Abstract
Activation of the tcpPH promoter on the Vibrio pathogenicity island by AphA and AphB initiates the Vibrio cholerae virulence cascade and is regulated by quorum sensing through the repressive action of HapR on aphA expression. To further understand how the chromosomally encoded AphA protein activates tcpPH expression, site-directed mutagenesis was used to identify the base pairs critical for AphA binding and transcriptional activation. This analysis revealed a region of partial dyad symmetry, TATGCA-N6-TNCNNA, that is important for both of these activities. Searching the V. cholerae genome for this binding site permitted the identification of a second one upstream of a penicillin V amidase (PVA) gene on the small chromosome. AphA binds to and footprints this site, which overlaps the pva transcriptional start, consistent with its role as a repressor at this promoter. Since aphA expression is under quorum-sensing control, the response regulators LuxO and HapR also influence pva expression. Thus, pva is repressed at low cell density when AphA levels are high, and it is derepressed at high cell density when AphA levels are reduced. Penicillin amidases are thought to function as scavengers for phenylacetylated compounds in the nonparasitic environment. That AphA oppositely regulates the expression of pva from that of virulence, together with the observation that PVA does not play a role in virulence, suggests that these activities are coordinated to serve V. cholerae in different biological niches.
A fascinating interplay exists between the virulence genes acquired by V. cholerae through horizontal gene transfer events and the ancestral chromosomal genes recruited to regulate them. This interplay serves to link virulence to the physiology of the cell, thereby maximizing the ability of the organism to cause disease when it is in an appropriate environmental niche and to conserve these resources when it is not. The acquisition of two pathogenicity elements into the large chromosome of V. cholerae, the Vibrio pathogenicity island (VPI) (16, 17) and the lysogenic CTX phage (45), conferred upon the organism the ability to cause the life-threatening diarrheal disease cholera. The VPI carries the genes required for the synthesis and assembly of the toxin-coregulated pilus (TCP) (43), the primary colonization factor, and CTX encodes cholera toxin (CT), a potent ADP-ribosylating toxin (15).
Once in the epithelium of the upper small intestine, a complex network comprised of regulators carried both within the VPI and the V. cholerae ancestral genome coordinates the expression of the TCP and CT genes. ToxT is an AraC-type regulator encoded by the VPI that directly activates the expression of tcp and ctx as well as other genes (4, 8, 13, 46). The activation of toxT expression is unusual in that it requires cooperation between two homologous pairs of transmembrane transcriptional activators, TcpP/TcpH and ToxR/ToxS (24). TcpP and TcpH (3, 12) are encoded by an operon within the VPI and have no known functions other than their role in toxT activation. ToxR and ToxS (31, 33) are encoded by a separate operon on the large chromosome, and in addition to their involvement in toxT activation they also play an important role in regulating the expression of the outer-membrane proteins OmpU and OmpT (7, 25, 26, 32).
The activation of the tcpPH operon on the VPI initiates the virulence cascade through the cooperation of two unlinked regulators located on the large chromosome, AphA and AphB (19, 41). AphA is a member of a small family of regulators that show homology to PadR, a repressor that controls the detoxification of phenolic acids (1), and AphB is a LysR-type regulator (19, 22). The specific roles of these proteins in V. cholerae aside from virulence gene regulation are not yet known.
Stimuli within the host environment typically serve as signals to allow pathogenic bacteria to mount a productive infection when they are in an appropriate biological niche. In V. cholerae, several chromosomally located genes that normally function to sense and regulate gene expression in response to environmental cues influence the expression of the virulence cascade. For example, H-NS, CRP, and PepA are transcriptional regulators that influence expression from various promoters within the cascade in response to factors such as temperature and pH (2, 35, 40). In addition, in certain strains of V. cholerae the virulence cascade is also responsive to cell density (47). Three quorum-sensing systems apparently function in parallel to control the expression of the cascade through the activity of the response regulator LuxO (30). At low cell density, LuxO functions to activate a putative repressor protein that reduces the expression of the V. cholerae LuxR homolog HapR, thus permitting high-level expression of the cascade. At high cell densities, LuxO fails to activate the putative repressor protein resulting in derepression of hapR, and HapR in turn shuts off the virulence cascade by binding to a site in the aphA promoter and repressing its expression (23). The regulation of aphA expression by HapR thus serves to link quorum sensing in V. cholerae to pathogenesis. However, not all virulent strains of V. cholerae have a fully functional quorum-sensing system that regulates virulence gene expression. Certain strains, such as El Tor N16961 and classical O395, possess a naturally occurring frameshift mutation in hapR (47), and other strains, such as classical CA401, have a naturally occurring point mutation in the aphA promoter that prevents HapR from binding even if it is functional (23).
AphA has previously been shown to bind to a region of the tcpPH promoter between −101 and −71 from the start of transcription (21). AphA appears to activate transcription by enhancing the ability of AphB to bind to its recognition site, a region of interrupted dyad symmetry from −69 to −53 that displays the LysR recognition motif (21, 22). It is the ability of AphB to bind to this motif, which differs by a single base pair in the classical and El Tor biotypes, that determines the biotype specificity of virulence gene expression in V. cholerae (20, 22). To gain further insights into how AphA contributes to the activation of the tcpPH promoter, a systematic mutational analysis of the AphA binding site from positions −75 to −98 was carried out. As shown here, this analysis identified nine base pairs absolutely critical for AphA to bind to the tcpPH promoter and activate transcription in the presence of AphB. These nine base pairs lie within a region of partial dyad symmetry that has the sequence TATGCA-N6-TNCNNA. Since aphA is located in the ancestral genome and its only known target, the tcpPH promoter, is present on an acquired pathogenicity element, it seemed likely that AphA might regulate the expression of other genes in V. cholerae and also link them to quorum sensing and pathogenesis by binding to a similar site. Searching the V. cholerae genome for matches to the 18-bp AphA binding site led to the identification of a gene on the small chromosome encoding a penicillin V amidase (PVA) that is regulated by AphA.
Penicillin amidases have been identified in a wide variety of different microorganisms, such as bacteria, yeast, and filamentous fungi (for a review see reference 44). These enzymes have had a significant impact on human health over the last 30 years because of their important role in the production of useful β-lactam antibiotics to aid in the control of infectious bacterial diseases. By hydrolyzing penicillins to yield 6-aminopenicillanic acid (6-APA), these enzymes simplified the procedure for generating a key intermediate that is used in the production of semisynthetic derivatives with a wide range of antimicrobial activities. Penicillin amidases have been classified based on their various substrate specificities. For example, the preferred substrate for type I enzymes is phenoxymethylpenicillin (penicillin V), for type II enzymes it is benzylpenicillin (penicillin G), and for type III enzymes (d-α-aminobenzylpenicillin) it is ampicillin. Although their physiological role is still not clear, it is thought that they serve as scavengers for phenylacetylated compounds in carbon-limiting environments (44).
