Abstract
The HAMP linker, a predicted structural element observed in many sensor kinases and methyl-accepting chemotaxis proteins, transmits signals between sensory input modules and output modules. HAMP linkers are located immediately inside the cytoplasmic membrane and are predicted to form two short amphipathic α-helices (AS-1 and AS-2) joined by an unstructured connector. HAMP linkers are found in the Escherichia coli nitrate- and nitrite-responsive sensor kinases NarX and NarQ (which respond to ligand by increasing kinase activity) and the sensor kinase CpxA (which responds to ligand by decreasing kinase activity). We constructed a series of hybrids with fusion points throughout the HAMP linker, in which the sensory modules of NarX or NarQ are fused to the transmitter modules of NarX, NarQ, or CpxA. A hybrid of the NarX sensor module and the CpxA HAMP linker and transmitter module (NarX-CpxA-1) responded to nitrate by decreasing kinase activity, whereas a hybrid in which the HAMP linker of NarX was replaced by that of CpxA (NarX-CpxA-NarX-1) responded to nitrate by increasing kinase activity. However, sequence variations between HAMP linkers do not allow free exchange of HAMP linkers or their components. Certain deletions in the NarX HAMP linker resulted in characteristic abnormal responses to ligand; similar deletions in the NarQ and NarX-CpxA-1 HAMP linkers resulted in responses to ligand generally similar to those seen in NarX. We conclude that the structure and action of the HAMP linker are conserved and that the HAMP linker transmits a signal to the output domain that ligand is bound.
Two-component regulatory systems are used by all domains of life to sense and respond to environmental changes (48). Many of the sensor components of two-component regulatory systems and most methyl-accepting chemotaxis proteins (MCPs) share a common molecular architecture. These proteins are dimers in the cytoplasmic membrane and have a modular structure. The sensory module of each monomer consists of a short, amino-terminal cytoplasmic tail, a transmembrane α-helix (TM-1), a periplasmic domain which may interact with a specific ligand, a second transmembrane α-helix (TM-2), and, in many but not all sensor proteins, a HAMP linker. The cytoplasmic output module interacts with a response regulator (14). Signal transduction requires that information about ligand binding in the exterior sensory input module of the protein cross the cytoplasmic membrane and regulate the activity of the output module in the interior of the cell. The output modules of MCPs interact with an associated soluble histidine kinase, CheA, whereas those of the two-component sensors are histidine kinases in equilibrium between kinase and phosphatase activities.
Many of these sensors appear to use a common structural element for signal transduction between external inputs and cytoplasmic outputs. The HAMP (histidine kinase, adenylyl cyclase, MCP, and phosphatase) linker or P-type linker is predicted to be a structure of approximately 50 aminoacyl residues connecting TM-2 and the cytoplasmic output domain (4, 53) (Fig. 1). Amino acid sequences characteristic of HAMP linkers are found in all MCPs and 15 of 30 known sensor kinases from Escherichia coli. There is little primary sequence identity among predicted HAMP linkers, but all have two segments of hydrophobic aminoacyl residues in a heptameric arrangement characteristic of amphipathic α-helices (AS-1 and AS-2; Fig. 1). Thus, the HAMP linker is postulated to consist of two short amphipathic α-helices joined by an unstructured connector; computer modeling suggests that the α-helices may be in a coiled-coil conformation (41).
FIG. 1.
HAMP linkers in this study. The single-letter amino acid sequences of the three HAMP linkers used in this study are shown. Sequences are aligned about the Pro at the N-terminal end of AS-1 and the Glu at the N-terminal end of AS-2; these residues are underlined. The predicted junction between TM-2 and cytoplasmic amino acid sequences is at the left, and the aminoacyl residue number at the C-terminal end of the HAMP linker is shown at the right for reference. The hydrophobic heptad repeat characteristic of HAMP linkers is shown, with hydrophobic positions capitalized and hydrophobic residues shown in boxes. The extents of the amphipathic sequences 1 and 2 (AS-1 and AS-2) are shown by bold lines, and the connector is shown by a light dotted line. It is not clear whether sequences upstream of the conserved Pro in AS-1 form a continuous α-helix with AS-1; these are indicated by a bold dashed line. Positions in AS-1 and AS-2 are numbered for reference, and deletions analyzed in this study are illustrated by black rectangles.
