Abstract
When cultured in broth, fresh clinical isolates of the gram-negative periodontal pathogen Actinobacillus actinomycetemcomitans form tenaciously adherent biofilm colonies on surfaces such as plastic and glass. These biofilm colonies release adherent cells into the medium, and the released cells can attach to the surface of the culture vessel and form new colonies, enabling the biofilm to spread. We mutagenized A. actinomycetemcomitans clinical strain CU1000 with transposon IS903φkan and isolated a transposon insertion mutant that formed biofilm colonies which were tightly adherent to surfaces but which lacked the ability to release cells into the medium and disperse. The transposon insertion in the mutant strain mapped to a gene, designated dspB, that was predicted to encode a secreted protein homologous to the catalytic domain of the family 20 glycosyl hydrolases. A plasmid carrying a wild-type dspB gene restored the ability of biofilm colonies of the mutant strain to disperse. We expressed A. actinomycetemcomitans DspB protein engineered to contain a hexahistidine metal-binding site at its C terminus in Escherichia coli and purified the protein by using Ni affinity chromatography. Substrate specificity studies performed with monosaccharides labeled with 4-nitrophenyl groups showed that DspB hydrolyzed the 1→4 glycosidic bond of β-substituted N-acetylglucosamine, which is consistent with the known functions of other family 20 glycosyl hydrolases. When added to culture medium, purified DspB protein, but not heat-inactivated DspB, restored the ability of the mutant strain to release cells and disperse. DspB protein also caused the detachment of cells from preformed biofilm colonies of strain CU1000 grown attached to plastic and the disaggregation of highly autoaggregated clumps of CU1000 cells in solution. We concluded that dspB encodes a soluble β-N-acetylglucosaminidase that causes detachment and dispersion of A. actinomycetemcomitans biofilm cells.
Actinobacillus actinomycetemcomitans is a gram-negative, nonmotile coccobacillus that colonizes the human oral cavity (20). A. actinomycetemcomitans has been implicated as the causative agent of localized juvenile periodontitis, a severe and rapid form of periodontal disease that affects adolescents (38). A. actinomycetemcomitans can also enter the submucosa and cause infective endocarditis and other nonoral infections (15). When cultured in broth, fresh clinical isolates of A. actinomycetemcomitans form tenacious biofilms on surfaces such as glass, plastic, and saliva-coated hydroxyapatite (5, 6, 9, 12-14, 16, 18, 20, 28). Nearly all of the cells grow attached to the surface, while the broth remains clear and is often sterile (5). The dense biofilm that forms on the surface is resistant to removal by agents such as detergents, proteases, heat, sonication, and vortex agitation and can be removed only by mechanical scraping or by treatment with the carbohydrate-modifying reagent periodic acid (5). A. actinomycetemcomitans biofilm cells exhibit increased resistance to antimicrobial agents compared to the resistance exhibited by cells grown in planktonic form (4). Tight adherence has been shown to play an important role in the ability of A. actinomycetemcomitans to colonize the mouths of rats (7) and probably plays an equally important role in its ability to colonize humans. Tight adherence to surfaces is dependent on the presence of long, bundled pili (fimbriae) that form on the surface of the cell (12, 28). Mutations in flp-1, which encodes the major pilin protein subunit, result in cells that fail to produce fimbriae or adhere to surfaces (14).
Kaplan and Fine showed that biofilm colonies of A. actinomycetemcomitans release cells into liquid medium and that these cells can attach to the surface of the culture vessel and form new colonies, enabling the biofilm to spread (16). Kaplan et al. isolated three A. actinomycetemcomitans transposon insertion mutants that formed biofilm colonies which were tightly adherent to surfaces but which failed to release cells into the medium or spread over the surface (18). All three of the transposon insertions mapped to genes required for the synthesis of the O-polysaccharide (O-PS) component of lipopolysaccharide. Microscopic analysis of the O-PS mutants indicated that they lacked a layer of nonaggregated cells that was present inside biofilm colonies of the wild-type parental strain. These findings led to the hypothesis that A. actinomycetemcomitans biofilm cell detachment occurs by means of a novel mechanism that involves the release of cells from inside the biofilm colony (18).
In this report we describe a fourth A. actinomycetemcomitans transposon insertion mutant that is deficient in biofilm cell detachment and biofilm dispersal. The transposon in this strain inserted into a novel gene, designated dspB, which encodes a protein homologous to the catalytic domain of the family 20 glycosyl hydrolases. By using DspB protein purified from an overexpressing strain of Escherichia coli, we obtained evidence that dspB encodes a soluble N-acetylglucosaminidase that causes the detachment of cells from A. actinomycetemcomitans biofilm colonies.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A. actinomycetemcomitans CU1000 (serotype f) is a clinical strain isolated from a 13-year-old patient with localized juvenile periodontitis (6). Strain CU1000N is a spontaneous nalidixic acid derivative of strain CU1000 that displays the same surface attachment, biofilm colony formation, and biofilm dispersal phenotypes as the parental strain (7, 13, 14, 18, 35). The bacteria were grown in Trypticase soy broth (BD Biosystems) supplemented with 6 g of yeast extract per liter and 8 g of glucose per liter at 37°C in the presence of 10% CO2. Mutagenesis of strain CU1000N with transposon IS903φkan was carried out as previously described (35).