We show here that AphA negatively regulates PVA gene expression in both the classical and El Tor biotypes of V. cholerae by binding to a site virtually identical to that at the tcpPH promoter which overlaps the pva transcriptional start site. In El Tor strain C6706, the pva gene is also regulated by quorum sensing such that its expression is reduced at low cell density when AphA levels are high and its expression increases at higher cell density when aphA is repressed by HapR. Since PVA does not appear to play a role in virulence, the ability of AphA to oppositely regulate pva expression from that of virulence gene expression may provide V. cholerae with advantages in different environmental niches.
MATERIALS AND METHODS
Bacterial strains and expression plasmids.
The V. cholerae strains and expression plasmids used in this study are listed in Table 1. Strains were maintained at −70°C in Luria-Bertani (LB) medium (28) containing 30% (vol/vol) glycerol. Antibiotics were used at the following concentrations in LB medium: ampicillin, 100 μg/ml; kanamycin, 45 μg/ml; polymyxin B, 50 U/ml; streptomycin, 1 mg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used in LB agar at 40 μg/ml.
TABLE 1.
Bacterial strains and expression plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Reference or source |
|---|---|---|
| V. cholerae | ||
| C6706 str2 | El Tor Inaba Smr | Laboratory collection |
| KSK262 | C6706 str2 ΔlacZ3 | 19 |
| O395 | classical Ogawa Smr | Laboratory collection |
| KSK618 | O395 ΔlacZ tcpP-lacZ | 41 |
| GK300 | KSK618 ΔtcpPH promoter | 20 |
| GK797 | GK300 + tcpPH promoter, −75 A to C | This work |
| GK798 | GK300 + tcpPH promoter, −76 T to A | This work |
| GK799 | GK300 + tcpPH promoter, −77 T to G | This work |
| GK800 | GK300 + tcpPH promoter, −78 A to C | This work |
| KSK1309 | GK300 + tcpPH promoter, −79 C to G | This work |
| GK834 | GK300 + tcpPH promoter, −80 T to G | This work |
| GK835 | GK300 + tcpPH promoter, −81 C to A | This work |
| GK801 | GK300 + tcpPH promoter, −82 T to G | This work |
| GK845 | GK300 + tcpPH promoter, −83 T to A | This work |
| GK846 | GK300 + tcpPH promoter, −84 G to T | This work |
| GK847 | GK300 + tcpPH promoter, −85 A to C | This work |
| GK848 | GK300 + tcpPH promoter, −86 G to C | This work |
| GK872 | GK300 + tcpPH promoter, −87 C to G | This work |
| GK873 | GK300 + tcpPH promoter, −88 T to A | This work |
| WL40 | GK300 + tcpPH promoter, −89 A to C | This work |
| WL35 | GK300 + tcpPH promoter, −90 A to C | This work |
| WL36 | GK300 + tcpPH promoter, −91 C to G | This work |
| WL37 | GK300 + tcpPH promoter, −92 G to T | This work |
| WL39 | GK300 + tcpPH promoter, −93 T to A | This work |
| WL38 | GK300 + tcpPH promoter, −94 A to C | This work |
| WL57 | GK300 + tcpPH promoter, −95 T to A | This work |
| WL52 | GK300 + tcpPH promoter, −96 T to A | This work |
| WL55 | GK300 + tcpPH promoter, −97 A to C | This work |
| WL56 | GK300 + tcpPH promoter, −98 T to A | This work |
| GK910 | C6706 ΔaphA | This work |
| GK925 | C6706 Δpva-lacZ | This work |
| GK927 | C6706 Δpva-lacZ ΔaphA | This work |
| GK930 | C6706 Δpva | This work |
| KSK1683 | O395 ΔlacZ ΔaphA | This work |
| KSK1863 | KSK262 ΔaphA | This work |
| KSK1918 | C6706 pva-lacZ ΔhapR | This work |
| KSK1920 | C6706 pva-lacZ ΔluxO | This work |
| Expression plasmid | ||
| pGKK268 | pMMB66EH pva | This work |
Construction of V. cholerae Δpva-lacZ chromosomal fusion.
The V. cholerae Δpva plasmid pGKK264 was constructed by amplifying two DNA fragments from C6706 with primer pair HydB (5′-GATCGGGATCCATGCAGGGATCGCAATGGGC) and Hyd2 (5′-GATCGGCGGCCGCGTGTTTCATCTTCATCTCTG) and primer pair HydN (5′-GATCGGCGGCCGCAGCAGAATGATCAGGATATTG) and HydE (5′-GATCGGAATTCTTTCTAATGTCTAATCATGC) and ligating them into pKAS154 (23). The Δpva-lacZ fusion plasmid was constructed by inserting a promoterless lacZ gene between the two fragments in pGKK264, generating pGKK265. The resulting fusion was introduced into KSK262 and its ΔaphA derivative KSK1863 by allelic exchange.
Construction of V. cholerae chromosomal deletions.
The construction of the classical biotype ΔaphA plasmid pKAS98 was previously described (41). The El Tor biotype ΔaphA plasmid pGKK259 was constructed by amplifying two products from C6706 with primer pair YF13 (23) and YF29 (5′-GATCGGCGGCCGCGATAACGTGTGGTAATGACATG) and primer pair YF4 and YF6 (41). These fragments were ligated into pKAS154. The construction of the ΔluxO plasmid pGKK249 and the ΔhapR plasmid pKAS187 was previously described (23). The mutations were introduced into V. cholerae by allelic exchange.
Introduction of AphA binding site changes into the tcpPH promoter.
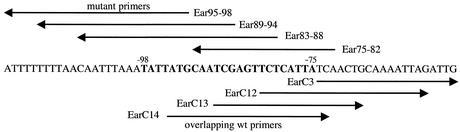
The AphA binding site mutations were constructed by amplifying fragments from the classical biotype by using overlapping primers containing the site for the type IIS restriction enzyme EarI (see Fig. 1). For each substitution (positions −75 to −98), fragments were generated by amplification with a specific mutant primer (Table 2) and I-Eco2 (20). These fragments were then ligated into pKAS32 (39) together with fragments generated by using overlapping wild-type primers and TP-Bam (41). The overlapping primer for mutants −75 to −82 was EarC3, for mutants −83 to −88 it was EarC12, for mutants −89 to −94 it was EarC13, and for mutants −95 to −98 it was EarC14. The mutations were introduced into the V. cholerae strain GK300 from the resulting plasmids (see Table 2) and were confirmed by sequencing.
FIG. 1.
Schematic showing the positions of the primers used to introduce the AphA binding site changes into the tcpPH promoter. The base pairs that were individually changed, from −75 to −98, are shown in bold. wt, wild type.
TABLE 2.
