.
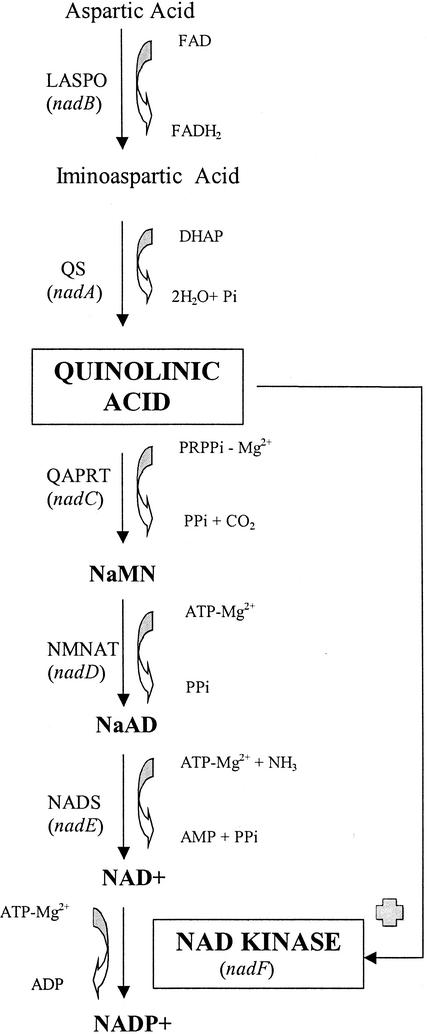
NAD can be synthesized de novo or through pyridine salvage pathways, and there are profound differences between prokaryotes and eukaryotes (2, 17). The key metabolite in de novo NAD biosynthesis in all living organisms is quinolinic acid (QA) (Fig. 1). Eukaryotes produce QA via trypthophan degradation, while in prokaryotes QA is obtained through the condensation of iminoaspartate with dihydroxyacetone phosphate in a reaction catalyzed by the quinolinate synthetase system (2, 17) (Fig. 1). Whereas the effect of a high concentration of QA in prokaryotes has never been analyzed, the neurotoxic action of QA in higher eukaryotes, through its ability to overstimulate N-methyl-d-aspartate receptors, is well documented (21). QA is efficiently removed from the cell by the last three steps of NAD biosynthesis, which are common to all organisms (Fig. 1) (2, 17). QA is transformed into nicotinic acid mononucleotide (NaMN) by QA phosphoribosyltransferase, after which NaMN adenylyltransferase catalyzes the adenylation of NaMN to nicotinic acid adenine dinucleotide. Finally, nicotinic acid adenine dinucleotide is converted into NAD through the reaction catalyzed by NAD synthetase (Fig. 1) (2, 17). In addition, all organisms have the ability to recycle NAD through different salvage pathways, all of which converge at the level of the pyridine mononucleotide, either nicotinamide mononucleotide or NaMN (2, 17).
FIG. 1.
Scheme of de novo NAD(P) biosynthesis in bacteria. The pathway has been described previously (7, 8). The enzymes (genes) involved in each step are abbreviated as follows: LASPO (nadB), l-aspartate oxidase; QS (nadA), quinolinate synthase; QAPRT (nadC), QA phosphoribosyltransferase; NMNAT (nadD), NaMN adenylyltransferase; NADS (nadE), NAD synthetase. Other abbreviations: FAD, flavin adenine dinucleotide; FADH2, reduced flavin adenine dinucleotide; DHAP, dihydroxyacetone phosphate; NaAD, nicotinic acid adenine dinucleotide. The role of QA as an activator of NAD kinase (this study) is indicated.