Cloning and sequencing of dspB.
The transposon insertion site in A. actinomycetemcomitans mutant strain JK1023 was cloned and sequenced by using an inverse PCR method as previously described (19). The DNA sequence of the inverse PCR product was compared to the genome sequence of A. actinomycetemcomitans strain HK1651 from the Actinobacillus Genome Sequencing Project (www.genome.ou.edu/act.html), and the transposon was found to have inserted into a long open reading frame which we designated dspB. Primers that hybridize to sequences upstream and downstream from HK1651 dspB were used to amplify by PCR the dspB coding region from A. actinomycetemcomitans strain CU1000 by using methods described previously (19). The forward primer (5-GCGCGCCATatgAATTGTTGCGTAAAAGGCAATTCC-3) introduced an NdeI restriction site (underlined) and an ATG initiation codon (lowercase) at codon positions 19 to 20 of dspB, and the reverse primer (5-GCGGTACCCTCATCCCCATTCGTCTTATGAATC-3) replaced the dspB stop codon with a KpnI restriction site (underlined). The PCR product (1,106 bp) was digested with NdeI and KpnI and was ligated into the NdeI-KpnI sites of plasmid pET29b (Novagen). The insert of the resulting plasmid (designated pRC1) was subjected to DNA sequence analysis as previously described (19).
Genetic complementation of the dspB mutation.
A plasmid containing the wild-type dspB gene for use in genetic complementation experiments was constructed as follows. First, the dspB gene from strain CU1000 was amplified by PCR by using forward primer 5-GCCGAATTCTTCTACAAGGAATTTTTatg-3, which introduced an EcoRI site (underlined) 18-bp upstream from the dspB start codon (lowercase), and reverse primer 5-GCCGCTGCAGtcaCTCATCCCCATTCGTCTTATG-3, which introduced a PstI site (underlined) immediately downstream from the dspB stop codon (lowercase). The PCR product (1,182 bp) was digested with EcoRI and PstI and ligated into the EcoRI and PstI sites of the broad-host-range vector plasmid pJAK16 (35), which placed dspB under control of an isopropyl-β-d-galactopyranoside (IPTG)-inducible tac promoter. The resulting plasmid, designated pJK618, was mobilized into the mutant strain by using the RK2 oriT-defective mutant plasmid pRK21761 as previously described (35). Plasmid-harboring strains were grown in broth supplemented with 3 μg of chloramphenicol/ml and 1 mM IPTG.
Expression and purification of recombinant DspB protein.
Plasmid pRC1 carried a gene that encoded amino acids 21 to 381 of dspB from strain CU1000 fused to a 32-amino-acid C-terminal tail that contained a hexahistidine metal-binding site and a thrombin protease cleavage site which could be used to cleave the C-terminal tail from the hybrid protein. This gene was located downstream from an IPTG-inducible tac promoter.
(i) Expression of DspB in E. coli.
A 2-liter Erlenmeyer flask containing 500 ml of Luria-Bertani broth supplemented with 30 μg of kanamycin per ml was inoculated with 5 ml of an overnight culture of E. coli strain BL21(DE3) (3) transformed with pRC1. The flask was incubated at 37°C with agitation (200 rpm) until the optical density of the culture at 600 nm reached 0.6 (ca. 3 h). IPTG was added to a final concentration of 0.2 mM, and the flask was incubated for an additional 5 h with agitation. The cells were harvested by centrifugation for 15 min at 6,000 × g, and the cell pellet was stored at −80°C.
(ii) Protein purification.
The cell pellet was thawed at room temperature and resuspended in 20 ml of lysis buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 0.1% Nonidet P-40) containing 2 mg of lysozyme/ml. The cell suspension was then sonicated on ice for 30 s at 30% capacity with a 30% duty cycle by using a Branson model 450 sonicator equipped with a microprobe. The cell debris was pelleted by centrifugation at 15,000 × g for 20 min at 4°C, and the supernatant was loaded onto a 3-ml (bed volume) activated Ni affinity column (catalog no. 154-0990; Pharmacia) according to the instructions supplied by the manufacturer. The column was washed with 50 ml of wash buffer (20 mM Tris [pH 7.5], 500 mM NaCl) containing 5 mM imidazole and then with 25 ml of wash buffer containing 50 mM imidazole. The protein was eluted with 25 ml of 20 mM Tris (pH 8.0)-500 mM NaCl-100 mM imidazole. Fractions of the eluate were collected and assayed for the presence of the hybrid protein by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Coomassie blue staining (29). Fractions containing the protein were pooled and dialyzed overnight against water by using a 10,000-kDa-cutoff dialysis membrane. The purified protein was digested with 5 U of thrombin (Novagen) per mg of protein for 1 h at room temperature, and the thrombin was removed with a thrombin cleavage capture kit (Novagen) used according to instructions supplied with the kit. Undigested protein was removed by loading the sample onto an Ni affinity column as described above and washing the column with 10 ml of wash buffer containing 5 mM imidazole. Fractions containing the protein were pooled, dialyzed against water, and stored at −20°C.