Primers used for mutational analysis of the AphA binding site in the tcpPH promotera
| Primer | Sequence | Plasmid |
|---|---|---|
| EAR75C | GATCGCTCTTCGTGAGAATGAGAACTCGATTG | pGKK208 |
| EAR76A | GATCGCTCTTCGTGATTATGAGAACTCGATTG | pGKK209 |
| EAR77G | GATCGCTCTTCGTGATACTGAGAACTCGATTG | pGKK211 |
| EAR78C | GATCGCTCTTCGTGATAAGGAGAACTCGATTG | pGKK212 |
| EAR79G | GATCGCTCTTCGTGATAATCAGAACTCGATTG | pKAS168 |
| EAR80G | GATCGCTCTTCGTGATAATGCGAACTCGATTG | pGKK237 |
| EAR81A | GATCGCTCTTCGTGATAATGATAACTCGATTG | pGKK238 |
| EAR82G | GATCGCTCTTCGTGATAATGAGCACTCGATTG | pGKK210 |
| EARC3 | GATCGCTCTTCGTCAACTGCAAAATTAGATTG | |
| EAR83A | GATCGCTCTTCGAGATCTCGATTGCATAATATTTAAATTG | pGKK243 |
| EAR84T | GATCGCTCTTCGAGAAATCGATTGCATAATATTTAAATTG | pGKK244 |
| EAR85C | GATCGCTCTTCGAGAACGCGATTGCATAATATTTAAATTG | pGKK245 |
| EAR86C | GATCGCTCTTCGAGAACTGGATTGCATAATATTTAAATTG | pGKK246 |
| EAR87G | GATCGCTCTTCGAGAACTCCATTGCATAATATTTAAATTG | pGKK247 |
| EAR88A | GATCGCTCTTCGAGAACTCGTTTGCATAATATTTAAATTG | pGKK248 |
| EARC12 | GATCGCTCTTCGTCTCATTATCAACTGCAAAATTAG | |
| EAR89C | GATCGCTCTTCGCGAGTGCATAATATTTAAATTGT | pWEL9 |
| EAR90C | GATCGCTCTTCGCGATGGCATAATATTTAAATTGTT | pWEL4 |
| EAR91G | GATCGCTCTTCGCGATTCCATAATATTTAAATTGTTA | pWEL5 |
| EAR92T | GATCGCTCTTCGCGATTGAATAATATTTAAATTGTTAA | pWEL6 |
| EAR93A | GATCGCTCTTCGCGATTGCTTAATATTTAAATTGTTAAA | pWEL8 |
| EAR94C | GATCGCTCTTCGCGATTGCAGAATATTTAAATTGTTAAAA | pWEL7 |
| EARC13 | GATCGCTCTTCGTCGAGTTCTCATTATCAACTG | |
| EAR95A | GATCGCTCTTCGCATTATATTTAAATTGTTAAAAAA | pWEL15 |
| EAR96A | GATCGCTCTTCGCATATTATTTAAATTGTTAAAAAAA | pWEL19 |
| EAR97C | GATCGCTCTTCGCATAAGATTTAAATTGTTAAAAAAAA | pWEL16 |
| EAR98A | GATCGCTCTTCGCATAATTTTTAAATTGTTAAAAAAAAT | pWEL17 |
| EaRC14 | GATCGCTCTTCGATGCAATCGAGTTCTCATTATC |
Substitution mutations are shown in bold.
Construction of the pva expression plasmid.
The pva expression plasmid pGKK268 was constructed by amplifying the pva coding region from C6706 with primers Hyd1 (5′-TAATCACTTTATTTCCATGG) and HydS (5′-GATCGGTCGACTATTAGTCACCGCAATATCC), digesting the resulting product with EcoRI and SalI, and ligating the resulting product into pMMB66EH (10).
Identification of the pva transcriptional start site.
Total RNA was isolated from C6706 after growth for 3.5 h under AKI conditions (14) by using RNA-WIZ (Ambion). The RNA was subjected to 5′ rapid amplification of cDNA ends (RACE) (Gibco BRL) as described previously (20), except that first-strand cDNA synthesis was carried out by using the pva-specific primer HydR1 (5′-CGTAGTTATCGAGAAAATAC), and the first and second nested primers were HydR2 (5′-GCCCAAGCACCAGCGGTCAG) and HydR3 (5′-AGGCTTTCCTGCCTTGGTTG).
Purification of AphA.
To obtain a purity of AphA greater than what was previously obtained (21), the IMPACT-CN protein fusion and purification system (New England Biolabs) was used. Amplification of the aphA gene from classical strain O395 was carried out with YF30 (GATCGCATATGTCATTACCACACGTTATC) and YF31 (GATCGGCTCTTCAGCATGCCATCGCGTTCAATTCTGC). After digestion of the fragment with NdeI and SapI, it was ligated into pTXB-1. The resulting plasmid, pWEL18, was introduced into Escherichia coli strain ER2566 and grown overnight in LB with ampicillin at 37°C. Cells were collected by centrifugation and were resuspended in column buffer (20 mM Tris [pH 8.0], 500 mM NaCl, 1 mM EDTA). The extract was sonicated and clarified by centrifugation at 14,000 rpm for 30 min in an SS34 rotor. The supernatant was then loaded onto a chitin column equilibrated with column buffer and was washed with 10 volumes of column buffer and high-salt column buffer (20 mM Tris [pH 8.0], 1 M NaCl, 1 mM EDTA). The column was then quickly washed with 3 volumes of cleavage buffer (column buffer containing 100 mM dithiothreitol [DTT]) and was left overnight at 16°C. AphA was then eluted off the column by using column buffer without DTT. The resulting protein was dialyzed overnight in a solution of 20 mM Tris (pH 7.5), 1 mM EDTA, 10 mM NaCl, and 0.1 mM DTT, glycerol was added to 10%, and it was frozen at −70°C. The final purity was estimated to be greater than 98%.
Gel shift experiments.
The 135-bp fragments from the tcpPH promoter were amplified by PCR from O395 or from the mutant plasmids listed in Table 2 by using TP-Eco5 (−175) (21) and TP-Bam5 (−40) (22). The 303- and 173-bp fragments from the pva promoter were amplified from either C6706 or O395, respectively, by using Hyd2 (+191) together with Hyd1 (−113) or with Hyd3 (+18) (5′-TGTGTTGTATTTGAATTCTC). The fragments were gel purified and 3′-end labeled with digoxigenin as previously described (21). Binding reactions with AphA were carried out as previously described (21).
DNaseI footprinting.
A 233-bp fragment was PCR amplified from C6706 with HydB3 (5′-GATCGGGATCCTAATCACTTTATTTCCATGG) (−113) and HydV4 (5′-GATCGGATATCCCTATAGTCCTAACGTGAGG) (+120) and was ligated into pBluescript (Stratagene), generating pWEL21. For upper-strand labeling the inserts were excised with BamHI and HindIII. For lower-strand labeling the inserts were excised with XbaI and EcoRV. The fragments were gel purified, treated with shrimp alkaline phosphatase, and kinased with [γ-33P]ATP (3,000 Ci/mmol; NEN). Singly end-labeled fragments were obtained by digestion with EcoRV (for the upper strand) and BamHI (for the lower strand). The conditions for AphA binding were as previously described (21, 22).
Antibody production.
