Abstract
Postreplication repair functions in gap-filling of a daughter strand on replication of damaged DNA. The yeast Saccharomyces cerevisiae Rad18 protein plays a pivotal role in the process together with the Rad6 protein. Here, we have cloned a human homologue of RAD18, hRAD18. It maps on chromosome 3p24–25, where deletions are often found in lung, breast, ovary, and testis cancers. In vivo, hRad18 protein binds to hHR6 protein through a conserved ring-finger motif. Stable transformants with hRad18 mutated in this motif become sensitive to UV, methyl methanesulfonate, and mitomycin C, and are defective in the replication of UV-damaged DNA. Thus, hRAD18 is a functional homologue of RAD18.
DNA lesions induced by mutagens, including UV light and chemicals, are efficiently removed by base or nucleotide excision repair. However, when the lesion is not removed by these repair systems, or DNA replication machinery meets the lesion before repair, the replication machinery stalls at the lesion, resulting in production of a gap in the newly synthesized strand across from the damaged site. This gap will be filled by postreplication repair, but the precise mechanisms are poorly understood. In the yeast Saccharomyces cerevisiae, RAD18 and RAD6, which belong to the same epistasis group, are required for this process (1). Mutants carrying rad18 or rad6 mutations are extremely sensitive to killing by various mutagens (2). A rad18 mutation causes an increased spontaneous mutation frequency (3), but does not affect UV-induced mutagenesis and sporulation (4, 5). In vivo, Rad18 protein forms a tight complex with Rad6 protein, which is a ubiquitin-conjugating enzyme (E2) (6). Rad18 protein binds single-stranded DNA (6). A homologue of RAD18 has been identified from the filamentous fungus Neurospora crassa. This gene, uvs-2, encodes a protein that shares 25.5% amino acid identity with Rad18 and interacts with a Rad6 homologue, MUS8 (7, 8). In contrast to the yeast rad18 mutant, the uvs-2 mutant shows high mutation frequencies under both spontaneous and induced conditions (9–11). Although Rad18 is crucial for maintaining genomic integrity, Rad18 homologous genes in higher eukaryotes, including human beings, have not been reported yet. In this study, we successfully isolated a human homologue of S. cerevisiae RAD18/N. crassa uvs-2, and we have shown its involvement in postreplication repair.
Materials and Methods
PCR.
Human expressed sequence tag (EST) clones homologous to S. cerevisiae Rad18 protein and N. crassa UVS2 protein were searched for by using the xrefdb program in the National Center for Biotechnology Information database. Oligonucleotide AA625–123 (5′-AGATGATTTGCTGCGGTGTG-3′) and AA625–481 (5′-GTAGAACCACTCATTTCTCT-3′) were derived from the EST AA625471 sequence. These PCR primers amplify a 378-bp fragment from +182 to +559 of the hRAD18 gene. This fragment was used as a probe to screen a human placental cDNA library by the colony hybridization method. For detection of the hRAD18 gene in a radiation hybrid panel (Gene Bridge 4, Research Genetics, Huntsville, AL), PCR primers were 5′-ACTGACATTTCACACAGGTA-3′ and 5′-TAACCCATACTCAACTATCC-3′, which amplify a 622-bp fragment in the 3′ untranslated region of hRAD18. Human hHR6A and hHR6B cDNA were obtained by PCR using a cDNA library prepared from normal human fibroblasts.
Cell Culture and Transfection.
Cells were cultured in DMEM supplemented with 10% FCS, 100 units/ml penicillin G, and 100 μg/ml streptomycin in a humidified 5% CO2 incubator. Mori-SV is a normal human skin fibroblast immortalized with simian virus 40. Mutant hRAD18 cDNAs were produced with a Quick-Change site-directed mutagenesis kit (Stratagene) and harbored in pcDNA3. Transformation was performed by the calcium phosphate precipitation method by using a Mammalian Transfection kit (Stratagene). Stable transformants of Mori-SV and Saos2 were selected with G418 at concentrations of 400 μg/ml and 500 μg/ml, respectively, and a polyclonal population was used in all experiments.
Yeast Two-Hybrid Assay.