N-terminal sequence analysis of the purified protein was carried out by using the Edman degradation procedure with a Beckman model 2300 protein sequencer. Mass spectra were determined by using a Hitachi model 4414 mass spectrometer.
Enzyme assays.
The synthetic substrates (purchased from Sigma Chemical Co.) were 4-nitrophenyl-N-acetyl-β-d-galactosaminide, 4-nitrophenyl-N-acetyl-α-d-galactosaminide, 4-nitrophenyl-N-acetyl-β-d-glucosaminide, and 4-nitrophenyl-N-acetyl-α-d-glucosaminide. Enzyme reactions were carried out in 1-ml mixtures containing 50 mM sodium phosphate buffer (pH 5.9), 100 mM NaCl, 5 mM substrate, and 3.7 μg of purified protein per ml in 1.5-ml polypropylene tubes placed in a 37°C water bath. The reactions were terminated at various times by adding 5 μl of 10 N NaOH. The increase in absorption resulting from the release of p-nitrophenolate in each tube was measured with a Shimadzu model 1240 UV-Mini spectrophotometer set to 405 nm.
Detachment of biofilm cells from polystyrene rods.
An assay to measure the detachment of cells from preformed biofilm colonies grown on polystyrene rods was carried out as previously described (18), except that 24-well microtiter plates (Falcon no. 353047) were used and the detached cells were destained in 400 μl of ethanol, 100 μl of which was transferred to a 96-well microtiter plate for spectrophotometric analysis at 590 nm. In some experiments polystyrene rods were treated with medium containing various amounts of purified DspB. The DspB enzymatic activity decayed over time (half-life, 3.3 h), which allowed detached cells to adhere to the bottom of the well by the end of the 24-h detachment period (18).
Ninety-six-well microtiter plate biofilm cell detachment assay.
The wells of a 96-well microtiter plate (Falcon no. 353072) were filled with 100 μl of medium containing 102 to 104 CFU of a single cell suspension of bacteria (16) and incubated at 37°C in 10% CO2 for 20 h. Ten microliters of enzyme solution (1 mg ml−1 in phosphate-buffered saline) or 10 μl of phosphate-buffered saline (for controls) was added to each well, and the plates were incubated for an additional 6 h. The wells were washed extensively under running tap water, and the bacteria remaining attached to the surface were stained with crystal violet, rewashed, and destained with ethanol as previously described (14). The optical density of the ethanol-dye solution was measured with a Bio-Rad Benchmark microtiter plate reader set to 590 nm.
Disaggregation assay.
A 100-mm-diameter tissue-culture-treated polystyrene petri dish (model 430167; Corning) containing 20 ml of broth was inoculated with 1 × 108 CFU of strain CU1000 and incubated at 37°C for 24 h. The medium was decanted, and the bacterial colonies growing attached to the surface of the dish were washed extensively under running tap water. The dish was filled with 20 ml of an ethidium bromide solution (0.5 μg/ml in water) and incubated at 37°C for 24 h. The cells were rewashed with tap water and then scraped from the surface into a small volume of water (2 to 3 ml) with a cell scraper. The resulting cell suspension was vortexed briefly and then dispensed into glass test tubes (10 by 75 mm; 0.5 ml per tube). Fifty microliters of a DspB enzyme solution (1.7 mg/ml in water) or 50 μl of water (for controls) was added, and the tubes were sealed with Parafilm and incubated at 37°C for 30 min with gentle rocking. Fifty microliters of the treated cell suspension was then pipetted onto a sheet of cellophane that had been placed on a UV transilluminator, and the preparation was photographed through an orange filter by using Polaroid type 667 film.
Nucleotide sequence accession number.
The DNA sequence of dspB from strain CU1000 has been deposited in the GenBank database under accession no. AY228551.
RESULTS
Isolation of A. actinomycetemcomitans biofilm detachment mutant JK1023.
