Abstract
Nonsense-mediated mRNA decay (NMD) is a surveillance mechanism that degrades mRNAs carrying premature translation termination codons. Generally, NMD is elicited if translation terminates >50–54 nucleotides (nt) upstream of an exon–exon junction. We have previously reported that human β-globin mRNAs carrying 5′-proximal nonsense mutations (e.g., β15) accumulate to normal levels, suggesting an exception to the “50–54-nt boundary rule.” In the present report, we demonstrate that the strength of the UPF1-dependent NMD of mutant β-globin mRNAs is specifically determined by the proximity of the nonsense codon to the initiation AUG. This conclusion is supported by a parallel effect of the short ORF size on NMD of nonsense-containing α-globin mRNAs. To determine whether the short-ORF effect on NMD response is conserved in heterologous transcripts, we assessed its effects on a set of β-globin/triosephosphate isomerase (TPI) hybrid mRNAs and on the TPI mRNA. Our data support the conclusion that nonsense mutations resulting in a short ORF are able to circumvent the full activity of the canonical UPF1-dependent NMD pathway.
Keywords: nonsense-mediated mRNA decay (NMD), UPF1-dependent NMD, 50–54-nt boundary rule, short open reading frame (ORF), translation reinitiation
INTRODUCTION
Post-transcriptional quality control of eukaryotic gene expression is critical to proper cell function. Nonsense-mediated mRNA decay (NMD) is an example of a post-transcriptional mechanism used to survey mRNA structure. By recognizing and degrading abnormal transcripts that prematurely terminate translation, NMD prevents the production of truncated proteins that could have a dominant-negative effect on the cell. NMD may also function in post-transcriptional control of selected physiologic transcripts with NMD features, such as mRNAs with upstream open reading frames (uORFs) and mRNAs containing an intron within the 3′-untranslated region (3′-UTR) (Mendell et al. 2004; Wittmann et al. 2006).
NMD targeting depends on the interaction of signals generated by transcript splicing and mRNA translation. This combination of determinants allows the translating ribosome to “recognize” where the termination codon is positioned relative to exon–exon junctions (Maquat 2004, 2005; Lejeune and Maquat 2005). In mammalian cells, an exon-junction protein complex (EJC) is deposited on the transcript during nuclear transcript splicing at a position 20–24 nucleotides (nt) upstream of each exon–exon junction (Le Hir et al. 2001). According to present models, translating ribosomes displace EJCs from the ORF during the “pioneer” round of translation (Ishigaki et al. 2001; Lejeune et al. 2002). If an mRNA contains a premature termination codon located >50–54 nt upstream of the last exon–exon junction, the ribosome will not extend far enough into the mRNA to displace all the EJCs, and one or more will be retained on the mRNA and trigger NMD. The NMD-linked destabilization mechanism appears to involve the recruitment and phosphorylation of UPF1 and its interaction both with the EJC and with the translation termination release factors eRF1 and eRF3 (Le Hir et al. 2000; Fribourg et al. 2003; Gehring et al. 2003; Lau et al. 2003; Kashima et al. 2006; Kunz et al. 2006). Thus, the UPF1 protein provides an essential link in the NMD pathway (Kashima et al. 2006). Consistent with this model, inhibition of UPF1 expression in a cell inhibits the NMD response (Sun et al. 1998; Mendell et al. 2002).
According to the literature cited above, nonsense codons elicit NMD if they are followed by at least one exon–exon junction located >50–54 nt downstream (Thermann et al. 1998; Zhang et al. 1998). Nevertheless, some exceptions to the “50–54-nt boundary rule” have been reported. These exceptions may each reflect different mechanisms that blunt or circumvent NMD. One exception has been reported in the context of the triosephosphate isomerase (TPI) transcript. Zhang and Maquat (1997) have shown that TPI transcripts nonsense mutated at codon 1, 2, or 10 circumvent NMD and that this NMD resistance results from translation reinitiation at codon 14Met (Zhang and Maquat 1997). In fact, it was proven that in these nonsense transcripts, translation reinitiation occurs, downstream from the nonsense codon, with high efficiency at 14Met (Zhang and Maquat 1997; Poyry et al. 2004). More recently, Stockklausner et al. (2006) have studied NMD for the human thrombopoietin (TPO) mRNA. This mRNA contains seven uORFs, all terminating >50 nt upstream of the last exon–exon junction (Ghilardi and Skoda 1999), and yet seems to avoid NMD. While these authors demonstrate that elongation of the 3′-most uORF from 27 to 40 codons decreases mRNA steady-state levels, this decrease did not appear to reflect the canonical human UPF1-dependent NMD pathway, and the underlying mechanism remains undefined (Stockklausner et al. 2006).
Studies from our laboratory indicate that human β-globin mRNAs containing nonsense mutations in the 5′ region of exon 1 accumulate to levels similar to those of normal β-globin transcripts (Romão et al. 2000). This resistance to NMD is erythroid independent and independent of promoter identity, and does not reflect translation reinitiation, abnormal RNA splicing, or impaired translation (Inácio et al. 2004). Instead, the observed NMD resistance appears to reflect the close proximity of the nonsense codon to an upstream determinant (Inácio et al. 2004). In this study, we demonstrate that the critical parameter for NMD inhibition in this setting is the proximity of the nonsense codon to the translation initiation codon. Our data further indicate that this AUG-proximity effect appears to be an attribute of the canonical UPF1-dependent NMD response that is independent of the transcript identity and sequence context.