Protein–protein interaction was examined in the yeast two-hybrid assay with the Matchmaker Gal4 II System (CLONTECH). Two plasmids, pAS2–1(GAL4 BD) and pACT2(GAL4 AD) containing full-length hRAD18 and hHR6A or hHR6B cDNA, respectively, were transfected into the yeast strain Y187. All combinations were at first tested for β-galactosidase activity by the colony filter-lift assay. To confirm the results, quantitative liquid assays for β-galactosidase activity were conducted. One unit of β-galactosidase activity was defined as the amount of enzyme that converts 1 μmol of o-nitrophenyl β-d-galactopyranoside to nitrophenol in 1 min at 30°C (pH 7.0), according to the manufacturer's protocol.
Immunoprecipitation.
COS-7 cells were transfected with 10 μg of plasmid by electroporation by using an Electro Cell Manipulator (BTX, San Diego) and cultured for 48 h. As control plasmids, a plasmid containing only glutathione S-transferase (GST)-tag or T7-tagged JBD was used. JBD encodes the c-Jun N-terminal kinase (JNK) binding domain of JNK interacting protein-1 (JIP-1). Cells were lysed in 1 ml of lysis buffer containing 50 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 0.2% Triton X-100, 0.5 mM DTT, and protease inhibitors (PMSF, aprotinin, pepstatin, and leupeptin) and sonicated with a Sonifier 450 (Branson). Ninety percent of the cell lysate was mixed with 2 μg of an anti-T7 mAb (Novagen) or an anti-GST polyclonal rabbit antibody (Santa Cruz Biotechnology) for 1 h at 4°C. After addition of 50 μl of a slurry of protein G Sepharose beads (Amersham Pharmacia), the lysate was mixed for another 1 h at 4°C. Fifty percent of the bound fraction and the remaining 10% of the original extract were resolved by SDS/12.5% PAGE. Separated proteins were detected by either anti-GST rabbit antibody or anti-MUS8 rabbit serum by using the enhanced chemiluminescence system (Amersham International). Anti-N. crassa MUS8 antiserum was raised in rabbits by immunizing with GST-MUS8 fusion protein. This antiserum crossreacted well with human hHR6A and hHR6B proteins.
Immunocytochemistry.
Polyclonal antiserum against hRad18 was raised by immunizing rabbits with GST-hRad18 (Leu-383 to Gln-495) fusion protein which was purified from Escherichia coli extracts through a glutathione-agarose column and a Sephadex G-100 column. Mutant and wild-type hRAD18 cDNA were harbored in eukaryotic expression vector pCAGGS (12). These plasmids (0.02 μg/μl) were microinjected into the nuclei of mouse p53−/− primary fibroblasts with glass needles (13). Two hours later, cells were prefixed with 3.5% formalin and then with 80% methanol. In some cases, cells were irradiated with UV (15 J/m2) 2 h after microinjection and cultured for another 1 h before fixation. These cells were stained with anti-hRad18 antiserum for 30 min at 4°C, and then with FITC-conjugated anti-rabbit-IgG (Cappel) for 30 min at room temperature.
Sedimentation Studies of Newly Synthesized DNA.
Postreplication repair was analyzed by the sedimentation velocity method (14). In brief, actively growing cells (105 cells per dish) were irradiated with UV at 8 J/m2, incubated for 30 min in 3 ml of prewarmed medium, and then pulse-labeled with 0.93 MBq/ml [methyl-3H]thymidine for 30 min. In pulse–chase experiments, the pulse-labeled cells were further incubated for 90 min in fresh medium containing 10 μM unlabeled thymidine. As a control, cells were treated in the same way without UV irradiation. The cells were lysed in the dishes by addition of 0.1 ml of lysis solution containing 0.2 M NaOH, and 20 mM EDTA, and the lysates were irradiated with 20 Gy of x-rays on ice. Aliquots of the cell suspension were layered on the top of 4.8 ml of 5–20% (wt/vol) alkaline sucrose density gradients containing 0.1 M NaOH, 0.1 M NaCl, and 10 mM EDTA, and centrifuged at 50,000 rpm for 2 h at 4°C in a SW55 rotor (Beckman). After centrifugation, drop fractions were collected directly onto GF/C glass filters (Whatman), and the acid-insoluble radioactivities were measured by a liquid scintillation counter.
Results
Isolation of the hRAD18 Gene.