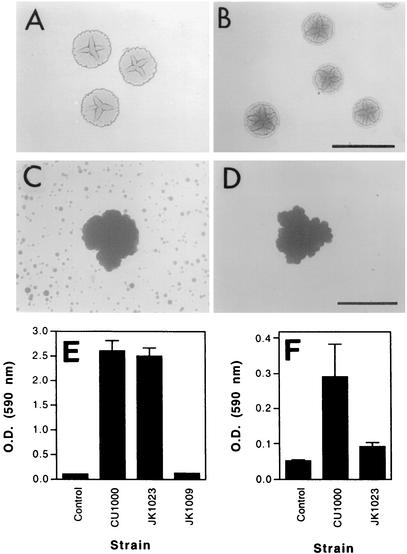
We mutagenized A. actinomycetemcomitans strain CU1000N with transposon IS903φkan and isolated a kanamycin-resistant mutant (designated JK1023) that displayed a colony morphology on agar that was rougher than the wild-type A. actinomycetemcomitans rough-colony phenotype (Fig. 1A and B) (6, 9, 12). Kaplan et al. previously showed that three other A. actinomycetemcomitans rougher-colony mutants were deficient in biofilm cell detachment (18). JK1023 colonies had a hard texture and were extremely difficult to remove from the agar surface. In a test tube, JK1023 cells aggregated and settled to the bottom of the tube much more rapidly than cells of wild-type strain CU1000 (data not shown). When cultured in broth, strain JK1023 produced biofilm colonies which were similar in size and shape to those of the wild-type strain but failed to produce satellite colonies on the surface of the culture vessel (Fig. 1C and D). The adherence of broth-cultured JK1023 cells to polystyrene was equivalent to that of wild-type strain CU1000N as measured by a 96-well microtiter plate binding assay (Fig. 1E).
FIG. 1.
Characterization of A. actinomycetemcomitans biofilm dispersal mutant JK1023. (A to D) Colony morphology on agar (A and B) and colony morphology in broth (C and D) of wild-type strain CU1000 (A and C) and mutant strain JK1023 (B and D). Bars = 1 mm. (E) Adherence to polystyrene as measured by a 96-well microtiter plate adherence assay (14). The optical density at 590 nm [O.D. (590 nm)] is proportional to cell adherence. Control wells contained no cells. Strain JK1009 carries a transposon insertion in flp-1 that causes complete loss of surface attachment (14). The values are mean ± standard deviations for triplicate samples. (F) Detachment of cells from biofilm colonies grown attached to polystyrene rods. The optical density at 590 nm is proportional to cell detachment. Control wells contained no cells. The values are means ± standard deviations for six wells of each sample.
To confirm that biofilm colonies of mutant strain JK1023 were deficient in biofilm cell detachment, we grew biofilm colonies for 24 h on polystyrene rods suspended in broth in the wells of a 24-well microtiter plate and quantified the amount of biofilm cell detachment by staining the bacteria growing on the bottoms of the wells with crystal violet (Fig. 1F). Colonization of the bottom of a well resulted from cells that detached from the biofilm colonies growing on the polystyrene rod and fell to the bottom of the well. In this assay, biofilm colonies of strain JK1023 produced significantly less growth on the bottoms of the wells than the wild-type strain produced (P < 0.01, as determined by an unpaired two-tailed t test). These data confirm that mutant strain JK1023 exhibited a decreased biofilm cell detachment phenotype when it was compared to the wild-type parental strain.
Characterization of the dspB gene.
The IS903φkan transposon in strain JK1023 inserted into a 1,143-bp open reading frame which we designated dspB. The dspB sequence of strain CU1000 (serotype f) was 99.1% identical to that of strain HK1651 (serotype b). Six of the ten observed base changes resulted in silent codon substitutions. The dspB gene from strain CU1000 was predicted to encode a protein having 381 amino acid residues with a molecular mass of 43.3 kDa. The 5′ end of dspB contained a predicted signal peptide (26), suggesting that DspB may be a secreted protein.
Residues 40 to 297 of the predicted DspB amino acid sequence were homologous to the catalytic domain of the family 20 glycosyl hydrolases (NCBI Conserved Domain Database accession number pfam00728). This family of enzymes includes bacterial chitinases, chitobiases and lacto-N-biosidases (30, 34, 36) and eukaryotic hexosaminidases (8). The protein most closely related to A. actinomycetemcomitans DspB was lacto-N-biosidase of Lactococcus lactis (GenBank accession no. AAK05592), which displayed 28% identity over 281 amino acid residues, not counting gaps and terminal extensions (Fig. 2). The similarity between DspB and lacto-N-biosidase was high in the regions surrounding Arg47 and the acidic amino acid pair Asp202 and Glu203. These residues have been shown to participate in substrate binding and catalysis in other family 20 glycosyl hydrolases (23, 24, 27). The C-terminal half of DspB contained three Trp residues that were conserved in L. lactis lacto-N-biosidase (at positions 236, 257, and 353). Multiple Trp residues are present in the C-terminal regions of the catalytic domains of all family 20 glycosyl hydrolases (8, 36). These Trp residues line the part of the substrate-binding pocket that is complementary to the hydrophobic surfaces of the hexosamine sugar ring (33).
FIG. 2.
Comparison of residues 21 to 381 of the predicted amino acid sequence of DspB from A. actinomycetemcomitans strain CU1000 (top line) with the amino acid sequence of lacto-N-biosidase from L. lactis (GenBank accession no. AAK05592) (bottom line). The hyphens indicate gaps inserted to maximize the similarity between the sequences. The lines between the sequences indicate identical residues. The asterisks indicate amino acid residues discussed in the text.
Genetic complementation of the dspB mutation.