Biochemical evidence supports this model for HAMP linker structure. Cross-linking and solvent accessibility studies have been performed on cysteinyl-substituted HAMP linkers from the Salmonella enterica aspartate-sensing MCP Tar. The results are consistent with AS-1 and AS-2 having α-helical structures with their hydrophobic faces protected from the aqueous environment (8). Also, these studies suggest that the AS-2 elements in a dimer are adjacent, whereas the AS-1 elements are somewhat separated.
Genetic analysis indicates that the HAMP linker plays an active role in intramolecular signal transduction. First, missense mutations in HAMP linkers from the E. coli nitrate sensor NarX, osmosensor EnvZ, and serine-responsive MCP Tsr result in sensor proteins biased toward one signaling mode (1, 3, 10, 22, 32). Second, deletions in the NarX HAMP linker reveal distinct roles in signal transduction for different HAMP linker components. NarX normally responds to the presence of its ligand, nitrate, by increasing the kinase activity of its cytoplasmic transmitter module (52). Deletions which shorten AS-1 or the connector of the NarX protein by 7 aminoacyl residues (corresponding to two α-helical turns) result in constitutive kinase activity [as monitored by expression of a Φ(narG-lacZ) operon fusion]. However, certain deletions which shorten AS-2 of NarX by 7 aminoacyl residues result in a reversed-response phenotype, in which kinase activity is significantly higher in the absence of ligand rather than in its presence (3).
Previously, hybrid sensors have been made in which the sensor module and HAMP linker of one protein communicate with the output module of another protein. Such hybrids have been made between different E. coli MCPs (16, 24, 40, 51), Bacillus subtilis MCPs (25), the MCP Tar or Trg and the sensor kinase EnvZ (5, 49), and the sensor kinase NarX and the MCP Tar (50). Most of these hybrids alter their output activity in response to ligand binding in their heterologous input domains. However, a few have responses that are attenuated or difficult to interpret. Nonetheless, the apparent ease with which functional hybrids have been generated using an arbitrary fusion point suggests that the HAMP linker is a commonly used signal-transducing structure and that the HAMP linker from one protein can communicate with a heterologous output module.
In this study, we have constructed a range of hybrid sensor proteins in order to determine whether different predicted HAMP linkers share common mechanisms of signal transduction as well as common predicted structures. We used the NarX and NarQ proteins, which respond to ligand (nitrate) by increasing output kinase activity (47), and the CpxA protein, which appears to respond to ligand (CpxP protein) by decreasing output kinase activity (39), as the substrates for making hybrid sensor kinases. Hybrids were made with junctions at the beginning and end of AS-1 and at the beginning and end of AS-2 in the HAMP linker. Some of these hybrids, including one which replaced just the HAMP linker of NarX with that of CpxA, responded to ligand, implying that different HAMP linkers respond to and transmit similar intramolecular signals. We also found that different HAMP linkers use AS-1 and AS-2 in similar ways in signal transduction, suggesting an operational test for HAMP linker candidates identified by sequence analysis. However, most of the hybrid sensor kinases that we generated failed to regulate transmitter activity in response to ligand, suggesting the importance of sequence-specific interactions within the HAMP linker. These observations may be useful in the future design of hybrid sensors.
MATERIALS AND METHODS
Strains, plasmids, and general methodology.
E. coli K-12 strains and plasmids used in this study are listed in Table 1. Standard methods were used for restriction endonuclease digestion, ligation, and transformation of DNA. Restriction enzymes and T4 DNA ligase were from New England Biolabs, Inc. (Beverly, Mass.). DNA for sequencing was isolated from purified plasmid minipreparations (Qiagen, Valencia, Calif.) or amplified by PCR from boiled lysates of bacterial colonies (27). The structures of all plasmid constructs used in this study were confirmed by automated DNA sequence analysis (DNA Sequencing Facility, University of California, Davis).
TABLE 1.
Strains
| E. coli straina | Genotype | Source or reference |
|---|---|---|
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150(Strr) relA1 flbB5301 deoC1 ptsF25 rbsR | 9 |
| TR8 | As MC4100 but cpxA1::cam | 38 |
| TR50 | As MC4100 but λRS88[cpxP-lacZ] | 38 |
| VJS632 | Prototroph | 45 |
| VJS676 | As VJS632 but Δ(argF-lac)U169 | Lab collection |
| VJS2197 | As VJS676 but λΦ(narG-lacZ)250 | 34 |
| VJS5054 | As VJS2197 but ΔnarX242 narQ251::Tn10d(Tc) pcnB1 zad-981::Tn10d(Km) | 54 |
| VJS7220 | As TR50 but cpxA1::cam | This work |
Strain and plasmid constructions are described in Materials and Methods.