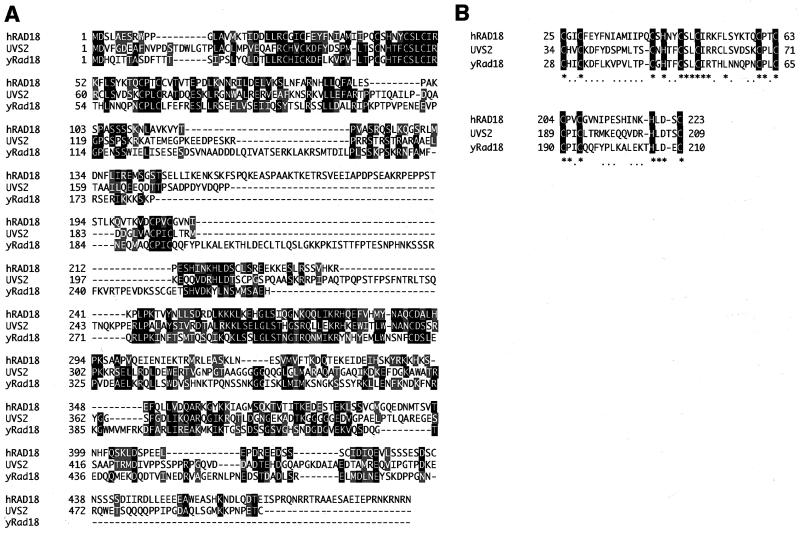
The human EST clone AA625471 (513 bp), which encodes a peptide with homology to the NH2 terminus of S. cerevisiae Rad18 protein and N. crassa UVS2 protein, was used to screen a human placental cDNA library. A single clone that contains a 2.8-kb cDNA insert was isolated. This gene, named hRAD18, encodes a protein of 495 aa with a calculated molecular mass of 56 kDa. The deduced amino acid sequence shows significant homology to the S. cerevisiae Rad18 and N. crassa UVS2 proteins (Fig. 1A). Human Rad18 protein shares 20% identical and 42% similar residues with yeast Rad18, whereas N. crassa UVS2 protein shares 23% and 43%, respectively. All of these proteins have one conserved ring-finger motif and one conserved zinc-finger motif in the NH2-terminal half (Fig. 1B). However, hRAD18 and uvs-2 do not have the nucleotide-binding motif that is found in yeast RAD18 (7, 8). Despite the high similarity between RAD18 and uvs-2, the uvs-2 gene cannot complement yeast rad18 mutant (7, 8). Similarly, hRAD18 could complement neither the S. cerevisiae rad18 mutant nor the N. crassa uvs-2 mutant (data not shown).
Figure 1.
Deduced amino acid sequence of the hRAD18 gene. (A) Alignment of human Rad18 (hRad18), N. crassa UVS2, and S. cerevisiae Rad18 (yRad18) protein sequences. Residues identical and similar to hRad18 are indicated in black and gray, respectively. (B) Alignment of aa sequences in the conserved ring-finger (Upper) and zinc-finger motif (Lower).
Chromosomal Localization of the hRAD18 Gene.
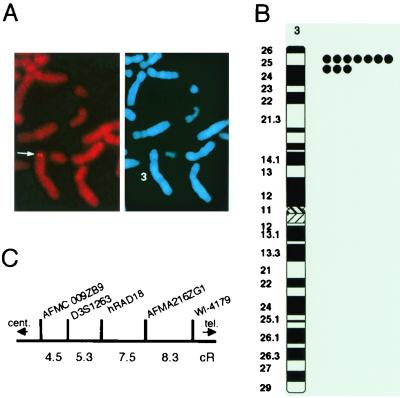
The locus of the hRAD18 gene was mapped to chromosome 3p24–25 by the fluorescence in situ hybridization (FISH) method (Fig. 2 A and B). No additional loci were detected by FISH. PCR mapping was performed by using a radiation hybrid panel. Comparison of the human chromosomal content of the hybrids exhibiting positive PCR signals localized the hRAD18 gene to chromosome 3 and placed it at 7.5 cR from AFMA216ZG1, and 5.3 cR from D3S1263 (Fig. 2C). Both of these polymorphic markers are located at chromosome 3p24.
Figure 2.