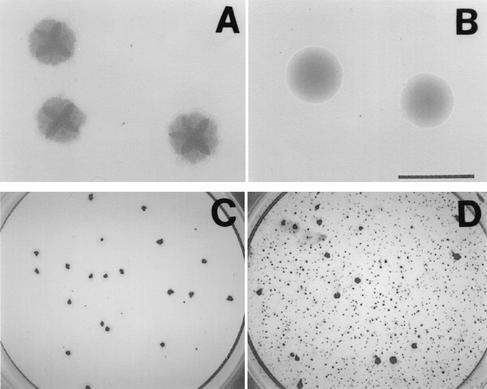
To confirm that expression of dspB is required for A. actinomycetemcomitans biofilm cell detachment, we cloned the wild-type dspB gene from strain CU1000 downstream from an inducible promoter on a broad-host-range plasmid and then introduced the recombinant plasmid (designated pJK618) into mutant strain JK1023 by conjugation with an E. coli donor strain. The colony morphology on agar of strain JK1023 harboring pJK618 was much smoother that that of strain JK1023 harboring the vector plasmid alone (Fig. 3A and B) or than that of wild-type strain CU1000N (Fig. 1A). Plasmid pJK618 restored the ability of biofilm colonies of strain JK1023 to disperse in broth (Fig. 3C and D).
FIG. 3.
Genetic complementation of the dspB mutation in A. actinomycetemcomitans strain JK1023: colony morphology on agar (A and B) and biofilm dispersal phenotype in broth (C and D) of strain JK1023 harboring vector plasmid pJAK16 (A and C) or the complementary plasmid pJK618 (B and D). Bar = 1 mm. The cells in panels C and D were grown in the wells of six-well microtiter plates (diameter, 35 mm; Falcon no. 353046) for 5 days and stained with crystal violet as previously described (16).
Expression, purification, and characterization of DspB protein.
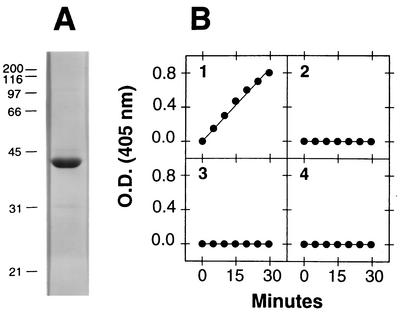
We expressed DspB protein engineered to contain a hexahistidine metal-binding site at its C terminus in E. coli, purified the protein by using Ni affinity chromatography, and cleaved the DspB portion from the hybrid protein by using thrombin. Figure 4A shows the results obtained when 5 μg of purified, thrombin-cleaved DspB protein was analyzed by SDS-polyacrylamide gel electrophoresis. The protein migrated with an apparent molecular mass of 42 kDa. The N-terminal sequence of purified DspB was XNXXVKGNSI (where X is an unidentified residue), which matched residues 21 to 29 of the deduced amino acid sequence encoded by HK1651 dspB. Analysis of purified, thrombin-cleaved DspB protein by mass spectrophotometry resulted in a single major peak at an apparent molecular mass of 41.5 kDa (data not shown), which is consistent with the predicted molecular mass of the thrombin-cleaved DspB protein (41.4 kDa). The yield of DspB expressed in E. coli was 10 mg of protein per liter of culture.
FIG. 4.
Purification and characterization of A. actinomycetemcomitans DspB protein expressed in E. coli. (A) Five micrograms of purified, thrombin-cleaved DspB was electrophoresed through an SDS-12% polyacrylamide gel and stained with Coomassie blue. The molecular masses (in kilodaltons) of protein standards electrophoresed in an adjacent lane are indicated on the left. (B) Glycosyl hydrolase activity of purified DspB with four 4-nitrophenyl-labeled synthetic substrates. The optical density at 405 nm [O.D. (405 nm)] is proportional to the amount of 4-nitrophenolate released in the reaction. The substrates tested were 4-nitrophenyl-N-acetyl-β-d-glucosaminide (graph 1), 4-nitrophenyl-N-acetyl-α-d-glucosaminide (graph 2), 4-nitrophenyl-N-acetyl-β-d-galactosaminide (graph 3), and 4-nitrophenyl-N-acetyl-α-d-galactosaminide (graph 4).
We tested whether DspB could cleave the glycosidic linkages of various 4-nitrophenyl-labeled synthetic hexosamine substrates in an in vitro enzyme assay (Fig. 4B). DspB showed specificity for the 1→4 glycosidic bond of β-substituted N-acetylglucosamine, which is consistent with the known functions of other family 20 glycosyl hydrolases (33). DspB showed no activity against α-substituted N-acetylglucosamine or against α- or β-substituted N-acetylgalactosamine.
Effects of DspB protein on A. actinomycetemcomitans biofilm cell detachment and disaggregation.