RESULTS
A critical proximity of the nonsense codon to the translation initiation codon inhibits the NMD response
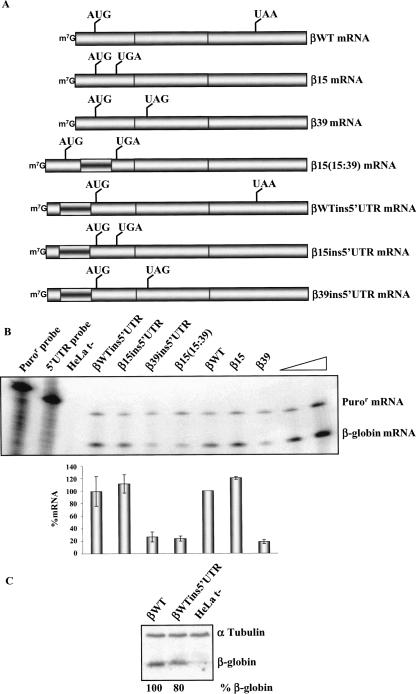
We have previously reported that human β-globin mRNAs carrying nonsense mutations near the initiation codon have a markedly blunted NMD response and are expressed at levels approaching those of the wild-type β-globin mRNA. Since these nonsense codons are situated well 5′ to the exon junctions of the β-globin transcript, they constitute an exception to the “50–54-nt boundary rule.” Thus, β-globin mRNAs carrying the nonsense mutation at codon 15 (β15) escape NMD, while transcripts carrying a nonsense mutation at codon 39 (β39) are effectively degraded via the NMD pathway (Romão et al. 2000). We have also shown that insertion of a 72-base-pair (bp) fragment between the AUG and the β15 nonsense mutation, which moves the nonsense codon to position 39, renders the mRNA NMD sensitive (Inácio et al. 2004). In this study, our first aim was to determine whether the blunting of NMD by this 72-bp sequence insertion reflected an alteration in the proximity of the nonsense codon to the initiation AUG or to a 5′-UTR determinant such as the cap. This was addressed by inserting the same 72-bp fragment into the 5′-UTR (at 21 bp upstream of the AUG) of the βWT, β15, and β39 genes and measuring the impact of each insertion on mRNA expression. This set of insertions created the βWTins5′UTR, β15ins5′UTR, and β39ins5′UTR genes, respectively (Fig. 1A). Each gene was transfected into HeLa cells, and the encoded mRNAs were quantified 20 h post-transfection (see Materials and Methods). Each expression level was normalized to the level of Puror mRNA encoded from the puromycin-resistance gene cloned into the β-globin expression vector. Results from three independent experiments revealed that the nonsense-containing β15ins5′UTR gene was expressed to the same level as the βWTins5′UTR gene and that the 72-bp insertion within the 5′-UTR had no adverse effect on the expression of either mRNA (Fig. 1B). As expected, expression of the NMD-sensitive β39 mRNA was markedly repressed, and the 5′-UTR insertion (β39ins5′UTR construct) (Fig. 1) had no effect on the β39 gene expression level. These results illustrate that insertion of a 72-bp spacer within the 5′-UTR fails to alter the expression of the NMD-competent and NMD-resistant mRNAs. In addition, since mammalian NMD is dependent on mRNA translation, and the 5′-UTR insertion failed to stabilize the β39 mRNA, we also conclude that the stability of the β15ins5′UTR mRNA did not reflect inhibition of translation by the 72-nt insertion. This conclusion was further validated assessing the translation of the βWTins5′UTR mRNA (βWT) (Fig. 1C). The translation efficacy of the βWTins5′UTR mRNA was 80% of the normal control (Fig. 1C). Therefore, the 72-bp spacer does not appear to result in a major alteration in translation and does not impair NMD. Thus, the 5′-UTR 72-bp insertion fails to destabilize the β15 mRNA, while insertion of the same sequence between the β15 nonsense mutation and the AUG, increasing the ORF from 15 to 39 codons [β15(15:39) mRNA], markedly decreases mRNA accumulation to a level comparable to that of β39 mRNA (24% and 18% of βWT, respectively) (Fig. 1B; Inácio et al. 2004). These results substantiate the role for AUG-proximity in the NMD mechanism.
FIGURE 1.
The proximity of the nonsense codon to the AUG, rather than the distance to a putative 5′-UTR determinant, inhibits mRNA decay. (A) Schematic representation of the studied human β-globin mRNAs. The position of initiation and termination (native or premature) codons is represented. The inverted-shadowed box represents the spacer insertion. The name of each transcript is indicated on the right. (B) Representative RPA of RNA isolated from HeLa cells untransfected (t−) or transfected with constructs specified above each lane. Increasing amounts of RNA (indicated by a triangle) from HeLa cells transfected with the βWT gene were also analyzed to demonstrate that the experimental RPA was carried out in probe excess; the amount of each of the input riboprobes used per analysis is also shown. The positions of the human β-globin or puromycin-resistance (Puror) mRNAs are indicated on the right of the autoradiograph. Levels of human β-globin mRNA were normalized relatively to Puror and then compared to wild type (WT). Average values from five independent experiments and standard deviations are plotted below the autoradiograph. (C) Western blot analysis of the HeLa cells extracts untransfected (t−) or transfected with the constructs specified above each lane. Immunoblotting was performed using a human β-globin-specific antibody and a α-tubulin-specific antibody to control for variations in protein loading.
Increasing the ORF length of the β-globin transcripts from 15 to 39 codons activates UPF1-dependent NMD
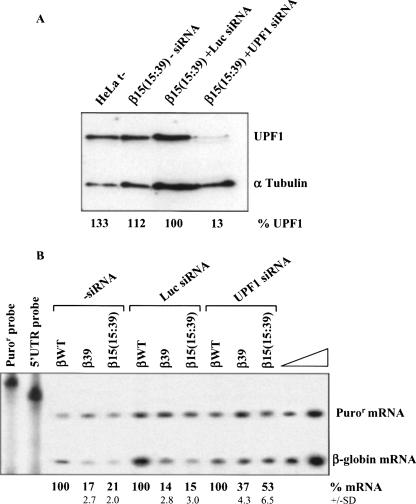
We next sought to demonstrate that the destabilization observed by increasing the ORF length of the β-globin transcripts from 15 to 39 codons was due to activation of the UPF1-dependent decay pathway. We analyzed the steady-state levels of β-globin nonsense-mutated mRNAs in HeLa cells depleted of human UPF1 protein by RNA interference (RNAi). UPF1 short interfering (si)RNA, or nonspecific control (Luciferase; Luc) siRNA, was cotransfected with each of three β-globin genes: βWT, β39, and β15(15:39). Two days after transfection, a Western blot analysis showed a decrease of 87% in UPF1 protein expression induced by siRNA [Fig. 2A, lane β15(15:39)+UPF1 siRNA versus lane β15(15:39)+Luc siRNA]. At this level of UPF1 down-regulation, mRNA was quantified by RPA, relatively to βWT. In untreated cells, the β15(15:39) mRNA accumulated to about the same level as β39 mRNA (21% and 17% of βWT mRNA, respectively), consistent with both mRNAs being targeted by NMD (Inácio et al. 2004). The same expression levels for β39 and β15(15:39) were measured in cells treated with the control siRNA (Luc siRNA; 14% and 15% of the βWT mRNA, respectively) (Fig. 2B). In contrast, depletion of UPF1 resulted in a 3.5-fold increase in the abundance of β15(15:39) mRNA and a 2.6-fold increase in β39 mRNA (Fig. 2B). These data confirm that the increase of β15(15:39) mRNA expression reflects the UPF1 depletion on the NMD pathway. These data further support the role of the short ORF (i.e., the AUG-proximity effect) in the inhibition of the UPF1-dependent NMD mechanism.
FIGURE 2.