AphA was purified as previously described (21) and was electrophoresed on a 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. AphA was excised from the gel and was electroeluted from the acrylamide as previously described (11). Antibodies were generated at the Pocono Rabbit Farms by using established protocols. Nonspecific antibodies to other V. cholerae proteins were removed by treatment of the antibody with an acetone powder (11) prepared from the O395 ΔaphA mutant strain KSK1683.
Immunoblot analysis.
Extracts were prepared from cultures grown in AKI conditions (14). The extracts were subjected to electrophoresis on a 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, transferred to nitrocellulose, probed with anti-AphA antibody, and visualized by using the ECL (enhanced chemiluminescence) detection system (Amersham).
PVA assays.
Cell extracts were prepared by growing the various strains in LB at 37°C for 7 h. The cells were collected by centrifugation and were resuspended in a 1/8 volume of buffer A (20 mM Na2HP04, 10 mM DTT, 1 mM EDTA [pH 7.0]). They were sonicated, spun in a microcentrifuge at 4°C for 30 min, and dialyzed against buffer A. Measurements of total protein were made by using the Bio-Rad Assay Reagent. Reaction mixtures contained 250 μg of protein in 0.1 M sodium citrate buffer, pH 5.8, with 2% potassium salt of penicillin V. Incubation was at 37°C for 30 min, and 6-APA was measured spectrophotometrically as previously described (18).
β-Galactosidase assays.
Assays with V. cholerae tcpP-lacZ fusions were carried out (28) after 3.5 h of shaking at 30°C and with pva-lacZ fusions after 3.5 or 7.5 h of growth under AKI conditions (14).
In vivo competition assays.
The infant mouse competition assays were performed essentially as described previously (43). Suckling CD-1 mice (3 to 5 days old; Charles River) were inoculated orally from cultures grown overnight in LB (pH 6.5) at 30°C, and the total CFU were obtained from the small intestine of four to six mice after 24 h by plating intestinal homogenates on streptomycin X-Gal plates.
RESULTS
Mutational analysis of the binding site for AphA at the tcpPH promoter.
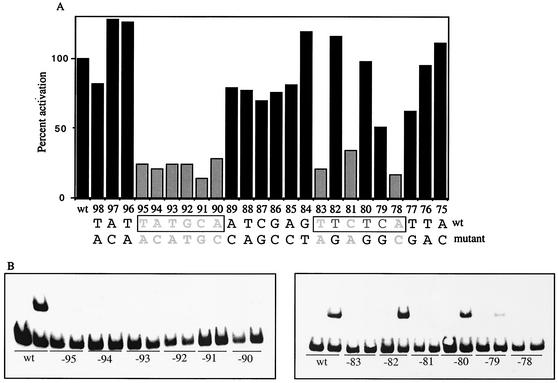
AphA cooperates with AphB to activate the transcription of tcpPH and to initiate the expression of the virulence cascade. To gain further insights into the mechanism by which this process occurs, the region of the tcpPH promoter previously shown to be footprinted by AphA (21) was systematically mutagenized to identify those base pairs important for AphA binding and transcriptional activation. Substitution mutations were individually introduced into each position from −98 to −75 by incorporating each into a primer with an EarI restriction site (see Fig. 1 and Table 2) and performing PCR together with wild-type primers. After ligating the products together to produce seamless junctions, the mutations were introduced into a classical biotype tcpP-lacZ fusion strain by allelic exchange. Out of the 24 positions that were analyzed, 9 were found to be critical for tcpPH expression in that they reduced expression of the promoter to less than 35% (Fig. 2A). In the upstream region, substitutions in a contiguous 6-bp stretch from position −95 to −90 reduced tcpPH expression to less than 30%. In the downstream region, only changes at positions −83, −81, and −78 showed a reduction to 34% or lower. Position −79 had an intermediate effect on transcription with a reduction of approximately 50%. The fact that the sequence comprising the base pairs that are critical for AphA to activate the expression of tcpPH (from −95 to −78) displays partial dyad symmetry (TATGCA-N6-TNCNNA) suggests that these base pairs serve as the binding site for AphA at the tcpPH promoter.
FIG. 2.
Mutational analysis of the AphA binding site at the tcpPH promoter. (A) V. cholerae strains were grown in LB (pH 6.5) at 30°C. From left to right: KSK618 (wild-type [wt] tcpP-lacZ); WL56 (−98A), WL55 (−97C), WL52 (−96A), WL57 (−95A), WL38 (−94C), WL39 (−93A), WL37 (−92T), WL36 (−91G), WL35 (−90C), WL40 (−89C), GK873 (−88A), GK872 (−87G), GK848 (−86C), GK847 (−85C), GK846 (−84T), GK845 (−83A), GK801 (−82G), GK835 (−81A), GK834 (−80G), KSK1309 (−79G), GK800 (−78C), GK799 (−77G), GK798 (−76A), and GK797 (−75C). Positions reducing expression of the promoter to 34% or lower are shaded in gray, and the corresponding bases are in gray. (B) AphA binding to various 135-bp mutant promoter fragments from −175 to −40. The first lane in each set has no protein added, and the second lane has 5 ng (15 nM) of AphA added.
To determine whether the base pair alterations that decreased tcpPH expression above were due to a reduction in the ability of AphA to bind to DNA, gel shift analysis was carried out with promoter fragments carrying the substitution mutations that comprise the region of dyad symmetry (−95 to −90 and −83 to −78). For these experiments, a preparation of AphA was utilized that was of greater purity than that which was previously achieved (21). This was accomplished by using an intein-mediated purification system (IMPACT) in which a self-cleaving intein and a chitin binding domain were fused to the carboxy-terminal end of AphA (see Materials and Methods). In the presence of DTT, AphA was released from the chitin-bound intein tag in highly purified form (greater than 98%). Comparison of the activity of AphA prepared by this method and that described previously by both gel shift and DNaseI footprinting (data not shown) indicated that their ability to bind to DNA was identical. As shown in Fig. 2B, chitin-purified AphA efficiently bound to a 135-bp tcpPH promoter fragment derived from a wild-type classical biotype strain but was unable to bind to similar fragments that contained one of the nine substitution mutations identified above that reduced transcription to less than 35%. In addition, the alteration at position −79, which decreased tcpPH expression to approximately 50%, also showed reduced AphA binding. However, the changes at positions −82 and −80, which did not significantly influence tcpPH expression, had no effect on AphA binding. These results demonstrate that there is a complete correlation between the influence of these various positions on the expression of tcpPH and their ability to support AphA binding. They also show that the sequence of the upstream AphA recognition region (from −95 to −90) is more stringent for AphA binding than that of the downstream region (−83 to −78).
Identification of an AphA binding site upstream of a gene encoding PVA.
Determination of the base pairs critical for AphA binding to the tcpPH promoter prompted us to investigate whether AphA might regulate other genes in V. cholerae by using a similar binding site. To address this, we searched the V. cholerae genome for the sequence TATGCA-N6-TNCNNA in both the forward and reverse orientations by using DNASTAR software and identified 11 additional sites on both the large and small chromosomes that matched this consensus. Of these sites, eight were located inside coding regions and only one of the remaining three was capable of efficiently binding AphA (see below). This site (TATGCA-N6-TACACA) was located on the small chromosome upstream of gene VCA0877 that showed homology to the cholylglycine hydrolase family of proteins. All of the base pairs shown above to be important for AphA binding to the site in the tcpPH promoter, including position −79, which was intermediate in phenotype, are completely conserved within this newly identified site (Fig. 3). In addition, the site displays better dyad symmetry than the one at tcpPH, suggesting that AphA might bind to it with even higher affinity.