Once NAD has been synthesized, there is only one obligate route for NADP synthesis in all living organisms, the magnesium-dependent phosphorylation of NAD catalyzed by the ubiquitous enzyme NAD kinase (2, 17) (Fig. 1). Since this reaction is the only biochemical event leading to the production of NADP from NAD, NAD kinase is the key enzyme for NADP synthesis and for the NADP-dependent anabolic and biosynthetic pathway in the cell. This enzyme was recently reported to be essential in Bacillus subtilis (12), further supporting the proposal that NAD kinase is a possible novel antibacterial drug target (8). Despite the fact that this enzyme is highly conserved in organisms, a number of differences have been observed among the enzymes from different sources. NADH and NADPH are potent allosteric negative modulators of E. coli NAD kinase (10, 24) but not for the enzymes from Bacillus licheniformis (23), Mycobacterium tuberculosis, Micrococcus flavus (11), and humans (16). Depending on the organism, the enzyme has been reported to be strictly nucleoside triphosphate dependent or to also utilize inorganic polyphosphate [poly(P)] as a phosphoryl donor for catalysis. poly(P) is a polymer of inorganic orthophosphate residues linked by high-energy phosphoanhydride bonds and is present in nearly all classes of living organisms, likely representing the primitive energy source for generation of phosphate group potential (13). poly(P)-dependent NAD kinase has been found in M. flavus and M. tuberculosis (11), whereas the enzyme from E. coli can utilize only nucleoside triphosphates (10) and human NAD kinase has been reported to be strictly ATP dependent (16). Interestingly, NAD kinase was very recently proposed to belong to a new superfamily of kinases, which includes 6-phosphofructokinases, diacylglyceride kinases, and sphingosine kinases (14). These enzymes are thought to have a common fold and a common strategy for catalysis and regulation of enzymatic activity (14).
In this paper, we report the cloning, expression, transcriptional analysis, purification, and characterization of B. subtilis NAD kinase (encoded by the yjbN gene). The enzyme consists of 266 residues and can use both ATP and poly(P) as phosphoryl donors. B. subtilis NAD kinase was found to be an allosteric enzyme with marked positive cooperativity for both ATP and poly(P). Moreover, we discovered that QA, the central metabolite in de novo NAD biosynthesis, is a potent allosteric activator of the enzyme.
MATERIALS AND METHODS
All the chemicals which we used were supplied by Sigma. The purification kit (GST gene fusion system) was purchased from Amersham Biosciences.
Cloning of yjbN.
yibN, the putative gene encoding NAD kinase in B. subtilis, was amplified from genomic DNA of B. subtilis strain 168 by PCR by using a reaction mixture (50 μl) containing 2 U of Taq polymerase, 1 ng of genomic DNA, 0.5 μmol of primer yjbnBamHI (5′-CGC GGA TCC ATG AAA TTT GCC GTA TCA TCA AAA GGA-3′), 0.5 μmol of primer yjbnXhoI (5′-CCC TCG AGT TAC TAT TCA CCT TTT CCA ATA AAC GAA TC-3′), each deoxynucleoside triphosphate at a concentration of 200 μM, 1.5 mM MgCl2, and 1× reaction buffer. The two primers contained BamHI and XhoI sites, respectively. The conditions for PCR were as follows: 94°C for 1 min, 68°C for 1 min, and 72°C for 1 min for 40 cycles. The PCR product (0.8 kb) was separated by 1% (wt/vol) agarose gel electrophoresis, isolated with a QIAquick gel extraction kit (Qiagen), and successively purified with a QIA PCR purification kit (Qiagen). The purified PCR product was digested with BamHI and XhoI and ligated into pGEX-6P-1, yielding plasmid pGex6p-yjbn.
Expression and purification of recombinant B. subtilis NAD kinase.
E. coli BL21(DE3) cells transformed with plasmid pGex6p-yjbn were grown overnight at 27°C in Luria-Bertani medium containing 100 μg of ampicillin per ml. The cells were then diluted (1/40) in the same medium and cultured at 27°C until the optical density at 600 nm reached 0.5 to 0.7. Expression was induced by addition of isopropyl-β-d-thiogalactoside (ITPG) to final concentration of 0.5 mM, and the temperature was maintained at 27°C. Cells were harvested by centrifugation after 16 h of induction.
Cells collected by centrifugation were sonicated in phosphate-buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4; pH 7.3), and cell debris was removed by ultracentrifugation at 30,000 rpm (Beckman LB-50 M/E) and 4°C for 90 min. The supernatant was loaded onto a column with 1 ml of glutathione Sepharose 4B resin equilibrated with PBS. The resin was extensively washed with PBS, equilibrated with PreScission protease (Amersham Biosciences) buffer (50 mM Tris-HCl [pH 7.0], 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol), and incubated with 80 U of PreScission protease for 16 h at 4°C. The NAD kinase was eluted in 20 ml of PreScission protease buffer and concentrated to a final concentration of 4 mg/ml by dialysis against a solution containing 20% polyethylene glycol 35000. Gel filtration of the pure enzyme was carried out by performing fast protein liquid chromatography on a Superdex S200 16/60 column equilibrated with 50 mM Tris-HCl (pH 7.0)-150 mM NaCl-1 mM EDTA-1 mM dithiothreitol.
Transcription analysis.