Down-regulating human UPF1 protein results in an up-regulation of the β15(15:39) transcripts. HeLa cells were transiently cotransfected with synthetic small-interfering RNA (siRNA) duplexes directed to human UPF1 or to a non-endogenous target (Luciferase; Luc) used as control, and plasmids with the β-globin gene variants [normal (βWT), β39, and β15(15:39); plasmid also contains the puromycin resistance (Puror) gene]; (t−) untransfected cells. About 24 h after siRNA treatment, cells were transfected with the β-globin constructs specified above each lane. Two days after transfection, protein and RNA were isolated. (A) Western blot analysis of the HeLa cell extracts transfected with human UPF1 siRNA. Immunoblotting was performed using a human UPF1-specific antibody and an α-tubulin-specific antibody to control for variations in protein loading. (B) Representative RPA to quantify the level of human β-globin transcript, normalized to the level of puromycin-resistance (Puror) mRNA. Increasing amounts of RNA (indicated by a triangle) from HeLa cells transfected with βWT gene were also analyzed to demonstrate that the experimental RPA was carried out in probe excess; the amount of each of the input riboprobes used per analysis is also shown. Levels of β39 and β15(15:39) transcripts were compared to βWT mRNA levels (defined as 100%) in the absence (−siRNA) or in the presence of each siRNA. The mRNA percentage average values and standard deviations from three independent experiments are indicated at the bottom.
The effect of a short ORF on NMD is also observed in human α-globin transcripts
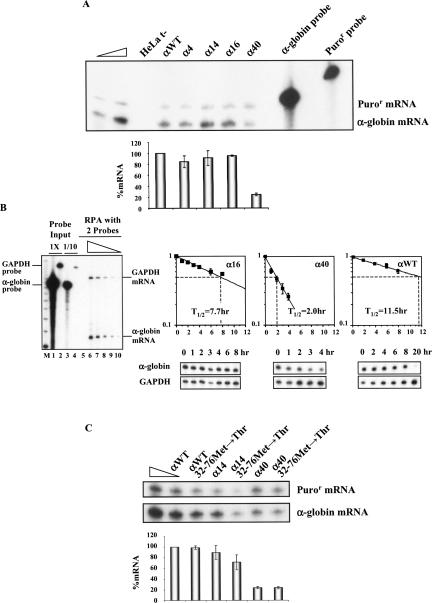
To determine the generality of our findings, we next tested the relationship between NMD and the ORF size in additional mRNAs. We first asked whether α-globin mRNAs show parallel NMD profiles to that seen for β-globin mRNA. Nonsense mutations at codons 4 (CCT→TAG), 14 (TGG→TAG), 16 (AAG→TAA), or 40 (AAG→TAG) (α4, α14, α16, or α40 genes, respectively) were introduced into the full-length α-globin gene cloned in the mammalian expression vector pTRE-αWT (see Materials and Methods). HeLa cells were then transfected with each plasmid, and the level of the encoded mRNA was determined and normalized (see Materials and Methods). The results reveal that α-globin mRNAs carrying nonsense mutations at codons 4, 14, or 16 accumulate to steady-state levels comparable to wild type (85%, 92%, and 95% of αWT, respectively) (Fig. 3A). In contrast, expression of the α40 mRNA was repressed to 25% of αWT levels, compatible with a full NMD response (Pereira et al. 2006). Thus, as was the case with the β-globin mRNA, the α-globin mRNA expression levels corresponded to the AUG-proximity of the nonsense codons.
FIGURE 3.
The AUG-proximity effect on NMD is observed in human α-globin transcripts. (A) HeLa cells were transfected with the α-globin constructs specified above each lane [the plasmid also contains the puromycin-resistance (Puror) gene]. Total RNA from either transfected or untransfected (t−) HeLa cells was isolated and analyzed by RPA. Levels of human α-globin mRNA were normalized relatively to the expression level of the Puror mRNA and compared with expression of the normal (αWT) gene. Increasing amounts of RNA (indicated by a triangle) from HeLa cells transfected with an αWT gene were also analyzed to demonstrate that the experimental RPA was carried out in probe excess; the amount of each of the input riboprobes used per analysis is also shown. The percentage mRNA values were plotted for each construct, and standard deviations from four independent experiments are shown. (B) MEL cells stably expressing the tet transactivator (MEL/tTA cells) were transiently transfected with αWT, α16, or α40 genes under the transcriptional control of a tTA-regulated promoter. Cells were pulsed with the respective α-globin mRNAs for 4 h by transfer to Tet(−) medium and then transferred back to Tet(+) medium for analysis of decay rates. Total mRNA was isolated at various time intervals during the transcriptional chase period and analyzed by RPA. On the left, a representative autoradiograph of a titration RPA is presented to show that experimental RPAs were carried out in probe excess and in the linear range of detection. The triangle above lanes 6–10 represents decreasing amounts (2×, 1×, 1/2, 1/4, and 1/8) of αWT RNA. Lanes 1 and 2 show the amount of each input α-globin or murine GAPDH probe used per RNA sample. Lanes 3 and 4 show 1/10 of each input probe. (M) Molecular weight marker. On the right, representative autoradiographs are presented to show the decay rate of each α-globin variant. RNase protection bands corresponding to human α-globin mRNA and to the constitutively expressed mouse GAPDH mRNA (loading control) are indicated. The intensities of the α-globin mRNA bands were quantified and normalized relatively to the GAPDH mRNA band. The data were recorded in the graphs presented. To calculate the α-globin mRNA half-lives, each data time point was expressed as a ratio of α-globin:GAPDH mRNA and normalized to the average value of all time points from a single transfection. The ratios were then renormalized to the average initial time point from all transfections (time 0 = 1). Each point represents the mean ± standard deviation from four independent experiments. Linear regression analysis was performed by standard techniques. The half-lives (T 1/2) of the mRNAs are indicated. (C) The two potential reinitiating AUGs at codons 32 and 76 were converted to ACG triplets in each of the index mRNAs. A representative RPA of RNA isolated from HeLa cells transiently transfected with αWT, α14, α40, αWT–32–76Met→Thr, α14–32–76Met→Thr, or α40–32–76Met→Thr genes is shown. Identification of the α-globin and Puror protected fragments is indicated to the right of the autoradiograph. Levels of α-globin mRNA were quantified relatively to the Puror mRNA, and these values are plotted beneath each respective lane (average and standard deviations) normalized to the expression level of the αWT gene. Increasing amounts of RNA (indicated by a triangle) from HeLa cells transfected with an αWT gene were also analyzed to demonstrate that the experimental RPA was carried out in probe excess. For each case, three independent experiments were performed.