FIG. 3.
Comparison of the AphA binding sites at the tcpPH and pva promoters. Conserved positions are shown in bold.
The cholylglycine family of hydrolases comprise both conjugated bile salt hydrolases and PVAs, since they share extensive sequence homology (5) and belong to the same group of N-terminal nucleophile hydrolases in which Cys-1 plays an essential role in the catalytic mechanism of the enzyme (42). VCA0877 is predicted to encode a 38-kDa protein with approximately 26% identity to the conjugated bile salt hydrolase from Clostridium perfringens (6) and 25% identity to PVA from Bacillus sphaericus (37). By employing assays for both of these activities we determined that the protein encoded by VCA0877 functions to hydrolyze penicillin V (see below), and we have designated the gene pva. Unlike the enzymes from C. perfringens and B. sphaericus, both of which are intracellular, V. cholerae PVA is predicted to have a 31-amino-acid signal sequence which directs it to the periplasm and which is cleaved to produce an N-terminal cysteine residue (34).
AphA represses the expression of the pva gene in response to cell density.
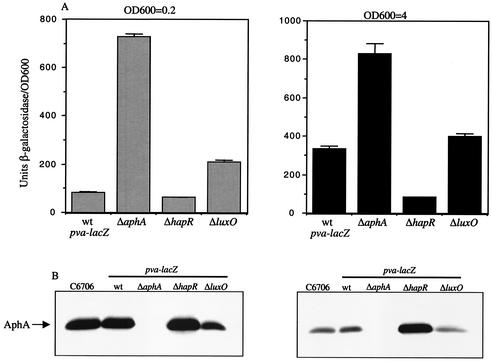
To assess whether AphA regulates the expression of the V. cholerae pva gene, a pva-lacZ fusion was constructed in the chromosome of C6706 (strain GK925). As shown in Fig. 4A, the presence of a ΔaphA mutation in GK925 increased expression from the pva-lacZ fusion almost ninefold compared to that of the wild-type fusion in AKI conditions at low cell density. At high cell density, due to the fact that the expression of the wild-type fusion itself increased approximately fourfold compared to that at low cell density, the influence of the ΔaphA mutation was somewhat less. These results showed that pva expression is regulated both by AphA and cell density.
FIG. 4.
Influence of ΔaphA, ΔhapR, and ΔluxO mutations on a C6706 pva-lacZ fusion at low and high cell density. Strains were grown in AKI conditions for 3.5 h (OD600 = 0.2) or for 7.5 h (OD600 = 4). (A) From left to right: GK925 (wild-type [wt] pva-lacZ); GK927 (ΔaphA); KSK1918 (ΔhapR); KSK1920 (ΔluxO). (B) Anti-AphA Western blot with strains grown as described for panel A. From left to right: C6706, GK925, GK927, KSK1918, and KSK1920.
Since HapR has previously been shown to directly influence the expression of aphA by binding to a recognition site in its promoter (23), it seemed likely that the influence of cell density on the expression of pva was the result of changes in the levels of AphA at different cell densities. To address this, Western blots were performed with anti-AphA antibodies by using extracts prepared from the strains shown in Fig. 4A, and the results are shown in Fig. 4B. At low cell density (optical density at 600 nm [OD600] = 0.2), the response regulator LuxO indirectly represses the expression of hapR. Since HapR levels are too low to strongly repress the expression of aphA under this condition, AphA levels are high in C6706 and in the wild-type pva-lacZ fusion strain GK925. This is consistent with the low level of pva expression observed in strain GK925 at this cell density (Fig. 4A). The presence of a ΔhapR mutation in the pva-lacZ fusion resulted in only a small increase in AphA levels and a small reduction in pva expression, since hapR is largely repressed under this condition. However, the presence of a ΔluxO mutation in the pva-lacZ fusion reduced the amount of AphA nearly in half due to increased expression of hapR, and coincident with this the expression of the pva-lacZ fusion doubled.
At high cell density (OD600 = 4) LuxO is no longer capable of indirectly repressing the expression of hapR, and HapR in turn represses the expression of aphA. As shown in Fig. 4B, AphA levels were considerably lower in both C6706 and the wild-type pva-lacZ fusion strain due to repression by HapR under this condition, consistent with the fourfold increase in pva expression observed relative to low cell density. A ΔhapR mutation significantly increased the levels of AphA protein in the pva-lacZ fusion strain and concomitantly decreased the expression of the fusion under this condition close to the levels observed at low cell density. In contrast, the ΔluxO mutation had only a small influence on AphA levels and pva-lacZ expression at high cell density, since hapR is not strongly repressed under this condition. These results show that pva expression is repressed by AphA and, as a result, is regulated in response to cell density.
The activity of PVA is increased in a ΔaphA mutant strain.
Since AphA represses the expression of the pva gene, it was of interest to determine whether the activity of PVA was also increased in V. cholerae strains lacking AphA. The enzyme PVA catalyzes the hydrolysis of penicillin V (phenoxymethlypenicillin) to 6-APA, and the resulting product can be measured spectrophotometrically by using a colorimetric assay (see Materials and Methods). Cell extracts from C6706 produced about 15 U of PVA activity/g when grown for 7 h at 37°C, and this value increased approximately sevenfold in a ΔaphA mutant, consistent with the increased transcription of pva observed in this background. C6706 containing a Δpva mutation, strain GK930, produced no detectable PVA activity under these conditions, but when the strain contained a plasmid, pGKK268, in which the V. cholerae pva gene was placed under the control of the tac promoter, PVA activity increased to 136 U/g in the absence of isopropyl-β-d-thiogalactopyranoside (IPTG) and to 1,883 U/g in the presence of 0.01 mM IPTG. Increasing the IPTG concentration to 0.1 mM increased the activity from pGKK268 even further (data not shown). These results clearly show that PVA activity correlates with the level of gene expression in the cell.
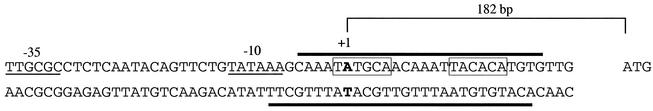
Identification of the transcriptional start of the pva promoter.
To determine the transcriptional start for the pva promoter, the technique of 5′ RACE (9) was carried out with wild-type C6706. A strong product corresponding to the size expected for the transcriptional start of pva was observed. Sequencing of this RACE product revealed that the start of the pva promoter is an A located 182 bp upstream of the ATG (Fig. 5). This positions the pva transcriptional start so as to lie within the AphA binding site identified above and is consistent with the role of AphA as a negative regulator at this promoter. The −10 sequence, TATAAA, has one mismatch from the consensus TATAAT, whereas the −35 sequence, TTGCGC, is less well conserved, with three mismatches from the consensus TTGACA.