Chromosomal localization of the hRAD18 gene. (A) An example of FISH mapping, showing the FISH signals on chromosome 3 (Left). The same mitotic figure was stained with 4′,6-diamidino-2-phenylindole to identify chromosome 3 (Right). FISH was performed by using normal human lymphocytes and a 2.8-kb cDNA for hRAD18 as a probe according to standard procedures (See DNA Biotech, Ontario, Canada). (B) Diagram of FISH mapping. Each dot represents the FISH signals detected on human chromosome 3. (C) Chromosome mapping by PCR with a radiation hybrid panel. PCR was carried out as described in Materials and Methods. The results of PCR patterns were analyzed at the Whitehead Institute/MIT Center for Genome Research (Cambridge, MA).
Interaction of hRad18 with hHR6.
In S. cerevisiae and N. crassa, Rad18 and UVS2 interact with Rad6 and MUS8, respectively. To examine whether hRad18 works in a similar manner, we obtained two RAD6-homologue genes, hHR6A and hHR6B (15), from a human cDNA library by PCR. Using these genes, two different yeast two-hybrid assays were conducted. The colony filter-lift assay showed a positive reaction in combinations of hRad18 and hHR6A, or hRad18 and hHR6B (data not shown). Quantitative liquid assay confirmed these results and indicated that interaction of hRad18-hHR6A is about twice that of hRad18-hHR6B (Fig. 3A). Interaction between hRad18 and hHR6s was further tested in mammalian cells. GST-tagged hRAD18 cDNA and T7-tagged hHR6 cDNA were cotransfected into COS-7 monkey cells, and immunoprecipitation experiments were performed. As shown in Fig. 3B, GST-hRad18 was specifically brought down by a T7-tag-specific mAb only when T7-hHR6A or T7-hHR6B was cotransfected with GST-hRAD18 (Fig. 3B Upper; lanes 10 and 11). Inversely, T7-hHR6A or T7-hHR6B was brought down by a GST-tag-specific polyclonal antibody only when GST-hRAD18 was transfected with T7-hHR6A or T7-hHR6B (Fig. 3B Lower, lanes 10 and 11). Hence, hRad18 and hHR6A/hHR6B can physically associate with each other in transfected mammalian cells.
Figure 3.
Interaction of hRad18 protein with human Rad6 (hHR6A or hHR6B) protein. (A) Yeast two-hybrid assay. Two-hybrid assay was conducted by using the Matchmaker Gal4 II System (CLONTECH). Protein–protein interaction was assessed by the β-galactosidase activity. The hRAD18(C28F), hRAD18(C28F, C60F), and hRAD18(C207F) constructs are mutant hRAD18 cDNAs with Cys-28, Cys-28 and Cys-60, and Cys-207 replaced by Phe, respectively. Cys-28 and Cys-60 are in the ring-finger motif, and Cys-207 is in the zinc-finger motif. (B) Transfection, immunoprecipitation, and Western blot assay. Cell extracts were prepared 48 h after transfection from COS-7 cells transfected with the indicated GST-tagged and/or T7-tagged expression plasmids. Transfected cell extracts (Left) and immunoprecipitated proteins (Right) were separated by SDS/PAGE and transferred to a nylon membrane. (Upper) Cell extracts (lanes 1–7) and proteins immunoprecipitated with an anti-T7 antibody (lanes 8–14) were immunoblotted with anti-GST antibody. (Lower) Cell extracts (lanes 1–7) and proteins immunoprecipitated with an anti-GST antibody (lanes 8–14) were immunoblotted with anti-MUS8 (N. crassa homologue of Rad6) antiserum.
Involvement of hRad18 in Postreplication Repair.