To confirm that the stability of the α-globin mRNAs with AUG-proximal nonsense mutations is maintained, we measured the absolute half-life of α16 mRNA and compared it to the αWT mRNA and to the NMD-sensitive α40 mRNA. Each of the three genes was cloned behind a tetracycline (tet) controlled promoter and transfected into a MEL cell line that stably expresses the tet-transcriptional transactivator (MEL/tTA cells). The cells were then transcriptionally pulsed for 4 h with each α-globin mRNA (see Materials and Methods) (Kong et al. 2003), and the rate of α-globin mRNA decay was measured over time (Fig. 3B). Data from four independent experiments show that the half-life of normal α-globin mRNA is 11.5 h. In contrast, the half-life of the α40 is 2.0 h, consistent with a full sensitivity to NMD, as previously reported (Pereira et al. 2006). The half-life of α16 mRNA (7.7 h) is intermediate to the αWT and α40 mRNAs (Fig. 3B). The observation that the α16 mRNA had a half-life almost fourfold higher than the α40 mRNA confirms that the AUG-proximal nonsense-mutated α-globin mRNAs carrying a short ORF escape the full impact of NMD.
Translation reinitiation downstream from the nonsense codon can blunt the NMD effect (Zhang and Maquat 1997). If the NMD resistance observed on the α-globin mRNAs with 5′-proximal nonsense mutations reflected translation reinitiation, then removing initiation codons 3′ to the nonsense mutation should restore NMD and destabilize the mRNA. To test this effect in the context of the α-globin mRNA, the AUG codons at positions 32 and 76 (in frame) were mutated to ACG (threonine) within the αWT, α14, and α40 genes. The 32Met and 76Met are located at 201 nt and 69 nt upstream of the last exon–exon junction, respectively. An additional out-of-frame AUG codon at position 24/25 was not tested because the potential reinitiation at this site would not extend far enough to displace the last EJC (the 24/25Met is in frame with a stop codon at position 48/49) and NMD would thus be triggered, as previously described (Pereira et al. 2006). The 32Met and 76Met AUG-to-ACG conversions created αWT–32–76Met→Thr, α14–32–76Met→Thr and α40–32–76Met→Thr genes. Expression of each of these genes was analyzed in HeLa cells (Fig. 3C). The α40–32–76Met→Thr mRNA was expressed at the same level as α40 mRNA (23% of αWT), consistent with both being fully committed to NMD. On the other hand, α14–32–76Met→Thr transcripts accumulate at ∼71% of αWT, and at ∼80% of α14 mRNA. Of note, expression of the α14 mRNA with the double mutation remains well above that of the α40 mRNA. Thus, we find that blocking potential translation reinitiation sites results in a minimal decrease in α14 mRNA expression. These results, in combination with our prior studies (Inácio et al. 2004), show that for both α-globin and β-globin mRNAs, the presence of the nonsense codon in close proximity to the AUG markedly reduces the effect of NMD on the mutant mRNA. In both cases, the effect of the short ORF appears to be the major determinant for NMD inhibition.
The effect of a short ORF on NMD is conserved in heterologous transcripts
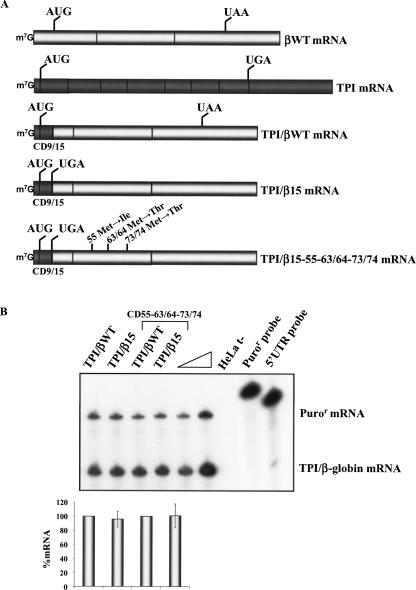
A well-documented exception to the “50–54-nt boundary rule” is represented by TPI mRNAs with AUG-proximal nonsense mutations at codons 1, 2, or 10 (Zhang and Maquat 1997). The NMD resistance in these cases has been shown to reflect translation reinitiation; these nonsense codons elicit NMD when translation reinitiation is inhibited at codon 14 (Zhang and Maquat 1997). Therefore, early nonsense codons on different transcripts and in different sequence contexts may manifest NMD resistance based on different mechanisms. To further explore this issue, we investigated the involvement of the AUG-proximity effect in a set of nonsense-mutated TPI/β-globin and β-globin/TPI hybrid genes (Figs. 4, 5). These genes were transiently transfected into HeLa cells, and the levels of the encoded mRNAs were quantified and normalized relative to the Puror mRNA and to the expression level of the corresponding nonsense-free genes (Figs. 4, 5).
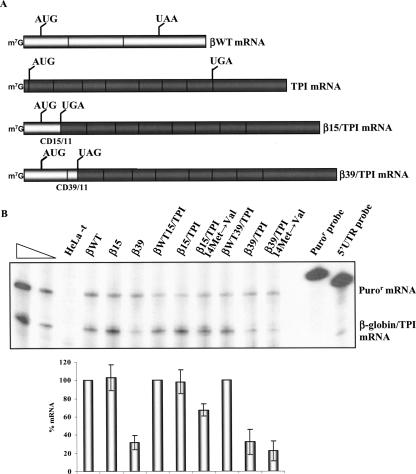
FIGURE 4.
Human β15 globin mRNA carrying the first nine-codon TPI ORF is resistant to NMD. (A) Physical maps of the hybrid TPI/β-globin constructs. The bright rectangles represent β-globin exons, and dark rectangles represent TPI exons. CD9/15 represents the position of the junction between the TPI and β-globin genes. The position of initiation and termination (premature or native) codons is indicated. The name of each transcript is indicated to the right. (B) HeLa cells were transfected with the constructs specified above each lane [the plasmid also contains the puromycin-resistance (Puror) gene]. Total RNA was isolated and analyzed by RPA. The levels of each nonsense-mutated TPI/β-globin mRNA were quantified relatively to the Puror mRNA and normalized to the expression level of the corresponding nonsense-free gene. Increasing amounts of RNA (indicated by a triangle) from HeLa cells transfected with a TPI/βWT gene were also analyzed to demonstrate that the experimental RPA was carried out in probe excess; the amount of each of the input riboprobes used per analysis is also shown. The percentage mRNA values were plotted for each construct, and standard deviations from three independent experiments are shown.
FIGURE 5.