FIG. 5.
Nucleotide sequence of the proximal region of the pva promoter. The transcriptional start site (+1), ATG codon, and −10 and −35 regions are shown. Boxes show the region of dyad symmetry involved in AphA recognition, and thick lines show the base pairs that are protected from DNaseI digestion by AphA.
AphA binds to and footprints the site in the pva promoter.
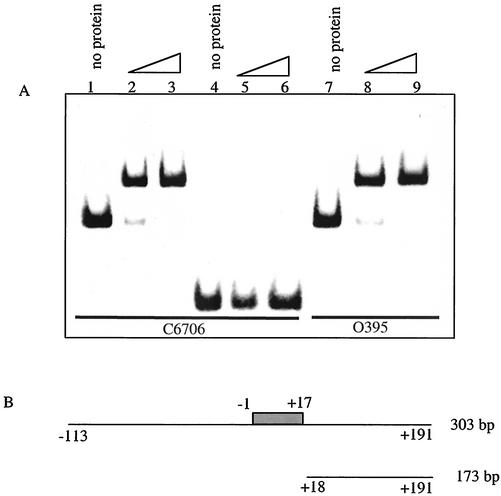
To assess whether AphA binds directly to the pva promoter, a 303-bp DNA fragment encompassing the region from −113 to +191 from the transcriptional start of the C6706 pva gene was incubated with purified AphA. AphA efficiently shifted the fragment, indicating that the protein is capable of binding directly to the pva promoter (Fig. 6, lanes 2 and 3). On a 173-bp fragment extending from +18 to +191 that lacks the AphA binding site, specific binding was lost (Fig. 6, lanes 5 and 6). When the 303-bp DNA fragment used above was amplified from the classical biotype strain O395 and incubated with purified AphA, a shift identical to that of C6706 was observed (Fig. 6, lanes 8 and 9). These results indicate that AphA binds to the pva promoter from strains representative of both biotypes.
FIG. 6.
A. Binding of purified AphA to the pva promoter. Lanes 1 to 3, a 303-bp fragment amplified from C6076; lanes 4 to 6, a 173-bp fragment amplified from C6706; lanes 7 to 9, the 303-bp fragment amplified from O395. The first lane in each set has no protein added, the second lane has 5 ng (15 nM) of AphA, and the third lane has 50 ng (150 nM) of AphA. (B) Diagrammatic representation of the binding fragments used for panel A. The gray box shows the AphA binding site.
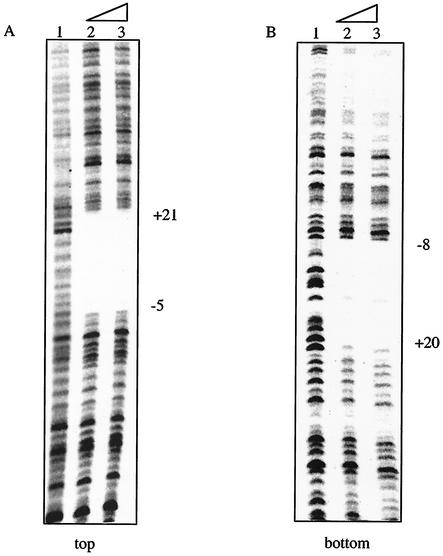
The ability of AphA to bind to the recognition sequence overlapping the pva transcriptional start was confirmed by DNaseI footprinting. As shown in Fig. 7A and B, on a 233-bp fragment extending from −113 to +120, the AphA footprint covers a region from approximately −5 to +21 on the top strand and from approximately −8 to +20 on the bottom strand. Thus, the footprint completely protects the identified AphA binding site (see Fig. 5) and places AphA in a position where it would likely interfere with transcription initiation.
FIG. 7.
DNaseI footprint for AphA at the pva promoter. (A) Top strand of a 233-bp fragment from −113 to +120. Lane 1, no protein; lane 2, 250 ng (2.5 μM) of AphA; lane 3, 300 ng (3 μM) of AphA. (B) Bottom strand of the 233-bp fragment. Lanes 1 to 3 are the same as those described for panel A.
The Δpva mutant does not influence virulence in V. cholerae.
Since AphA negatively regulates the pva gene while simultaneously activating tcpPH and the rest of the genes that comprise the virulence cascade, it might not be expected that pva would have a role in virulence, since its expression is opposite from that of the other virulence genes. To assess this, competition experiments in infant mice were carried out with the C6706 Δpva mutant GK930 and the Δlac wild-type strain KSK262. These experiments revealed no difference in the ability of these strains to colonize infant mouse intestines, suggesting that the product of the pva gene plays a role for V. cholerae in a different biological niche, most likely under conditions in which the ability of AphA to repress its promoter is reduced.
DISCUSSION
AphA was initially identified as a chromosomally encoded activator of the VPI tcpPH operon (41). Since the ability of AphA to activate the expression of tcpPH appears to have been acquired at a point in evolution subsequent to integration of the VPI element into the large chromosome, it seemed likely that AphA would have other regulatory roles in V. cholerae, possibly coordinating the expression of those genes together with or opposite from those with roles in virulence. We show here that by determining the base pairs critical for AphA binding and transcriptional activation at the tcpPH promoter, we were able to identify a second binding site for AphA on the small chromosome upstream of a gene encoding a PVA.
All of the base pairs in the tcpPH binding site that are important for AphA binding (from −95 to −90 and −83, −81, −79, and −78) are conserved within the site upstream of the pva gene. In fact, the pva binding site displays better dyad symmetry than that at tcpPH, suggesting that AphA may bind with even higher affinity to this site. A total of 11 other sites were found in the V. cholerae genome that contained the base pairs most important for AphA binding at tcpPH, and the majority of these sites were found to lie within coding regions. It is possible that AphA regulates other genes in V. cholerae by recognizing binding sites that deviate from those at tcpPH and pva, and this is presently under investigation. Although there are two base pair differences, positions −86 and −87, in the AphA binding site between the classical strain O395 and the El Tor strain C6706, these changes lie between the regions of dyad symmetry and do not influence AphA binding or transcriptional activation (20, 22). From the symmetry of the AphA recognition sites at tcpPH and pva, it appears that AphA is binding as a dimer. It is interesting that at tcpPH all of the contacts in the upstream half site (from −95 to −90) are essential for AphA binding and transcriptional activation, whereas in the downstream half site (−83 to −78) the most important positions are those which maintain the symmetry (−83, −81, and −78). Thus, the downstream half site appears to be less stringent for AphA binding than the upstream half site. A variety of data suggested that at tcpPH, AphB plays an active role in RNA polymerase recruitment, whereas AphA plays an accessory role, possibly by altering the conformation of the DNA to facilitate AphB binding (21). The finding that one of the normal roles for AphA in V. cholerae is as a negative regulator at pva further supports the notion that this protein functions through steric mechanisms as opposed to direct interactions with RNA polymerase itself.