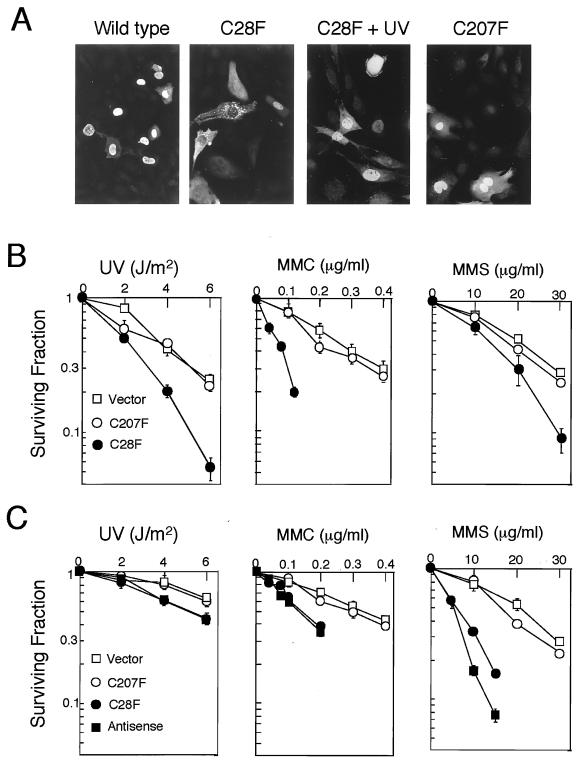
We constructed two plasmids containing mutations in hRAD18 cDNA: hRad18(C28F) which contained a mutation to Phe at the second conserved Cys (Cys-28) in the ring-finger motif, and hRad18(C207F) in which the second conserved Cys (Cys-207) in the zinc-finger motif was replaced with Phe. Such mutations are known to abolish protein–protein interaction through the ring-finger motif or DNA-binding activity of the zinc-finger motif in other cases (16, 17). Indeed, hRad18(C28F) no longer associated with hHR6 (hHR6A or hHR6B) as determined by the yeast two-hybrid assay, whereas hRad18(C207F) associated with hHR6 protein (Fig. 3A). These results indicate that Cys-28 in the ring-finger motif is essential for interaction with hHR6A or hHR6B. Wild-type and mutant hRad18 (C207F) localized predominantly in the nucleus, whereas mutant hRad18 (C28F) localized in both the cytoplasm and the nucleus (Fig. 4A). Interestingly, mutant hRad18 (C28F) localized prominently in the nucleus after UV irradiation, suggesting that it binds to damaged sites on DNA.
Figure 4.
Enhanced sensitivity of human cells transformed with mutant hRAD18 to various DNA-damaging agents. (A) Intracellular localization of wild-type and mutant hRad18. Plasmids containing hRAD18 cDNA were microinjected into the nuclei of mouse p53−/− primary fibroblasts. The cells were cultured for 2 h. Localization of overexpressed hRad18 protein was visualized by the indirect immunostaining method by using polyclonal rabbit serum against hRad18. From left to right, wild-type hRad18, hRad18(C28F), hRad18(C28F) 1 h after UV irradiation (15 J/m2), and hRad18(C207F). (B) Sensitivity of Mori-SV-stable transformants with mutant hRAD18 cDNA to various DNA-damaging agents. Cells were irradiated with UV (Left), or treated with mitomycin C (Center) or methyl methanesulfonate (Right) for 1 h, and sensitivity was determined by the colony-formation assay. Mean values of triplicate dishes are shown with standard deviations. (C) Sensitivity of Saos2 stable transformants to different DNA-damaging agents. Saos2 antisense is a stable transformant with pcDNA3 containing hRAD18 cDNA in the opposite direction.
If hRad18 is involved in postreplication repair, human cells with the impaired hRad18 function should be sensitive to different types of DNA damage. Because hRad18(C28F) localized in the nucleus without association with hHR6 (hHR6A or hHR6B), overexpression of such mutant hRad18 would compete with wild-type hRAD18 for DNA-damaged sites. Expecting a dominant-negative effect, stable transformants of a human fibroblastic cell line (Mori-SV) were isolated after transfection with these mutant hRAD18 cDNAs. C28F transformants (Mori-SV C28F) showed significantly increased sensitivity to UV, the alkylating agent methyl methanesulfonate, and the DNA cross-linker mitomycin C, whereas C207F transformants (Mori-SV C207F) showed almost the same sensitivity as control cells transformed with the vector (Mori-SV Vec) (Fig. 4B). These three types of stable transformants grew at similar rates. We have also obtained stable transformants of the human osteosarcoma cell line Saos2 with these mutant hRAD18 plasmids. Again, C28F transformants (Saos2 C28F) became sensitive to UV, methyl methanesulfonate, and mitomycin C, whereas sensitivity of C207F transformants (Saos2 C207F) remained at the control cell (Saos2 Vec) levels (Fig. 4C). Furthermore, Saos2 transformants with antisense hRAD18 cDNA (Saos2 anti) also showed levels of sensitivity to these agents comparable to those of C28F transformants. These results suggest that the enhanced sensitivity to multiple DNA-damaging agents of C28F transformants was not caused by some unknown events associated with the transformation process, but by impaired postreplication repair caused by coexpression of mutant hRad18 protein.