Human TPI mRNA carrying the first 15 codons of the β15 globin ORF escapes NMD. (A) Physical maps of the hybrid β-globin/TPI mRNAs. The bright rectangles depict β-globin exons, and dark rectangles depict TPI exons. CD15/11 and CD39/11 indicate the junction position between the β-globin and TPI genes. The vertical lines represent the initiation and termination (native or premature) codons. The identification of each mRNA is indicated to the right. (B) HeLa cells were transfected with the constructs specified above each lane [the plasmid also contains the puromycin-resistance (Puror) gene]. Total RNA was isolated and analyzed by RPA. Levels of each nonsense-mutated β-globin/TPI mRNA were quantified relative to the Puror mRNA and normalized to the expression level of the corresponding nonsense-free gene. Increasing amounts of RNA (indicated by a triangle) from HeLa cells transfected with a βWT gene were also analyzed to demonstrate that the experimental RPA was carried out in probe excess; the amount of each of the input riboprobes used per analysis is also shown. The percentage mRNA values were plotted for each construct, and standard deviations from three independent experiments are shown.
The first chimeric mRNA to be assessed contains a fusion between the 5′ terminus of the TPI mRNA up to codon 9 with the β15 mRNA beginning with the nonsense UGA at codon 15 (Fig. 4A). This TPI/β15 mRNA is expressed at a level comparable to the control TPI/βWT chimeric mRNA that lacks the nonsense mutation. This comparison demonstrates that the TPI/β15 mRNA is fully resistant to NMD. To investigate whether this resistance is due to translation reinitiation, all potential reinitiating AUGs that could inhibit NMD were inactivated in the TPI/β15 and TPI/βWT genes (Fig. 4A). These AUG codons occur at positions 55, 63/64, and 73/74 (located at 149, 124, and 94 nt upstream of the last exon–exon junction, respectively). The normalized expression level of the TPI/β15–55–63/64–73/74 mRNA was similar to the control TPI/βWT–55–63/64–73/74 mRNA (Fig. 4B). Thus, elimination of all the potential translation reinitiation sites failed to destabilize the TPI/β15 mRNA. These data demonstrate that the AUG-proximity effect is sufficient to fully inhibit NMD in the context of a heterologous ORF sequence.
We next altered the context of the nonsense codon in a reciprocal manner by constructing hybrid β-globin/TPI genes in which the 5′ terminus of the β15 or β39 globin mRNAs up to and including the respective nonsense codons or the corresponding normal codons were fused to the TPI mRNA beginning at codon 11 (Fig. 5A). These genes were transfected into HeLa cells, and the impact of the β15 and β39 nonsense codons in the chimeric sequence context was assessed. The β39/TPI mRNA accumulates at 32% of the corresponding wild-type βWT39/TPI mRNA control (Fig. 5B). This repressed level of expression is identical to that of the β39 mRNA (Fig. 5B), showing that the β39/TPI mRNA is fully committed to NMD. Introducing a missense mutation at TPI codon 14 of the β39/TPI gene (β39/TPI–14Met→Val construct) to block potential translation reinitiation did not alter the accumulation of the β39/TPI mRNA (Fig. 5B). In contrast, the mRNA generated by shortening the ORF length from 39 to 15 codons in the same chimeric mRNA context (β15/TPI mRNA) is expressed at levels equivalent to those of the βWT15/TPI mRNA (Fig. 5B). These results indicate that NMD of β15/TPI mRNA is fully inhibited. Based on the results published by Zhang and Maquat (1997), we next investigated if the observed β15/TPI mRNA NMD resistance could reflect translation reinitiation at codon 14 of TPI mRNA, as this has been previously shown to be an effective reinitiation site (Zhang and Maquat 1997; Poyry et al. 2004). A Met→Val substitution at codon 14 of the TPI was introduced into the β15/TPI gene. It must be noted that this amino acid substitution leaves intact the TPI sequence (three codons) between the nonsense codon and 14Met, which parallels the condition of the TPI10Ter–14Met→Val construct studied by Zhang and Maquat (1997). Expression in HeLa cells showed that β15/TPI–14Met→Val mRNA accumulates at ∼ 67% of the βWT15/TPI mRNA level (Fig. 5B). Thus, if translation reinitiation is abrogated at codon 14 of TPI, the β15/TPI–14Met→Val mRNA accumulation is partially repressed. This result is consistent with the analysis of the α14 transcript (Fig. 3C). Taken together, these results illustrate that while translation reinitiation can play a role in NMD inhibition of β15/TPI transcripts, this is only partially responsible for the resistance of this mRNA to NMD because the β15/TPI–14Met→Val mRNA does not reach low levels comparable to those of the controls for full NMD response: β39, β39/TPI, and β39TPI–14Met→Val mRNAs (Fig. 5B). Therefore, while NMD inhibition can be partially attributed to translation reinitiation, as previously shown by Zhang and Maquat (1997), our data demonstrate that the presence of a short ORF is independently sufficient to circumvent NMD. Although we cannot completely rule out that translation reinitiation may occur at another downstream AUG, namely, at codon 82, which is the next putative reinitiation codon that would inhibit NMD, this hypothesis seems unlikely based on results described by Zhang and Maquat (1997).
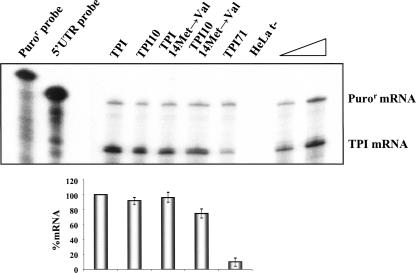
The above-mentioned conclusions were further tested in the context of the native TPI transcript (Fig. 6). Our results show that TPI mRNA carrying a nonsense mutation at codon 71 (TPI71 mRNA) is expressed at ∼10% of normal, demonstrating that it is fully committed to NMD. However, if the nonsense codon is moved to an AUG-proximal location at codon 10, the corresponding mRNA (TPI10 mRNA) accumulates at ∼92% of normal TPI. This NMD resistance is consistent with prior results (Zhang and Maquat 1997). By analyzing the mRNA TPI sequence, it is observed that downstream from codon 10 of the TPI mRNA, there are several putative reinitiation AUG codons. While the AUG at codons 14, 82, and 190/191 (located at 586, 382, and 56 nt upstream of the last exon–exon junction, respectively) would allow the ribosome to extend far enough to displace all downstream EJCs and block NMD, the studies of Zhang and Maquat (1997) indicate that there are no effective reinitiation sites downstream from the 14Met codon. Based on these data, we studied the effect of blocking translation reinitiation at codon 14 on the TPI10 mRNA (TPI10–14Met→Val construct) (Fig. 6). Results show that expression of the TPI10–14Met→Val mRNA is decreased to 75% of wild-type TPI mRNA, while normal TPI transcript levels are unaffected by the same missense mutation (TPI–14Met→Val mRNA). Thus, inhibition of translation reinitiation at codon 14 fails to fully commit TPI10 mRNA to NMD, as the AUG-proximity effect is acting in parallel. In this respect, our results differ from a previous analysis of TPI NMD by Zhang and Maquat (1997). These authors reported a decrease in the expression level of nonsense TPI10, from 82% to 45% of normal, when translation reinitiation at codon 14 is blocked. Although puzzling, this discrepancy may reflect differences in the experimental settings: variations in NMD proficiency within the two cellular systems (HeLa cells versus mouse L cells) or deviations inherent to the different quantification methodologies, for example, in this study, the control for both transfection efficiency and RNA loading was an independent transcriptional unit, present in the plasmid carrying the test gene, whereas Zhang and Maquat (1997) used a cotransfected transgene.