The location of the AphA binding site overlapping the pva transcriptional start is consistent with its negative mode of function at this promoter. It seems likely that by binding to this site, AphA interferes with the ability of RNA polymerase to access the promoter. It is interesting that the family of phenolic acid decarboxylase regulators (PadR), of which AphA is a member, appears to exhibit a similar mode of repression (1). Inverted repeats which potentially serve as the target sites for PadR binding are located in the vicinity of the transcriptional start of the padA genes of Pediococcus pentosaceus, Lactobacillus plantarum, Bacillus subtilis, and Bacillus pumilus. Although V. cholerae appears to encode a phenolic acid decarboxylase (VC2240) that shows a high degree of homology to those of P. pentosaceus (48%) and L. plantarum (47%), we have not been able to show that AphA binds to the promoter of this gene or significantly influences its expression (data not shown). Thus, it does not appear that AphA regulates the expression of the phenolic acid decarboxylase gene in V. cholerae.
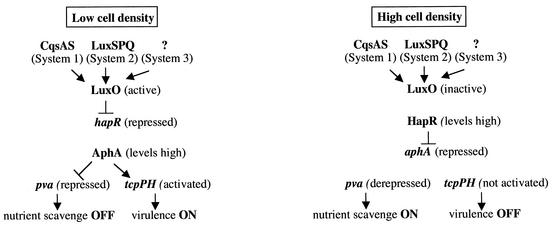
The expression of aphA is controlled by quorum sensing through the repressive action of HapR (23). By virtue of the ability of AphA to activate the expression of tcpPH, this places the rest of the virulence cascade also under quorum-sensing control (47). Likewise, the ability of AphA to repress pva expression also imparts quorum-sensing regulation on this gene. In fact, VCA0877 was previously identified in a microarray screen as a gene upregulated in a luxO mutant (47). Based on the work presented here it is now known that this is an indirect effect due to the repressive action of HapR on aphA expression. At low cell densities, the response regulator LuxO is phosphorylated by a relay from at least three quorum-sensing systems (30). According to the present model (Fig. 8 and references 23, 30, and 47), under this condition LuxO activates the expression of a putative repressor which decreases the expression of hapR, thereby preventing HapR from binding to the aphA promoter and repressing its expression. As confirmed here by Western analysis, AphA levels are high in wild-type C6706 under this condition, and the expression of the pva gene is maximally repressed. In contrast, at high cell densities LuxO is no longer able to activate the putative hapR repressor, and HapR in turn represses the expression of aphA. Under this condition AphA levels are reduced considerably and the expression of pva is increased. That pva expression increases even further in the ΔaphA mutant indicates that some repression by AphA still occurs even at high cell density. Whether there are other conditions that allow full derepression of pva expression is not yet known and is presently under investigation.
FIG. 8.
Contrasting pathways of gene expression regulated by AphA in response to cell density. At least three sensory circuits, CqsAS (System 1), LuxSPQ (System 2), and the unknown components of proposed System 3 function in parallel to control the activity of LuxO in response to the presence of autoinducers. At low cell density, activated LuxO indirectly represses the expression of hapR. The resulting high levels of AphA repress the expression of pva and, in the presence of AphB, activate tcpPH expression and the rest of the virulence cascade. At high cell density, inactive LuxO no longer represses hapR expression. HapR then reduces AphA levels by repressing expression from the aphA promoter. This derepresses pva expression while simultaneously preventing the activation of tcpPH and the rest of the virulence cascade.
AphA also regulates the expression of pva in the classical biotype strain O395. The DNA sequence of the pva gene and its promoter was found to be identical between O395 and C6706, and AphA shifted the pva promoter in a similar manner in both biotypes. In addition, an O395 pva-lacZ fusion was found to be repressed by AphA between 14- and 17-fold at both low and high cell densities (data not shown). Since O395 does not have a functional quorum-sensing system due to the presence of a hapR frameshift mutation and a point mutation in the aphA promoter that renders HapR incapable of binding (23), the levels of AphA do not change in response to cell density as in the C6706 strain background (data not shown). This finding confirms that the presence of a functional quorum sensing system is necessary for the derepression of pva expression that is observed in C6706 at high cell density.
Two different types of penicillin amidases, V and G, have been identified in a wide variety of microorganisms and are responsible for virtually all of the industrial production of 6-aminopenicillinic acid, the β-lactam precursor used for the synthesis of penicillins with a wide range of antimicrobial activities. These amidases have distinct substrate preferences, with PVA being active on phenoxyacetylated derivatives, such as penicillin V, and penicillin G amidase (PGA) being active on phenylacetylated derivatives, such as penicillin G. Although the two enzymes have no detectable sequence homology, common structural patterns have been identified between them by X-ray crystallography (42). PVA from B. sphaericus is a tetrameric enzyme consisting of four identical subunits with a monomer molecular size of 35 kDa (36). The PVA from V. cholerae shows 25% identity to the B. sphaericus enzyme and has a predicted monomer molecular size of 38 kDa. One obvious difference between the two proteins is that the V. cholerae enzyme has a 31-amino-acid leader sequence (34) which most likely directs it to the periplasm.
The activity of PGA from E. coli is controlled by a variety of mechanisms, such as catabolite repression, induction by phenylacetic acid, and thermoregulation (44). Although less information is available regarding the regulation of PVA enzymes, it has been reported for some organisms that expression is similarly repressed by sugars and is induced by phenoxyacetic acid (38). It is not yet known whether the V. cholerae pva gene is regulated by catabolite repression, but pva expression did not appear to be induced at low cell densities in AKI medium by addition of phenoxyacetic acid, and the enzyme was found to be active from cultures grown at both 30 and 37°C. In addition, this is the first report of penicillin amidase activity being regulated by quorum sensing. Thus, there appears to be considerable variation in the mechanisms that are involved in the regulation of penicillin amidases.
Although penicillin amidases are produced by a wide range of microorganisms, their physiological role is still somewhat unclear. The finding that these enzymes are capable of cleaving other phenyl and phenoxyacetic acid derivatives in addition to penicillins, coupled with the regulatory mechanisms that involve induction and repression by different carbon sources, suggests that penicillin amidases are involved in the assimilation of phenyl and phenoxyacetylated compounds in the environment (27, 44). These compounds, such as phenolic and caffeic acids and flavonoids, are abundant in the environment since they are produced by the degradation of plant cell material by microorganisms. Since E. coli PGA is active at 30°C or lower, it has been suggested that temperature serves as a cue for the expression of this gene in the environment where it could function in scavenging phenylacetylated compounds. Although the V. cholerae enzyme is active at both 30° and 37°C, a similar role for this enzyme might exist in the nonparasitic environment. The finding that AphA regulates the expression of pva opposite from those genes involved in virulence (see Fig. 8) supports this hypothesis.