To evaluate the expression of nadF (yjbN), we used reverse transcription (RT)-PCR. Cells of B. subtilis 168 were grown in Schaeffer sporulation medium, and samples were harvested during exponential growth, at the transition point between the exponential and postexponential growth phases, and two hours after the transition point. Total RNA was isolated by the method of Caldwell et al. (3), except that samples (5 ml) of cultures were rapidly frozen by dripping into liquid nitrogen in a 50-ml Falcon tube. The tubes were stored at −80°C overnight before the preparations were ground in a hand-held coffee grinder in the presence of dry ice. RT cDNA synthesis and PCR were performed with the oligonucleotides 5′-GCC GTA TCA TCA AAA GGA GAT CAA G-3′ and 5′-AAA TGG AAA CGG ACG GAA TCT CG-3′. RT-PCRs in the absence of reverse transcriptase were performed in parallel to check for the absence of DNA contamination.
Enzyme assay.
NAD kinase activity was assayed by measuring the increase in absorbance at 340 nm caused by the reduction of NADP to NADPH by glucose-6-phosphate dehydrogenase. The reaction was carried out in a solution containing 100 mM Tris-HCl (pH 7.8), 100 mM KCl, 100 mM NaCl, 5 mM MgCl2, 5 mM ATP, 5 mM NAD, 1 mM glucose 6-phosphate, and 1 U of glucose-6-phosphate dehydrogenase. The reaction was started by adding the enzyme solution. One unit of enzyme activity was defined as production of 1.0 μmol of NADP in 1 min at 37°C. The source of poly(P) was phosphate glass from Sigma (practical grade) containing 13 to 18 phosphoryl residues and having an estimated molecular weight of 3,286. To rule out the possibility that the experimental variables had any effect on the glucose-6-phosphate dehydrogenase, a control was included for each experiment performed, in which the enzymatic activity of only glucose-6-phosphate dehydrogenase was measured in the absence of NAD kinase.
Kinetic analysis.
Enzymatic activity was assayed at 25°C by using various concentrations of NAD and ATP or poly(P) under conditions identical to those described above except for the substrates and effector. Kinetic parameters were determined as follows: for ATP, 5 mM (fixed concentration) NAD in the absence and in the presence of 0.1 mM QA; for NAD, 5 mM ATP in the absence and in the presence of 0.1 mM QA; and for poly(P), 5 mM NAD in the absence and in the presence of 0.2 mM QA. In all cases, the enzyme activity was assayed at 10 different concentrations of substrate. All measurements were obtained in triplicate, and a Lineweaver-Burk plot was used to determine the apparent Km and Vmax. A Hill plot was used to determine the apparent substrate concentration that resulted in one-half of Vmax (S0.5) and the Hill coefficient (nH).
RESULTS
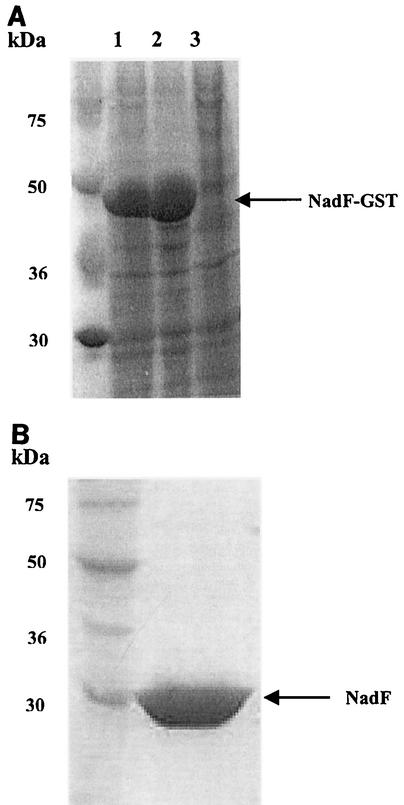
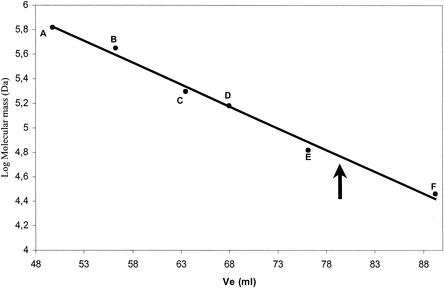
The B. subtilis gene yjbN, coding for a polypetide chain containing 266 residues, was considered a potential candidate for encoding NAD kinase based on the high sequence similarity between its product and several NAD kinases (data not shown). To obtain a sufficient quantity of the gene product for full characterization, we cloned the yjbN gene in plasmid pGEX-6P-1, a vector suitable for expression of foreign genes in E. coli. The complete coding sequence of yjbN was cloned downstream of the IPTG-inducible promoter to obtain plasmid pGEX6p1-yjbN. E. coli BL21(DE3) cells transformed with pGex6p1-yjbN produced a large