FIGURE 6.
TPI transcripts bearing a short ORF escape NMD. HeLa cells were transfected with the constructs specified above each lane [the plasmid also contains the puromycin-resistance (Puror) gene]. Total RNA was isolated and analyzed by RPA. Levels of each TPI mRNA were quantified relative to the Puror mRNA and normalized to the expression level of the wild-type TPI mRNA. Increasing amounts of RNA (indicated by a triangle) from HeLa cells transfected with a normal TPI gene were also analyzed to demonstrate that the experimental RPA was carried out in probe excess; the amount of each of the input riboprobes used per analysis is also shown. The percentage mRNA values relative to normal were plotted for each construct, and standard deviations from three independent experiments are shown.
DISCUSSION
Nonsense codons that are located >50–54 nt 5′ to an exon–exon junction generally elicit NMD. We have previously reported that human β-globin mRNAs bearing nonsense mutations in the 5′ region of exon 1 accumulate to levels similar to those of wild-type β-globin mRNA (Romão et al. 2000). Functional analyses of these mRNAs with 5′-proximal nonsense mutations demonstrated that their resistance to NMD does not reflect translation reinitiation. Moreover, NMD inhibition is independent of promoter identity and erythroid specificity, and it is not a result of abnormal RNA splicing or impaired translation. In fact, analysis of the polysome profile of AUG-proximal nonsense mRNAs revealed their association with 1–1.5 ribosomes (Inácio et al. 2004). These studies demonstrated that the AUG-proximity of the nonsense codon may comprise the basis of the NMD resistance, overriding the “50–54-nt boundary rule” in establishing the overall efficiency of NMD. In this study, we confirm in an independent sequence context that the length of a translated ORF modulates NMD efficiency and extend our conclusions by demonstrating that the NMD inhibition is, in fact, ruled by the proximity of the nonsense codon to the AUG, rather than the distance to a putative 5′-UTR determinant. The presence of a short ORF on a given mRNA inhibits the canonical UPF1-dependent NMD, and this inhibition is independent of the transcript sequence context. While our results show that AUG proximity is a major determinant of the NMD effect, the data also indicate that it operates in parallel with other modifying influences. Specifically, translation reinitiation 3′ to the shortened ORF may contribute to alleviation of the NMD effect. These two parameters can independently contribute to the net overall NMD sensitivity of a nonsense-containing mRNA.
Knowing that NMD is translation dependent, the present data may suggest that the observed NMD resistance of mRNAs with a short ORF resulting from a nonsense codon could reflect ineffective translation. In this context, it is interesting to note that the smallest naturally occurring eukaryotic ORF that produces significant levels of protein product comprises 24 codons (Yu and Warner 2001). We have described a set of human β-globin gene nonsense mutations at codons 5, 15, and 17 that are not committed to NMD (Romão et al. 2000). On the other hand, a nonsense codon at position 21/22, resulting from a β-thalassemic frameshift mutation, effectively elicits NMD (Zhang et al. 1998). Therefore, our results and those published by Zhang et al. (1998) suggest that for human β-globin mRNAs, the boundary between the 3′-most mutation that fails to trigger full NMD and the 5′-most mutation able to fully commit mRNA to the NMD pathway maps between codons 17 and 21/22 (Zhang et al. 1998; Romão et al. 2000). These data, when combined with data in the present report, allow us to conclude that mRNAs carrying nonsense mutations that create ORFs shorter than 18–20 codons may be NMD resistant, independent of the sequence background. The apparent concordance between the minimal length of an ORF for effective protein synthesis and the minimal length for effective NMD suggests that the cell may not require a quality-control mechanism to clear inefficiently translated mRNAs that encode relatively short peptide fragments.
It has been shown that nonsense surveillance contributes to the regulation of mRNAs containing uORF(s) (Mendell et al. 2004; Whittmann et al. 2006). However, it should be noted that, in general, the analyzed transcripts that were proposed to be naturally NMD-regulated seem to contain at least one uORF >20 codons. Thus, based on our results, it is possible to hypothesize that only those transcripts with at least one uORF containing >20 codons are NMD targets, and those naturally occurring with the uORF(s) smaller than ∼20 codons could be NMD resistant.
The mechanisms of the observed NMD resistance of mRNAs with AUG-proximal nonsense codons and corresponding short ORFs may relate to critical aspects of the translational machinery. The short ORF may result in a temporal overlap between translational initiation and termination. The ribosome may arrive at the premature termination (nonsense) codon before it has had time to discharge its initiation factors and/or stabilize its interactions with elongation/termination factors. This might result in interference with elongation/termination reactions that may be central to the NMD pathway. Present evidence specifically identifies functional linkages between the translation termination process and the NMD pathway. Studies by others have revealed that during the process of translation of short upstream ORFs, interactions involving ribosomal or ribosome-associated factors, including, but not limited to releasing factors, can inhibit translation elongation as well as termination (Morris and Geballe 2000; Janzen et al. 2002). In the case of NMD, UPF1 must interact with eRF1 and eRF3 to terminate translation and, in the proper setting, to interact with additional factors to trigger NMD (Gaba et al. 2005; Kashima et al. 2006). Therefore, it is possible that the dynamics of the mRNP remodeling necessary for NMD cannot occur, or is defective, in this compressed environment of the mRNAs carrying a short ORF. This study illustrates that NMD may be modulated by the way a short ORF is translated.