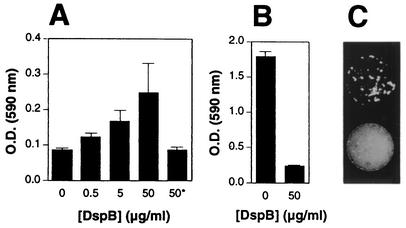
We tested whether adding DspB protein to growth medium could restore the biofilm detachment phenotype in mutant strain JK1023 (Fig. 5A). Polystyrene rods containing preformed biofilm colonies of strain JK1023 were suspended in broth containing various amount of DspB, and the level of biofilm cell detachment was measured by staining the bacteria growing on the bottom of each well with crystal violet as described above (Fig. 1F). Purified DspB restored the ability of mutant strain JK1023 to release cells into the medium and colonize the bottom of a microtiter plate well in a dose-dependent manner (Fig. 5A). Heat-inactivated DspB had no effect on strain JK1023 biofilm cell detachment.
FIG. 5.
DspB-induced detachment and disaggregation of A. actinomycetemcomitans biofilm cells. (A) Detachment of cells from preformed biofilm colonies of mutant strain JK1023 grown attached to polystyrene rods. The rods were suspended for 24 h in broth media containing different concentrations of purified DspB. There was no detachment in the presence of heat-inactivated DspB (100°C, 3 min) (asterisk). The optical density at 590 nm [O.D. (590 nm)] was proportional to biofilm cell detachment. The values are means ± standard deviations for triplicate wells. (B) Detachment of preformed biofilm colonies of strain CU1000 grown in the wells of a 96-well microtiter plate. Biofilm colonies were treated for 6 h with different amounts of DspB, and the cells remaining attached to the surface were stained with crystal violet. The optical density at 590 nm was proportional to the amount of attached cells. The values are means ± standard deviations for triplicate wells. (C) Disaggregation of ethidium bromide-stained clusters of CU1000 cells by purified DspB. Stained clusters of cells were mock treated (top) or treated with 170 mg of purified DspB per ml (bottom) for 30 min, and then 50-μl portions of the treated cell clusters were pipetted onto a sheet of cellophane that was placed on a UV transilluminator and photographed through an orange filter. Clusters of cells appear as white spots on a dark background.
We also tested whether DspB could cause the detachment of preformed biofilm colonies of wild-type strain CU1000 (Fig. 5B). Addition of DspB (50 μg/ml) caused >85% reduction in the amount of surface-associated bacteria after 6 h. DspB also caused disaggregation of highly autoaggregated clumps of CU1000 biofilm cells in solution (Fig. 5C).
DISCUSSION
In the present study we investigated an A. actinomycetemcomitans IS903φkan mutant that was deficient in biofilm cell detachment. The transposon in this strain inserted into a gene that we designated dspB. Our findings indicate that expression of dspB is required for dispersal of A. actinomycetemcomitans biofilms in vitro and that dspB encodes an N-acetylglucosaminidase which causes detachment of cells from A. actinomycetemcomitans biofilm colonies and disaggregation of highly autoaggregated clusters of A. actinomycetemcomitans cells in solution.
Previous studies have shown that biofilm colonies of A. actinomycetemcomitans develop complex architectural features, including a layer of highly autoaggregated cells on the outside of the colony and a layer of nonadherent cells and large, transparent cavities on the inside of the colony (18). These studies have also shown that A. actinomycetemcomitans strains that are deficient in the synthesis of the O-PS component of lipopolysaccharide are also deficient in biofilm cell detachment. Microscopic analyses of these O-PS mutants indicated that they produce biofilm colonies which lack a layer of nonaggregated cells inside the colony. These findings led to the hypothesis that detachment of cells from A. actinomycetemcomitans biofilm colonies occurs by means of a novel mechanism that involves the release of nonaggregated cells located inside the colony (18). It is possible that DspB plays a role in the production of nonaggregated cells inside the biofilm colony. Expression of dspB could be induced inside the colony in response to a physiologically altered microenvironment within the colony. This microenvironment could result from changes in oxygen tension, pH, temperature, or the concentration of nutrients in the center of the colony. DspB could then be secreted into the cavity within the colony and cause cells on the inside of the colony to disaggregate, resulting in the layer of nonaggregated cells observed inside A. actinomycetemcomitans biofilm colonies (18). Continued DspB activity might eventually result in a breach in the colony wall and a sudden release of large numbers of nonaggregated cells into the surrounding medium. This model is consistent with the observation that biofilm cell detachment in A. actinomycetemcomitans may be a rapid and transient process (16).
A possible substrate for DspB is a type IV pilus that has been shown to be required for the A. actinomycetemcomitans surface adherence and autoaggregation phenotypes (14). This pilus is encoded by a cluster of 14 chromosomal genes (13), the first of which, flp-1, encodes the major pilin subunit (14). A. actinomycetemcomitans Flp-1 protein been shown to be posttranslationally modified, probably by glycosylation (11). It is possible that this posttranslational glycosylation includes an N-acetylglucosamine component and that cleavage of this sugar by DspB alters the adhesive properties of the pili, thereby releasing cells from the aggregate. The putative glycosylation sites on Flp-1 are conserved among phylogenetically diverse strains of A. actinomycetemcomitans (17), which is consistent with the finding that DspB causes detachment of cells from biofilm colonies of various A. actinomycetemcomitans strains (Kaplan, unpublished data). Homologous type IV pili have been shown to be involved in the adherence, autoaggregation, and dispersal of enteropathogenic E. coli (21). It is also possible that DspB activity alters a component of the A. actinomycetemcomitans cell surface that interacts with the adherent pili (31).