amount of a soluble protein with a molecular mass of approximately 50 kDa, a size corresponding to the size of the fusion of glutathione S-transferase (GST) to YjbN (Fig. 2A). The YjbN protein was purified from a cell extract of E. coli BL21(DE3) in a single purification step by affinity chromatography and proteolytic release from the GST moiety (Fig. 2B). The pure protein, whose identity was confirmed by N-terminal sequencing (data not shown), migrated during sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as a single band at a molecular mass of about 30 kDa (Fig. 2B), which is in agreement with the expected protein size. The pure enzyme showed NAD kinase activity and had a specific activity of 2 U/mg. Thus, yjbN was identified as the nadF gene (NAD kinase) of B. subtilis. Gel filtration experiments performed with the pure enzyme showed that the native molecular mass is about 60 kDa, indicating that the native protein exists as a dimer (Fig. 3). The expression of nadF in B. subtilis was evaluated by an RT-PCR-based transcriptional analysis. The experiment revealed the presence of a transcript corresponding to about 800 bp, in agreement with the expected size of NAD kinase, in all of the samples investigated during exponential and postexponential growth, as well as in the transition state between the two phases (see Materials and Methods).
FIG. 2.
Expression and purification of B. subtilis NAD kinase. (A) SDS-10% PAGE of total (lane 1), soluble (lane 2), and insoluble (lane 3) E. coli BL21(DE3) cell extract harboring plasmid pGEX6p1-yjbN. (B) SDS-10% PAGE of 10 μg of purified recombinant B. subtilis NAD kinase (released from the affinity column after in-column proteolytic cleavage of the GST moiety carried out with PreScission protease).
FIG. 3.
Estimation of the molecular mass of B. subtilis NAD kinase. Gel filtration was performed as described in Materials and Methods. The arrow indicates the elution volume (Ve) of pure B. subtilis NAD kinase. The following protein standards (Sigma) were used: thyroglobulin (669 kDa) (A), apoferritin (443 kDa) (B), β-amylase (200 kDa) (C), alcohol dehydrogenase (150 kDa) (D), albumin (66 kDa) (E), and carbonic anhydrase (29 kDa) (F).
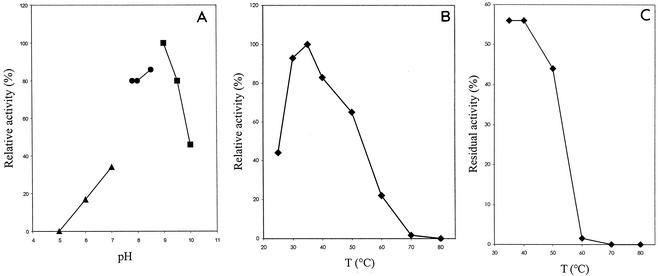
The enzyme could be assigned to the subfamily containing the poly(P)/ATP NAD kinases since it was able to efficiently use ATP, other nucleoside triphosphates, or various polyphosphates as phosphoryl donors, while no activity was detected with nucleoside mono- or diphosphates (Table 1). B. subtilis NAD kinase had an optimum pH of 9.0 in glycine-NaOH buffer (Fig. 4A). The enzyme was most active at 35°C (Fig. 4B), and one-half of the activity was lost after treatment at 40°C for 15 min (Fig. 4C). As observed for several other NAD kinases, the B. subtilis enzyme is strictly dependent on bivalent cations, such as Mg2+, Mn2+, and Ca2+, for activity. Both Ca2+ and Mn2+ are more effective activators than Zn2+, Co2+, Cu2+, and Mg2+ at low concentrations, whereas no activity was detected in the presence of Ni2+ and Fe2+ (Table 2). Strong inhibition was observed with HgCl2, indicating that an SH group of the enzyme may play an important role in catalysis (Table 3). B. subtilis NAD kinase is inhibited by its product, NADP, whereas no effect on enzymatic activity was observed in the presence of NADH or NADPH, nicotinamide mononucleotide, nicotinamide, and nicotinic acid (Table 3).
TABLE 1.