MATERIALS AND METHODS
Construction of expression vectors
The wild-type β-globin gene (βWT), as well as the previously described human β-globin variants β15, β39, and β15(15:39) (Inácio et al. 2004), were subcloned into the ClaI/BspLU11I sites of the pTRE2pur vector (BD Biosciences) by PCR amplification of the 1806-bp ClaI/BspLU11I fragment, using primers with linkers for ClaI and BspLU11I (primers #1 and #2; Table 1). The βWTins5′UTR, β15ins5′UTR, and β39ins5′UTR constructs were made by inserting the HindIII/ApaI fragment from β15(15:39) into the HindIII/ApaI restriction sites, which were previously created at 21 bp upstream of the ATG codon of βWT, β15, and β39 constructs, using the QuikChange Site-Directed Mutagenesis Kit (Stratagene), with mutagenic primers #3 and #4 (Table 1), as indicated by the manufacturer.
TABLE 1.
DNA oligonucleotides used in the cloning strategies
The 1677-bp EcoRI/RcaI fragment containing the whole wild-type α2-globin gene was obtained from plasmid pTet-αWT (Kong et al. 2003) and subcloned into EcoRI/BspLU11I sites of the pTRE2pur vector, originating the pTRE-αWT plasmid. Constructs α4, α14, α16, and α40, carrying nonsense mutations at codon positions 4 (CCT→TAG), 14 (TGG→TAG), 16 (AAG→TAA), and 40 (AAG→TAG), respectively, were created by site-directed mutagenesis, with mutagenic primers #5–#12 (Table 1), using the plasmid pTRE-αWT as DNA template. Potential reinitiation sites in α-globin codons 32 and 76 were sequentially mutated in cis by site-directed mutagenesis with primers #13–#16 (Table 1), using pTRE-αWT, α14, or α40 as template, creating the constructs αWT–32–76Met→Thr, α14–32–76Met→Thr, and α40–32–76Met→Thr.
The human TPI gene sequence from 39 bp upstream of the initiation codon to 720 bp downstream from the stop codon was amplified by PCR, from plasmid pMT-TPI (Zhang and Maquat 1997), using primers with linkers for ClaI and BspLU11I (primers #17 and #18; Table 1), and inserted into pTRE2pur ClaI/BspLU11I sites. This construct was used as the parental vector for the TPI10, TPI10–14Met→Val, TPI–14Met→Val and TPI71 mutants, which were originated by site-directed mutagenesis as previously described, using primers #19–#24 (Table 1).
The hybrid genes βWT15/TPI, β15/TPI, βWT39/TPI, and β39/TPI were created by subcloning β-globin fragments amplified from the corresponding pTRE2purβ-globin variants, using specific primers with ClaI and EcoRV linkers (primers #25–#29; Table 1). These fragments were inserted into the ClaI/EcoRV sites of the pTRE2purTPI construct (the EcoRV site was previously inserted into this original construct between TPI codons 10 and 11 by site-directed mutagenesis with primers #30 and #31) (Table 1). Then, this EcoRV site was deleted from the finished constructs using the same procedure with primers #32–#39 (Table 1). In addition, β15/TPI as well as β39/TPI constructs were used to introduce the mis-sense mutation (ATG→GTT) at codon 14 of TPI by site-directed mutagenesis using specific primers #21 and #22 (Table 1), originating the constructs β15/TPI–14Met→Val and β39/TPI–14Met→Val.
The hybrid genes TPI/βWT and TPI/β15 were created by overlapping PCR using primers #2, #17, and #40–#47 (Table 1) to fuse the TPI 5′-UTR and the first 10 codons containing fragments of the β-globin gene sequence from codon 16 to the 3′-UTR. The putative reinitiation codons at codons 55 (ATG→ATA), 63/64 (ATG→ACG), and 73/74 (ATG→ACG) of these hybrid genes were mutated as previously described in Inácio et al. (2004) originating TPI/βWT–55–63/64–73/74 and TPI/β15–55–63/64–73/74 constructs.
Cell culture and transfection
HeLa cells, stably expressing the tet transactivator (HeLa/tTA) (described by Kong et al. 2003), were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Transient transfections were performed using Lipofectamine 2000 Transfection Reagent (Invitrogen), following the manufacturer's instructions, in 35-mm plates using 250 ng of the test construct DNA and 1750 ng of pEGFP vector (BD Biosciences) DNA as a control for transfection efficiency. Cells were harvested after a 20-h transcription pulse.
Mouse erythroleukemia (MEL) cells stably expressing the tet transactivator (MEL/tTA) (Kong et al. 2003) were used for conditional expression of human α-globin genes (previously cloned into the pTRE2pur vector). For transient transfections, MEL/tTA cells were split 1 d before transfection and cultured in minimal essential medium (MEM) supplemented with 10% fetal bovine serum, and 100 ng/mL tetracycline. Cells were transfected with 2 μg of pTRE-αWT, or each variant, along with 18 μg of carrier DNA, as previously described. After an overnight recovery in Tet+ medium, cells were split into 60-mm-diameter dishes, and pulsed with α-globin mRNA for 4 h by growth in Tet− media. Following this period, transcription from the plasmid was blocked by the addition of Tet to the media. Cells from each culture dish were harvested at different time points for further analysis.
Transient transfection of siRNA
Transient transfections of siRNAs were carried out using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions in 35-mm plates using 100 pmol of siRNA oligonucleotides and 4 μL of transfection reagent. Twenty-four hours later, cells were transfected again with 50 pmol of siRNAs, 250 ng of the test construct DNA, and 1000 ng of pEGFP vector. Twenty-four hours later, cells were harvested for analysis of RNA and protein expression. The siRNA oligonucleotides used for transfections [Luciferase (AA-CGUACGCGGAAUACUUCGA) and hUPF1 (AA-GAUGCAGUUCCGCUCCAUU)] were purchased as annealed, ready-to-use duplexes from Dharmacon.
RNA isolation
Total RNA from transfected cells was prepared using the RNeasy minikit (QIAGEN) following the manufacturer's indications. RNA samples were treated with RNase-free DNase I (Ambion) and purified by phenol:chloroform extraction. Before further analyses, mRNA samples were tested by RT-PCR to reject the hypothesis of activation of cryptic splicing pathway(s), with consequent alteration in mRNA sequence and possible circumvention of the premature termination codon. From all transcript species a single full-length product was amplified (data not shown), demonstrating that the studied nonsense transcripts present a normal splicing pattern.
Ribonuclease protection assays (RPA)
The probes used were generated by in vitro transcription, using a Maxiscript T7 kit (Ambion), under conditions recommended by the manufacturer. The 5′-UTR probe is the 180-bp StuI/HindIII fragment of pTRE2pur inserted into HindIII/EcoRV sites of pcDNA3 (Invitrogen) that protects the 150 nt of the 5′ part of the 5′-UTR common to all genes cloned into the pTRE2pur ClaI site. The α-globin probe is a 174-bp fragment encompassing the 3′ part of exon 3 inserted into the polylinker region of pTRI-amp-18 (Ambion). The Puror probe is a 280-bp puromycin-resistance gene fragment cloned into pGEM-3 (Promega) and protects a 197-nt sequence of the puromycin-resistance mRNA. The α-globin probe for half-life analysis is a 244-nt fragment protecting the 132-nt exon 1. The murine GAPDH probe protects a 316-nt fragment (Ambion) (Kong et al. 2003). Samples were processed as described in Inácio et al. (2004).