Another plausible substrate for DspB is exopolysaccharide. Previous studies performed by using chemical and microscopic techniques have shown that A. actinomycetemcomitans produces an exopolysaccharide matrix and that this matrix may play a role in biofilm colony formation (5, 6, 10, 25). The chemical structure of A. actinomycetemcomitans exopolysaccharide is unknown. N-Acetylglucosamine has been shown to be a component of the exopolysaccharides produced by other biofilm bacteria, including Staphylococcus epidermidis (22), Staphylococcus aureus (2), and Helicobacter pylori (32). It is possible that A. actinomycetemcomitans exopolysaccharide also contains an N-acetylglucosamine component and that this sugar is hydrolyzed by DspB, causing degradation of the exopolysaccharide and release of cells from the biofilm aggregate. This mechanism is analogous to that of other biofilm-releasing enzymes, such as Pseudomonas aeruginosa alginate lyase (1) and Methanosarcina mazei disaggregatase (37).
It is tempting to speculate that DspB plays a role in the dissemination of A. actinomycetemcomitans in the human oral cavity and therefore in the pathogenesis of periodontitis, endocarditis, and other chronic infections. A better understanding of biofilm detachment in A. actinomycetemcomitans could lead to improved strategies for treatment and prevention of these diseases.
Acknowledgments
We thank David Figurski and Paul Planet for helpful comments and Bruce Roe, Fares Najar, Sandy Clifton, Tom Ducey, Lisa Lewis, and Dave Dyer of the University of Oklahoma for the use of unpublished nucleotide sequence data from the Actinobacillus Genome Sequencing Project.
This work was supported in part by Public Health Service award DE12585 to N.R.
REFERENCES
- 1.Boyd, A., and A. M. Chakrabarty. 1994. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 60:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesin (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubendorff, J. W., and F. W. Studier. 1991. Creation of a T7 autogene. Cloning and expression of the gene for bacteriophage T7 RNA polymerase under control of its cognate promoter. J. Mol. Biol. 219:61-68. [DOI] [PubMed] [Google Scholar]
- 4.Fine, D. H., D. Furgang, and M. L. Barnett. 2001. Comparative antimicrobial activities of antiseptic mouthrinses against isogenic planktonic and biofilm forms of Actinobacillus actinomycetemcomitans. J. Clin. Periodontol. 28:697-700. [DOI] [PubMed] [Google Scholar]
- 5.Fine, D. H., D. Furgang, J. B. Kaplan, J. Charlesworth, and D. H. Figurski. 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral Biol. 44:1063-1076. [DOI] [PubMed] [Google Scholar]
- 6.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 145:1335-1347. [DOI] [PubMed] [Google Scholar]
- 7.Fine, D. H., P. Goncharoff, H. Schreiner, K. M. Chang, D. Furgang, and D. H. Figurski. 2001. Colonization and persistence of rough and smooth colony variants of Actinobacillus actinomycetemcomitans in the mouths of rats. Arch. Oral Biol. 46:1065-1078. [DOI] [PubMed] [Google Scholar]
- 8.Graham, T. R., H. P. Zassenhaus, and A. Kaplan. 1988. Molecular cloning of the cDNA which encodes beta-N-acetylhexosaminidase A from Dictyostelium discoideum. Complete amino acid sequence and homology with the human enzyme. J. Biol. Chem. 263:16823-16829. [PubMed] [Google Scholar]
- 9.Haase, E. M., J. L. Zmuda, and F. A. Scannapieco. 1999. Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect. Immun. 67:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holt, S. C., A. C. R. Tanner, and S. S. Socransky. 1980. Morphology and ultrastructure of oral strains of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. Infect. Immun. 30:588-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue, T., H. Ohta, I. Tanimoto, R. Shingaki, and K. Fukui. 2000. Heterogeneous post-translational modification of Actinobacillus actinomycetemcomitans fimbrillin. Microbiol. Immunol. 44:715-718. [DOI] [PubMed] [Google Scholar]
- 12.Inouye, T., H. Ohta, S. Kokeguchi, K. Fukui, and K. Kato. 1990. Colonial variation and fimbriation of Actinobacillus actinomycetemcomitans. FEMS Microbiol. Lett. 69:13-18. [DOI] [PubMed] [Google Scholar]