Phosphoryl donor specificity of B. subtilis NAD kinasea
| Phosphoryl donor | Relative activity (%) |
|---|---|
| ATP | 100 |
| GTP | 69 |
| dATP | 77 |
| dGTP | 77 |
| dCTP | 30 |
| dTTP | 35 |
| ADP | ND |
| AMP | ND |
| Phosphoenolpyruvate | ND |
| Pyrophosphate | 10 |
| poly(P) | 50 |
NAD kinase activity was assayed as described in Materials and Methods with each of the phosphoryl donors at a concentration of 5.0 mM. ND, not detected.
FIG. 4.
Effects of pH and temperature on the activity of B. subtilis NAD kinase. (A) Effect of pH on NAD kinase activity assayed as described in Materials and Methods with 100 mM sodium acetate (▴), Tris-HCl (•), and glycine/NaOH (▪). (B) Effect of temperature (T) on NAD kinase activity assayed as described in Materials and Methods. (C) Thermal stability of NAD kinase. The purified enzyme was incubated for 15 min at different temperatures, and the residual activity was assayed as described in Materials and Methods.
TABLE 2.
Effects of metal ions on activity of B. subtilis NAD kinasea
| Metal | Relative activity (%) |
|---|---|
| None | ND |
| MgCl2 | 100 |
| MnCl2 | 180 |
| CaCl2 | 104 |
| ZnCl2 | 89 |
| MgCl2 | 68 |
| CoCl2 | 15 |
| Cu(CH3COO)2 | 8 |
| Ni(NO3)2 | ND |
| FeSO4 | ND |
The effects of metal ions were investigated by assaying the activity in the reaction mixture described in Materials and Methods, in which 5 mM MgCl2 was replaced by a metal ion at a concentration of 1.0 mM. ND, not detected.
TABLE 3.
Effects of various compounds on activity of B. subtilis NAD kinasea
| Compound | Concn (mM) | Relative activity (%) |
|---|---|---|
| None | 100 | |
| NADPH | 0.01 | 100 |
| NADH | 0.01 | 100 |
| NADP | 0.01 | 40 |
| 0.05 | 25 | |
| Nicotinamide mononucleotide | 0.10 | 100 |
| Nicotinamide | 0.10 | 100 |
| Nicotinic acid | 0.10 | 100 |
| QA | 0.10 | 150 |
| HgCl2 | 0.50 | 50 |
| 1.0 | 25 | |
| β-Mercaptoethanol | 1.0 | 100 |
| Dithiothreitol | 1.0 | 100 |
The effects of various compounds were studied by assaying the activity in the reaction mixture described in Materials and Methods containing various compounds.
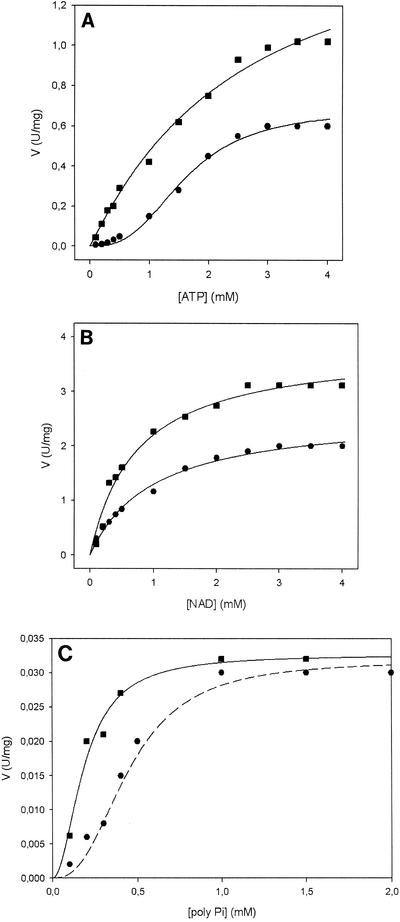
B. subtilis NAD kinase exhibited sigmoidal kinetics with respect to the substrates ATP and poly(P) and an hyperbolic saturation curve for NAD (Fig. 5) Indeed, calculation of the nH values for ATP (nH = 2.0) and poly(P) (nH = 2.0) revealed a strong allosteric behavior (Table 4). Positive cooperativity could be completely eliminated by QA, a metabolite of NAD(P) biosynthesis (Fig. 1). In fact, in the presence of QA the ATP and poly(P) saturation curves were actually transformed from sigmoidal to hyperbolic and nH changed from 2.0 in absence of QA to 1 (no cooperativity) (Fig. 5) (Table 4). QA also had a remarkable effect on the maximum turnover rate, and there were significant increases in ATP, poly(P), and NAD (Fig. 5 and Table 4). Therefore, B. subtilis NAD kinase was found to be an allosteric enzyme with mixed S- and V-type allostery (Fig. 5 and Table 4).