Quorum sensing has been found to control a wide variety of different physiological activities in both gram-positive and gram-negative bacteria, such as bioluminescence, biofilm formation, virulence, sporulation, conjugation, and antibiotic production (29). Regulation of penicillin amidase activity can now be added to this growing list of activities controlled by quorum sensing. Regulating PVA expression by quorum sensing may allow V. cholerae to gain access to additional nutrient sources once the cells have achieved a certain cell density in a particular niche and the preferred substrates have been utilized. Although it is not yet known whether pva expression can be induced by other conditions in strains that have a hapR frameshift mutation or a point mutation in the aphA promoter that prevents HapR from repressing its expression, it appears that these strains might be at a disadvantage in certain environments relative to strains with a functional quorum-sensing system.
Acknowledgments
We thank James Coleman for helpful advice and Ronald Taylor for critical reading of the manuscript.
This work was supported by Public Health Service Grant AI-41558 to K.S.
REFERENCES
- 1.Barthelmebs, L., B. Lecomte, C. Divies, and J.-F. Cavin. 2000. Inducible metabolism of phenolic acids in Pediococcus pentosaceus is encoded by an autoregulated operon which involves a new class of negative transcriptional regulator. J. Bacteriol. 182:6724-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behari, J., L. Stagon, and S. B. Calderwood. 2001. pepA, a gene mediating pH regulation of virulence genes in Vibrio cholerae. J. Bacteriol. 183:178-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll, P. A., K. T. Tashima, M. B. Rogers, V. J. DiRita, and S. B. Calderwood. 1997. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol. Microbiol. 25:1099-1111. [DOI] [PubMed] [Google Scholar]
- 4.Champion, G. A., M. N. Neely, M. A. Brennan, and V. J. DiRita. 1997. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol. Microbiol. 23:323-331. [DOI] [PubMed] [Google Scholar]
- 5.Christiaens, H., R. J. Leer, P. H. Pouwels, and W. Verstraete. 1992. Cloning and expression of a conjugated bile acid hydrolase gene from Lactobacillus plantarum by using a direct plate assay. Appl. Environ. Microbiol. 58:3792-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman, J. P., and L. L. Hudson. 1995. Cloning and characterization of a conjugated bile acid hydrolase gene from Clostridium perfringens. Appl. Environ. Microbiol. 61:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford, J. A., J. B. Kaper, and V. J. DiRita. 1998. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol. Microbiol. 29:235-246. [DOI] [PubMed] [Google Scholar]
- 8.DiRita, V. J., C. Parsot, G. Jander, and J. J. Mekalanos. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:5403-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fürste, J. P., W. Pansegrau, R. Frank, H. Blöcker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 11.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Häse, C. C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 95:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hulbert, R. R., and R. K. Taylor. 2002. Mechanism of ToxT-dependent transcriptional activation of the Vibrio cholerae tcpA promoter. J. Bacteriol. 184:5533-5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwanaga, M., K. Yamamoto, N. Higa, Y. Ichinose, N. Nakasone, and M. Tanabe. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 30:1075-1083. [DOI] [PubMed] [Google Scholar]
- 15.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karaolis, D. K. R., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 95:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karaolis, D. K. R., S. Somara, D. R. Maneval, Jr., J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 18.Kornfeld, J. M. 1978. A new colorimetric method for the determination of 6-aminopenicillanic acid. Anal. Biochem. 86:118-126. [DOI] [PubMed] [Google Scholar]
- 19.Kovacikova, G., and K. Skorupski. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J. Bacteriol. 181:4250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovacikova, G., and K. Skorupski. 2000. Differential activation of the tcpPH promoter by AphB determines biotype specificity of virulence gene expression in Vibrio cholerae. J. Bacteriol. 182:3228-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovacikova, G., and K. Skorupski. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol. Microbiol. 41:393-407. [DOI] [PubMed] [Google Scholar]
- 22.Kovacikova, G., and K. Skorupski. 2002. Binding site requirements of the virulence gene regulator AphB: differential affinities for the Vibrio cholerae classical and El Tor tcpPH promoters. Mol. Microbiol. 44:533-547. [DOI] [PubMed] [Google Scholar]
- 23.Kovacikova, G., and K. Skorupski. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 46:1135-1147. [DOI] [PubMed] [Google Scholar]
- 24.Krukonis, E. S., R. R. Yu, and V. J. DiRita. 2000. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol. Microbiol. 38:67-84. [DOI] [PubMed] [Google Scholar]
- 25.Li, C. C., J. A. Crawford, V. J. DiRita, and J. B. Kaper. 2000. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol. Microbiol. 35:189-203. [DOI] [PubMed] [Google Scholar]
- 26.Li, C. C., D. S. Merrell, A. Camilli, and J. B. Kaper. 2002. ToxR interferes with CRP-dependent transcriptional activation of ompT in Vibrio cholerae. Mol. Microbiol. 43:1577-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merino, E., P. Balbás, F. Recillas, B. Becerril, F. Valle, and F. Bolivar. 1992. Carbon regulation and the role in nature of the Escherichia coli penicillin acylase (pac) gene. Mol. Microbiol. 6:2175-2182. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 30.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 31.Miller, V. L., V. J. DiRita, and J. J. Mekalanos. 1989. Identification of toxS, a regulatory gene whose product enhances ToxR-mediated activation of the cholera toxin promoter. J. Bacteriol. 171:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, V. L., R. K. Taylor, and J. J. Mekalanos. 1987. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell 48:271-279. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 35.Nye, M. B., J. D. Pfau, K. Skorupski, and R. K. Taylor. 2000. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J. Bacteriol. 182:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsson, A., T. Hagström, B. Nilsson, M. Uhlén, and S. Gatenbeck. 1985. Molecular cloning of Bacillus sphaericus penicillin V amidase gene and its expression in Escherichia coli and Bacillus subtilis. Appl. Environ. Microbiol. 49:1084-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsson, A., and M. Uhlén. 1986. Sequencing and heterologous expression of the gene encoding penicillin V amidase from Bacillus sphaericus. Gene 45:175-181. [DOI] [PubMed] [Google Scholar]
- 38.Shewale, J. G., and H. SivaRaman. 1989. Penicillin acylase: enzyme production and its application in the manufacture of 6-APA. Proc. Biochem. 24:146-154. [Google Scholar]
- 39.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 40.Skorupski, K., and R. K. Taylor. 1997. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 94:265-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skorupski, K., and R. K. Taylor. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 31:763-771. [DOI] [PubMed] [Google Scholar]
- 42.Suresh, C. G., A. V. Pundle, H. SivaRaman, K. N. Rao, J. A. Brannigan, C. E. McVey, C. S. Verma, Z. Dauter, E. J. Dodson, and G. G. Dodson. 1999. Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members. Nat. Struct. Biol. 6:414-416. [DOI] [PubMed] [Google Scholar]
- 43.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valle, F., P. Balbás, E. Merino, and F. Bolivar. 1991. The role of penicillin amidases in nature and in industry. Trends Biochem. Sci. 16:36-40. [DOI] [PubMed] [Google Scholar]
- 45.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 46.Yu, R. R., and V. J. DiRita. 2002. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol. Microbiol. 43:119-134. [DOI] [PubMed] [Google Scholar]
- 47.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]