- 13.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in Bacteria and Archaea. J. Bacteriol. 182:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, D. H. Figurski, and J. B. Kaplan. 2001. flp-1, first representative of a new pilin gene subfamily, is required for nonspecific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40:542-554. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan, A. H., D. J. Weber, E. Z. Oddone, and J. R. Perfect. 1989. Infection due to Actinobacillus actinomycetemcomitans: 15 cases and review. Rev. Infect. Dis. 11:46-63. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan, J. B., and D. H. Fine. 2002. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Appl. Environ. Microbiol. 68:4943-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan, J. B., S. Kokeguchi, Y. Murayama, and D. H. Fine. 2002. Sequence diversity in the major fimbrial subunit gene (flp-1) of Actinobacillus actinomycetemcomitans. Oral Microbiol. Immunol. 17:374-379. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan, J. B., M. F. Meyenhofer, and D. H. Fine. 2003. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J. Bacteriol. 185:1399-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan, J. B., M. B. Perry, L. L. MacLean, D. Furgang, M. E. Wilson, and D. H. Fine. 2001. Structural and genetic analyses of O polysaccharide from Actinobacillus actinomycetemcomitans serotype f. Infect. Immun. 69:5375-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King, E. O., and H. W. Tatum. 1962. Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. J. Infect. Dis. 111:85-94. [DOI] [PubMed] [Google Scholar]
- 21.Knutton, S., R. K. Shaw, R. P. Anantha, M. S. Donnenberg, and A. A. Zorgani. 1999. The type IV bundle-forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol. Microbiol. 33:499-509. [DOI] [PubMed] [Google Scholar]
- 22.Mack, D., W. Fisher, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mark, B. L., D. J. Vocadlo, S. Knapp, B. L. Triggs-Raine, S. G. Withers, and M. N. G. James. 2001. Crystallographic evidence for substrate-assisted catalysis in a bacterial β-hexosaminidase. J. Biol. Chem. 276:10330-10337. [DOI] [PubMed] [Google Scholar]
- 24.Mark, B. L., G. A. Wasney, T. J. S. Salo, A. R. Khan, Z. Cao, P. W. Robbins, M. N. G. James, and B. L. Triggs-Raine. 1998. Structural and functional characterization of Streptomyces plicatus β-N-acetylhexosaminidase by comparative molecular modeling and site-directed mutagenesis. J. Biol. Chem. 273:19618-19624. [DOI] [PubMed] [Google Scholar]
- 25.Meyer, D. H., and P. M. Fives-Taylor. 1993. Evidence that extracellular components function in adherence of Actinobacillus actinomycetemcomitans to epithelial cells. Infect. Immun. 61:4933-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 27.Prag, G., Y. Papanikolau, G. Tavlas, C. E. Vorgias, K. Petratos, and A. B. Oppenheim. 2000. Structures of chitobiase mutants complexed with the substrate di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540. J. Mol. Biol. 300:611-617. [DOI] [PubMed] [Google Scholar]
- 28.Rosan, B., J. Slots, R. J. Lamont, M. A. Listgarten, and G. M. Nelson. 1988. Actinobacillus actinomycetemcomitans fimbriae. Oral Microbiol. Immunol. 3:58-63. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sano, M., K. Hayakawa, and I. Kato. 1993. Purification and characterization of an enzyme releasing lacto-N-biose from oligosaccharides with type 1 chain. J. Biol. Chem. 268:18560-18566. [PubMed] [Google Scholar]
- 31.Sheikh, J., J. R. Czeczulin, S. Harrington, S. Hicks, I. R. Henderson, C. Le Bouguénec, P. Gounon, A. Phillips, and J. P. Nataro. 2002. A novel dispersin protein in enteroaggregative Escherichia coli. J. Clin. Investig. 110:1329-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stark, R. M., G. J. Gerwig, R. S. Pitman, L. F. Potts, N. A. Williams, J. Greenman, I. P. Weinzweig, T. R. Hirst, and M. R. Millar. 1999. Biofilm formation by Helicobacter pylori. Lett. Appl. Microbiol. 28:121-126. [DOI] [PubMed] [Google Scholar]
- 33.Tews, I., A. Perrakis, A. Oppenheim, Z. Dauter, K. S. Wilson, and C. E. Vorgias. 1996. Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat. Struct. Biol. 3:638-648. [DOI] [PubMed] [Google Scholar]
- 34.Tews, I., R. Vincentelli, and C. E. Vorgias. 1996. N-Acetylglucosaminidase (chitobiase) from Serratia marcescens: gene sequence, and protein production and purification in Escherichia coli. Gene 170:63-67. [DOI] [PubMed] [Google Scholar]
- 35.Thomson, V. J., M. K. Bhattacharjee, D. H. Fine, K. M. Derbyshire, and D. H. Figurski. 1999. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J. Bacteriol. 181:7298-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsujibo, H., N. Hatano, T. Mikami, Y. Izumizawa, K. Miyamoto, and Y. Inamori. 1998. Cloning, characterization and expression of a β-N-acetylglucosaminidase gene from Streptomyces thermoviolaceus OPC-520. Biochim. Biophys. Acta 1425:437-440. [DOI] [PubMed] [Google Scholar]
- 37.Xun, L., R. A. Mah, and D. R. Boone. 1990. Isolation and characterization of disaggregatase from Methanosarcina mazei LYC. Appl. Environ. Microbiol. 56:3693-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed] [Google Scholar]