FIG. 5.
Kinetic behavior of B. subtilis NAD kinase. Enzyme activity was assayed at 25°C and pH 7.8 as described in Materials and Methods. (A) ATP titration curve in the absence of QA (•) and in the presence of 100 μM QA (▪). (B) NAD titration curve in the absence of QA (•) and in the presence of 100 μM QA (▪). (C) poly(P) titration curve in the absence of QA (•) and in the presence of 200 μM QA (▪).
TABLE 4.
Kinetic parameters of B. subtilis NAD kinasea
| Compound | QA concn (μM) | S0.5 (mM) | Km (mM) | Vmax (U/mg) | kcat (s−1) | nH | kcat/S0.5 (s−1/mM) | kcat/km (s−1/mM) |
|---|---|---|---|---|---|---|---|---|
| ATPb | 0 | 1.1 ± 0.08 | 0.65 ± 0.08 | 1.36 ± 0.09 | 2.0 ± 0.089 | 1.24 | ||
| 100 | 2.2 ± 0.08 | 1.6 ± 0.07 | 3.36 ± 0.098 | 1.0 ± 0.06 | 1.53 | |||
| poly(P)b | 0 | 0.45 ± 0.08 | 0.032 ± 0.08 | 0.064 ± 0.09 | 2.0 ± 0.10 | 0.14 | ||
| 200 | 0.20 ± 0.08 | 0.032 ± 0.09 | 0.064 ± 0.088 | 1.2 ± 0.07 | 0.32 | |||
| NADc | 0 | 1.0 ± 0.09 | 2.6 ± 0.075 | 5.46 ± 0.09 | 1.0 ± 0.05 | 5.46 | ||
| 100 | 0.73 ± 0.088 | 3.82 ± 0.098 | 8.0 ± 0.09 | 1.0 ± 0.06 | 10.95 |
NAD kinase activity was assayed as described in Materials and Methods. The data are means ± standard errors for three determinations with three different protein preparations.
Kinetic parameters for ATP and poly(P) were obtained by fitting data to the Hill equation and were determined at a fixed NAD concentration of 5 mM.
Determined at a fixed ATP concentration of 5 mM.
DISCUSSION
Pyridine nucleotides are considered not only guardians for the maintenance of the redox state of the cell but also key cofactors for several other biochemical processes, including signaling and transcription modulation (25). Such observations raise intriguing questions, such as how is the role of NAD(P) in transcription and signaling linked to its role in cellular metabolism? How are variations in the NAD/NADH and NADP/NADPH ratios sensed by the cell and translated into information for transcription modulation or regulation of signaling pathways? It is evident that the relationships among the different biological functions played by NAD(P) require strict regulation of its biosynthesis.
NAD kinase is a ubiquitous and highly conserved enzyme that catalyzes the only enzymatic route for NADP synthesis from NAD and therefore is an attractive candidate for regulation of the NADP metabolic flux. B. subtilis NAD kinase showed marked positive cooperativity for ATP and poly(P) (Fig. 5 and Table 4). The ability to efficiently adjust the rate of NADP synthesis as a function of ATP concentration indicates that NAD kinase is a key enzyme for regulation of the NADP metabolic flux. In fact, a high ATP concentration, which is the major signal of energy availability, requires efficient NADP synthesis to sustain anabolic metabolism, whereas NADP production must be severely slowed when energy availability is low to avoid unbalancing the NADP/NADPH ratio, a vital parameter for cell viability. Moreover, the observed remarkable NADP enzyme inhibition (Table 3) emphasizes the role of B. subtilis NAD kinase in the regulation of NADP synthesis, which could be completely eliminated at high NADP concentrations through a product inhibition mechanism.