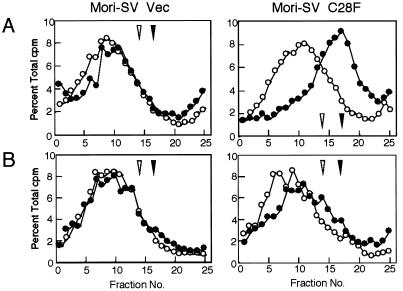
To confirm this conclusion, we measured replicated DNA sizes after UV irradiation. After a 30-min incubation of UV-irradiated C28F transformants (Mori-SV C28F), the cells were pulse labeled with [3H]thymidine with or without chase incubation. Lysates of these pulse-labeled cells were analyzed by alkaline sucrose density gradient centrifugation. As shown in Fig. 5A, the size of replicated DNA in UV-irradiated C28F transformants was smaller than that of nonirradiated cells, whereas the profile from the control cells (Mori-SV Vec) was very similar to that from nonirradiated cells. After 90 min of the chase period, the size of pulse-labeled DNA in UV-irradiated C28F transformants increased but still remained small when compared with that of nonirradiated cells (Fig. 5B). These results indicate that coexpression of hRad18 protein containing a mutation in the ring-finger motif manifests a dominant-negative effect on postreplication repair, resulting in enhanced sensitivity to multiple DNA-damaging agents.
Figure 5.
Impaired postreplication repair of human cells transformed with mutant hRAD18. (A) Cells (Mori-SV C28F or Mori-SV Vec) were irradiated with UV (8 J/m2), incubated for 30 min, and then pulse labeled with [3H]thymidine (0.93 MBq/ml) for 30 min. Samples were sedimented on 5–20% alkaline sucrose gradients from right to left, and the profile of UV-irradiated cell sample (●) was compared with that of the nonirradiated control cell sample (○). (Left) Mori-SV-stable transformant with vector alone. (Right) Mori-SV-stable transformant with hRAD18(C28F). (B) After the pulse label, cells were chased for 90 min in fresh medium containing 10 μM unlabeled thymidine. Samples were analyzed by the same method used in A. Closed and open arrowheads indicate the positions of bacteriophage λ DNA (42 kb) and bacterial artificial chromosome DNA (100 kb), respectively.
Discussion
A gene, named hRAD18, has been cloned by screening a human placental cDNA library with a human EST clone that encodes a peptide with homology to the NH2 terminus of S. cerevisiae Rad 18 protein and N. crassa UVS2 protein. This gene was mapped on a single locus and no other signal was detected by FISH, suggesting that this is a unique gene. The amino acid sequence deduced from the cDNA shows significant homology to the S. cerevisiae Rad18 and N. crassa UVS2 proteins. Interestingly, all of these proteins have one conserved ring-finger motif and one conserved zinc-finger motif in the NH2-terminal half. hRad18 localizes in the nucleus and physically associates with hHR6A/hHR6B in vivo. Stable transformants with hRad18 mutated in the ring-finger motif manifested sensitivity to UV, mitomycin C, and methyl methanesulfonate, and showed smaller sizes of replicated DNA compared with those of control cells after UV irradiation. From these results, we concluded that hRAD18 is a functional human homologue of the yeast RAD18 and N. crassa uvs-2 involved in postreplication repair. Because hRad18(C28F) mutant protein localized in the nucleus after DNA damage without association with hHR6, this mutant protein may compete with endogenous wild-type hRad18 for the damaged sites, and thus inhibit the initial step of postreplication repair.