Western blot analysis
Protein lysates were resolved, according to standard protocols, in 10% or 14% SDS-PAGE for hUPF1, or β-globin detection, respectively, and transferred to PVDF membranes (Bio-Rad). Membranes were probed using mouse monoclonal anti-α-tubulin (Roche) at 1:10000 dilution and goat polyclonal anti-hUPF1 (Bethyl Labs) at 1:500 dilution or rabbit polyclonal anti-hβ-globin (Cappel) at 1:1000 dilution. Detection was carried out using secondary peroxidase-conjugated anti-mouse IgG (Bio-Rad), anti-rabbit IgG (Bio-Rad), or anti-goat IgG (Sigma) antibodies followed by chemiluminescence.
ACKNOWLEDGMENTS
We thank Lynne Maquat for a plasmid carrying the human TPI gene. This work was partially supported by Fundação para a Ciência e a Tecnologia (POCTI/MGI/48296/2002, POCTI/SAU-MMO/57573/2004 and Programa Plurianual/CIGMH) and Programa Operacional de Saúde–Saúde XXI. A.L.S., F.J.C.P., A.M., and R.M. were supported by fellowships from Fundação para a Ciência e a Tecnologia. Funding was also made available from NIH grant R37-HL 65449–MERIT (to S.A.L.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.201406.
REFERENCES
- Fribourg, S., Gatfield, D., Izaurralde, E., Conti, E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat. Struct. Biol. 2003;10:433–439. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- Gaba, A., Jacobson, A., Sachs, M.S. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell. 2005;20:449–460. doi: 10.1016/j.molcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Gehring, N.H., Neu-Yilik, G., Schell, T., Hentze, M.W., Kulozik, A.E. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- Ghilardi, N., Skoda, R.C. A single-base deletion in the thrombopoietin (TPO) gene causes familial essential thrombocythemia through a mechanism of more efficient translation of TPO mRNA. Blood. 1999;94:1480–1482. [PubMed] [Google Scholar]
- Inácio, A., Silva, A.L., Pinto, J., Ji, X., Morgado, A., Almeida, F., Faustino, P., Lavinha, J., Liebhaber, S.A., Romão, L. Nonsense mutations in close proximity to the initiation codon fail to trigger full nonsense-mediated mRNA decay. J. Biol. Chem. 2004;279:32170–32180. doi: 10.1074/jbc.M405024200. [DOI] [PubMed] [Google Scholar]
- Ishigaki, Y., Li, X., Serin, G., Maquat, L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- Janzen, D.M., Frolova, L., Geballe, A.P. Inhibition of translation termination mediated by an interaction of eukaryotic release factor1 with a nascent peptidyl-tRNA. Mol. Cell. Biol. 2002;22:8562–8570. doi: 10.1128/MCB.22.24.8562-8570.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima, I., Yamashita, A., Izumi, N., Kataoka, N., Morishita, R., Hoshino, S., Ohno, M., Dreyfuss, G., Ohno, S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes & Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, J., Ji, X., Liebhaber, S.A. The KH-domain protein αCP has a direct role in mRNA stabilization independent of its cognate binding site. Mol. Cell. Biol. 2003;23:1125–1134. doi: 10.1128/MCB.23.4.1125-1134.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz, J.B., Neu-Yilik, G., Hentze, M.W., Kulozik, A.E., Gehring, N.H. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA. 2006;12:1–8. doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, C.K., Diem, M.D., Dreyfuss, G., Van Duyne, G.D. Structure of the Y14-Magoh core of the exon junction complex. Curr. Biol. 2003;13:933–941. doi: 10.1016/s0960-9822(03)00328-2. [DOI] [PubMed] [Google Scholar]
- Le Hir, H., Izaurralde, E., Maquat, L.E., Moore, M.J.2000The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions EMBO J. 196860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Gatfield, D., Izaurralde, E., Moore, M.J. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune, F., Maquat, L.E. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lejeune, F., Ishigaki, Y., Li, X., Maquat, L.E. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat, L.E. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. Nonsense-mediated mRNA decay in mammals. J. Cell Sci. 2005;118:419–422. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- Mendell, J.T., ap Rhys, C.M., Dietz, H.C. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- Mendell, J.T., Sharifi, N.A., Meyers, J.L., Martinez-Murillo, F., Dietz, H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Morris, D.R., Geballe, P. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, F.J.C., Silva, M.C., Picanço, I., Seixas, M.T., Ferrão, A., Faustino, P., Romão, L. Human α2-globin nonsense-mediated mRNA decay induced by a novel α-thalassaemia frameshift mutation at codon 22. Br. J. Haematol. 2006;133:98–102. doi: 10.1111/j.1365-2141.2006.05971.x. [DOI] [PubMed] [Google Scholar]
- Poyry, T.A., Kaminski, A., Jackson, R.J. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes & Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romão, L., Inácio, A., Santos, S., Ávila, M., Faustino, P., Pacheco, P., Lavinha, J. Nonsense mutations in the human β-globin gene lead to unexpected levels of cytoplasmic mRNA accumulation. Blood. 2000;96:2895–2901. [PubMed] [Google Scholar]
- Stockklausner, C., Breit, S., Neu-Yilik, G., Echner, N., Hentze, M.W., Kulozik, A.E., Gehring, N.H. The uORF-containing thrombopoietin mRNA escapes nonsense-mediated decay (NMD) Nucleic Acids Res. 2006;34:2355–2363. doi: 10.1093/nar/gkl277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Perlick, H.A., Dietz, H.C., Maquat, L.E. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl. Acad. Sci. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann, R., Neu-Yilik, G., Deters, A., Frede, U., Wehr, K., Hagemeier, C., Hentze, M.W., Kulozik, A.E. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann, J., Hol, E.M., Jack, H.-M. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X., Warner, J.R. Expression of a micro-protein. J. Biol. Chem. 2001;276:33821–33825. doi: 10.1074/jbc.M103772200. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Maquat, L.E. Evidence that translation re-initiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J. 1997;16:826–833. doi: 10.1093/emboj/16.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Sun, X., Qian, Y., Maquat, L.E. Intron function in the nonsense-mediated decay of β-globin mRNA: Indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]