To further prove the central role played by NAD kinase in the regulation of NAD(P) biosynthesis, we investigated the effects that intermediates along the biosynthetic pathways had on the enzyme activity. QA is a key metabolite in NAD(P) biosynthesis, representing the common point of the two major biosynthetic pathways in living organisms (i.e., trypthophan degradation in eukaryotes and de novo biosynthesis in prokaryotes) (2, 17) (Fig. 1). A possible regulatory mechanism involving QA for controlling NAD(P) levels was previously proposed for S. enterica serovar Typhimurium, in which QA was shown to inhibit NAD kinase (5). QA is a powerful allosteric activator of B. subtilis NAD kinase (Fig. 5 and Table 4). It is possible that activation of NAD kinase by QA causes a decrease in the NAD level, which is compensated for by an increase in NAD synthesis by the biosynthetic enzymes upstream of NAD kinase (Fig. 1). The net effect is therefore reequilibration of the NAD level and a rapid decrease in the QA concentration. Considering that NAD(P) can also be synthesized by means of recycling routes (2, 17), we investigated the possible regulatory role of B. subtilis NAD kinase in salvage pathways. All recycling pathways converge at the level of NaMN, which is then rechannelled into de novo biosynthesis (2, 17). Two key metabolites of recycling pathways are represented by nicotinic acid and nicotinamide (2, 17). We tested the effects of both metabolites on B. subtilis NAD kinase, but even at a relatively high concentration (0.1 mM) no effect on the enzyme activity was observed (Table 3). To assess the significance of our observations for B. subtilis metabolism, we performed a transcriptional analysis, which demonstrated that NAD kinase is constantly expressed during different bacterial growth phases. We therefore concluded that at least in B. subtilis, NAD kinase is a key regulatory enzyme for de novo NAD(P) biosynthesis and not for salvage recycling pathways.
Another relevant observation that supports the central role of NAD kinase as a highly regulated enzyme is the reported activation of plant and human enzymes by calmodulin (6, 15). Calmodulin is a major intracellular calcium receptor in eukaryotes, and upon calcium binding it can interact with a variety of target enzymes to modulate their activity (4). In bacteria, calcium is involved in a wide range of cellular processes, including the cell cycle and cell division (18). A number of calmodulin-like proteins have been identified in bacteria (9, 18), and, interestingly, a calmodulin-like protein from sporulating cells of B. subtilis was reported to activate plant NAD kinase (7). This observation prompted us to hypothesize that B. subtilis NAD kinase can be activated by B. subtilis calmodulin-like proteins, suggesting that the enzyme plays a role in calcium metabolism. In this respect it is worth noting that a relationship between oxidative stress and calcium signaling, mediated by calmodulin-like proteins, was recently suggested for B. subtilis (9). Since NAD(P) is an important constituent of cellular defense mechanisms against oxidative stress (1, 20), it is tempting to speculate about a possible role of NAD kinase in oxidative stress and calcium homeostasis in B. subtilis.
Acknowledgments
We thank Gianna Valentini and Laurent Chiarelli (University of Pavia, Pavia, Italy) for several helpful suggestions and Robert van den Heuvel (University of Pavia) for critical reading of the manuscript. Alessandro Coda and Andrea Mattevi (University of Pavia) are acknowledged for their constant support.
This work was supported in part by grants from MIUR [Project Biologia strutturale di enzimi coinvolti nella biosintesi del NAD(P) per lo sviluppo di nuovi farmaci antibatterici] and the Agenzia Spaziale Italiana (project number IR/167/01).
REFERENCES
- 1.Antelman, H., R. Schimd, and M. Hecker. 1997. The NAD synthetase NadE (OutB) of Bacillus subtilis is a σB-dependent general stress protein. FEMS Microbiol. Lett. 153:405-409. [DOI] [PubMed] [Google Scholar]
- 2.Begley, T. P., C. Kinsland, R. A. Mehl, A. Osterman, and P. Dorrestein. 2001. The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam. Horm. 61:103-119. [DOI] [PubMed] [Google Scholar]