It has been hypothesized that the complex of Rad18 and Rad6 binds to damaged sites by the single-stranded DNA-binding activity of Rad18, and that it ubiquitinates some components of the stalled DNA replication machinery by the ubiquitin- conjugating enzyme activity of Rad6 (6). Through stimulating degradation of these blocking components, the complex may facilitate DNA repair by different repair systems (6). In this model, Rad6–Rad18 complex might interact with the DNA replication machinery and/or DNA polymerases specifically involved in translesion DNA synthesis. In a preliminary experiment, we found that hRad18 did not interact with the recently found DNA polymerase η, defects of which are the cause of xeroderma pigmentosum variant (18) (data not shown).
hRAD18 has been mapped on chromosome 3p24–25. In this region, loss of heterozygosity is often recognized in lung, breast, ovary, and testis cancers (19). Coincidentally, some fractions of these cancers are susceptible to chemotherapy. In S. cerevisiae and N. crassa, the rad18 and uvs-2 mutants show high mutation frequency under spontaneous conditions. Therefore, it is plausible that mutation in the hRAD18 gene enhances subsequent mutation rates in some oncogenes and tumor suppressor genes, thus promoting progression of malignant transformation. These cancer cells with impaired postreplication repair might be sensitive to different types of DNA-damaging agents. Information on the hRAD18 gene status of individual cancer patients will be important for determining the likely success of chemotherapy.
Acknowledgments
We thank M. Ohkubo and T. Itoh for valuable discussions, K. Suzuki for antiserum production, and A. Schroeder for critical reading of the manuscript. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan.
Abbreviations
- FISH
fluorescence in situ hybridization
- EST
expressed sequence tag
- GST
glutathione S-transferase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB035274).
References
- 1.Prakash L. Mol Gen Genet. 1981;184:471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- 2.Haynes R H, Kunz B A. In: The Molecular Biology of the Yeast Saccharomyces. Strathern J N, Jones E W, Broach J R, editors. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1981. pp. 371–414. [Google Scholar]
- 3.Kunz B A, Kang X, Kohalmi L. Mol Cell Biol. 1990;11:218–225. doi: 10.1128/mcb.11.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawrence C W, Christensen R. Genetics. 1976;82:207–232. doi: 10.1093/genetics/82.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones J S, Weber S, Prakash L. Nucleic Acids Res. 1988;16:7119–7131. doi: 10.1093/nar/16.14.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 7.Tomita H, Soshi T, Inoue H. Mol Gen Genet. 1993;238:225–233. doi: 10.1007/BF00279551. [DOI] [PubMed] [Google Scholar]
- 8.Soshi T, Sakuraba Y, Kafer E, Inoue H. Curr Genet. 1996;30:224–231. doi: 10.1007/s002940050125. [DOI] [PubMed] [Google Scholar]
- 9.de Serres F J. Mutat Res. 1980;71:181–191. doi: 10.1016/0027-5107(80)90069-x. [DOI] [PubMed] [Google Scholar]
- 10.Inoue H, Ong T, de Serres F J. Mutat Res. 1981;80:27–41. doi: 10.1016/0027-5107(81)90175-5. [DOI] [PubMed] [Google Scholar]
- 11.Inoue H. Mutat Res. 1999;437:121–133. doi: 10.1016/s1383-5742(99)00079-4. [DOI] [PubMed] [Google Scholar]
- 12.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 13.Nakagawa K, Taya Y, Tamai K, Yamaizumi M. Mol Cell Biol. 1999;19:2828–2834. doi: 10.1128/mcb.19.4.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehmann A R, Kirk-Bell S, Arlett C F, Paterson M C, Lohman P H M, de Weerd-Kastelein E A, Bootsma D. Proc Natl Acad Sci USA. 1975;72:219–223. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koken M H M, Reynolds P, Jaspers-Dekker I, Prakash L, Prakash S, Bootsma D, Hoeijmakers J H J. Proc Natl Acad Sci USA. 1991;88:8865–8869. doi: 10.1073/pnas.88.20.8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brzovic P S, Meza J, King M C, Klevit R E. J Biol Chem. 1998;273:7795–7799. doi: 10.1074/jbc.273.14.7795. [DOI] [PubMed] [Google Scholar]
- 17.Asahina H, Kuraoka I, Shirakawa M, Morita E H, Miura N, Miyamoto I, Ohtsuka E, Okada Y, Tanaka K. Mutat Res. 1994;315:229–237. doi: 10.1016/0921-8777(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 18.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. Nature (London) 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 19.Bieche I, Lidereau R. Genes Chromosomes Cancer. 1995;14:227–251. doi: 10.1002/gcc.2870140402. [DOI] [PubMed] [Google Scholar]