- 3.Caldwell, R., R. Sapolsky, W. Weyler, R. R. Maile, S. C. Causey, and E. Ferrari. 2001. Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcription analysis. J. Bacteriol. 183:7329-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carafoli, E. 1987. Intracellular calcium homeostasis. Annu. Rev. Biochem. 56:395-433. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, W., and J. R. Roth. 1994. Evidence for two NAD kinases in Salmonella typhimurium. J. Bacteriol. 176:4260-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epel, D., C. Patton, R. W. Wallace, and W. Y. Cheung. 1981. Calmodulin activates NAD kinase of sea urchin eggs: an early event of fertilization. Cell 23:543-549. [DOI] [PubMed] [Google Scholar]
- 7.Fry, I. J., M. Becker-Hapak, and J. H. Hageman. 1991. Purification and properties of an intracellular calmodulin-like protein from Bacillus subtilis cells. J. Bacteriol. 173:2506-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerdes, S. Y., M. D. Scholle, M. D'Souza, A. Bernal, M. V. Baev, M. Farrell, O. V. Kurnasov, M. D. Daugherty, F. Mseeh, B. M. Polanuyer, J. W. Campbell, S. Anantha, K. Y. Shatalin, S. A. Chowdhury, M. Y. Fonstein, and A. L. Osterman. 2002. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 184:4555-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbaud, M. H., A. Guiseppi, F. Denizot, J. Haiech, and M. C. Kilhofer. 1998. Calcium signalling in Bacillus subtilis. Biochim. Biophys. Acta 1448:212-226. [DOI] [PubMed] [Google Scholar]
- 10.Kawai, S., S. Mori, T. Mukai, W. Hashimoto, and K. Murata. 2001. Molecular characterization of Escherichia coli NAD kinase. Eur. J. Biochem. 268:4359-4365. [DOI] [PubMed] [Google Scholar]
- 11.Kawai, S., S. Mori, T. Mukai, S. Suzuki, T. Yamada, W. Hashimoto, and K. Murata. 2000. Inorganic polyphosphate/ATP-NAD kinase of Micrococcus flavus and Mycobacterium tuberculosis H37Rv. Biochem. Biophys. Res. Commun. 276:57-63. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi, K., S. D. Ehrlich, A. Albertini, A. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Mason, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scalan, W. Schumann, J. F. M. L. Seegers, J. Sekiguchi, A. Sekowska, S. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornberg, A., N. N. Rao, and D. Ault-Riche. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 14.Labesse, G., D. Douguet, L. Assairi, and A. M. Gilles. 2002. Diacylglyceride kinases, sphingosine kinases and NAD kinases: distant relatives of 6-phosphofructokinases. Trends Biochem. Sci. 27:273-275. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S. H., H. Y. Seo, J. C. Kim, W. D. Heo, W. S. Chung, K. J. Lee, M. C. Kim, Y. H. Cheong, J. Y. Choi, C. O. Lim, and M. J. Cho. 1997. Differential activation of NAD kinase by plant calmodulin isoforms. J. Biol. Chem. 272:9252-9259. [DOI] [PubMed] [Google Scholar]
- 16.Lerner, F., M. Niere, A. Ludwig, and M. Ziegler. 2001. Structural and functional characterization of human NAD kinase. Biochem. Biophys. Res. Commun. 288:69-74. [DOI] [PubMed] [Google Scholar]
- 17.Magni, G., A. Amici, M. Emanuelli, N. Raffaelli, and S. Ruggieri. 1999. Enzymology of NAD+ synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 73:135-182. [DOI] [PubMed] [Google Scholar]
- 18.Michiels, J., C. Xi, J. Verhaert, and J. Vanderleyden. 2002. The functions of Ca2+ in bacteria: a role for EF-hand proteins? Trends Biochem. Sci. 10:87-93. [DOI] [PubMed] [Google Scholar]
- 19.Penfound, T., and J. W. Foster. 1999. NAD-dependent DNA-binding activity of the bifunctional NadR regulator of Salmonella typhimurium. J. Bacteriol. 181:648-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scharf, C., S. Riethdorf, H. Ernst, S. Engelmann, U. Volker, and M. Hecker. 1998. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J. Bacteriol. 180:1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone, T. W., and J. I. Addae. 2002. The pharmacological manipulation of glutamate receptors and neuroprotection. Eur. J. Pharmacol. 447:285-296. [DOI] [PubMed] [Google Scholar]
- 22.Tritz, G. J., and J. L. Chandler. 1973. Recognition of a gene involved in the regulation of nicotinamide adenine dinucleotide biosynthesis. J. Bacteriol. 114:128-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zerez, C. R., D. E. Moul, and A. J. Andreoli. 1986. NAD kinase from Bacillus licheniformis: inhibition by NADP and other properties. Arch. Micobiol. 144:313-316. [DOI] [PubMed] [Google Scholar]
- 24.Zerez, C. R., D. E. Moul, E. G. Gomez, V. M. Lopez, and A. J. Andreoli. 1987. Negative modulation of Escherichia coli NAD kinase by NADPH and NADH. J. Bacteriol. 169:184-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziegler, M. 2000. New functions of a long-known molecule. Emerging roles of NAD in cellular signalling. Eur. J. Biochem. 267:1550-1564. [DOI] [PubMed] [Google Scholar]