Plasmids encoding hybrid proteins were made by a modification of the QuickChange PCR protocol (Stratagene, La Jolla, Calif.), with high-fidelity thermostable DNA polymerase (Accuzyme; Bioline USA, Reno, Nev.). Site-directed mutagenesis was performed by one of two similar techniques. In both methods, PCR products were generated from one mutagenic primer and another primer which annealed to the cDNA strand some 100 to 200 bp distant. The resulting double-stranded DNA fragment, carrying the desired mutation, provided a primer for a second PCR. In the first method, this second PCR utilized high-fidelity thermostable DNA polymerase to replicate a small (<1,500-bp) fragment, which was subsequently digested with restriction enzymes to produce a smaller (<400-bp) fragment. This fragment, carrying the desired mutation, was subcloned into the unmutagenized parent plasmid. In the second method, the second PCR utilized high-fidelity, high-processivity thermostable DNA polymerase (Pfu Turbo; Stratagene) to replicate the entire plasmid. The resulting nicked plasmid DNA was used to transform E. coli strain DH5α. Plasmid DNA was isolated from each of several transformants and digested with restriction enzymes which allowed subcloning of a small (<400-bp) fragment carrying the desired mutation into unmutagenized parent plasmid. In both instances, the entire subcloned region of the resulting mutagenized plasmid was sequenced.
Construction of hybrid sensor kinases.
Hybrids of the E. coli NarX, NarQ, and CpxA proteins were made by restriction digestion followed by ligation of the narX, narQ, and cpxA genes containing restriction endonuclease sites introduced as described above. Restriction sites were designed to be silent with respect to at least one of the protein sequences.
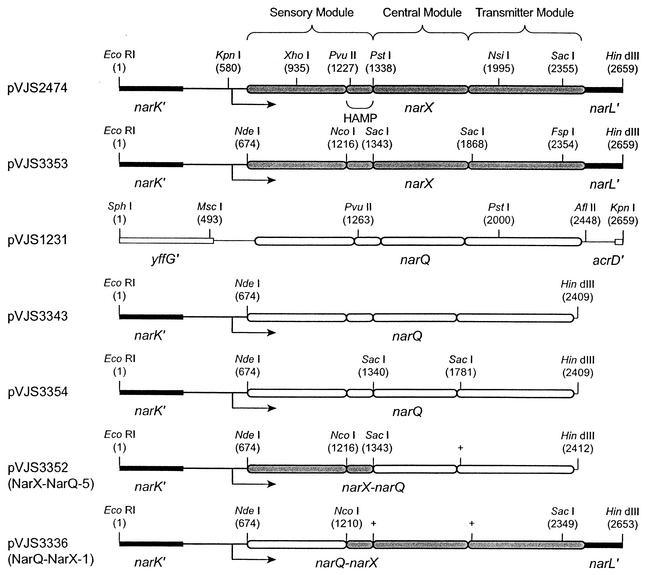
The starting point for constructions involving the narX gene was plasmid pVJS1241, which previously was engineered to contain silent restriction sites for the enzymes KpnI, PvuII, and SacI (54). This plasmid was modified in two steps to form plasmid pVJS2474 (Fig. 2). First, plasmid pVJS1241 was digested with the enzyme AgeI, which cleaves within codon 110 of the narK gene. The overhang was made blunt by filling in with Klenow polymerase, and the plasmid was cleaved with the enzyme SmaI (which cuts in the pUC8-derived polylinker of the vector plasmid, pHG165 [43]) and self ligated. This deleted 607 bp from the narK gene and in the process eliminated one of the two PstI sites in plasmid pVJS1241. Second, a BamHI-cleaved Ω interposon (15) was cloned into the BglII site at codon 59 of the narL gene, which placed a HindIII site immediately adjacent to the BamHI-BglII junction. Digestion with HindIII deleted 1,110 bp of narL and ychO sequence downstream of the narX gene.
FIG. 2.
Structures of narX and narQ plasmids. See Materials and Methods for details of construction. The narX (gray) and narQ (white) genes are depicted as rounded rectangles to show the coding regions for the corresponding modules and the HAMP linker. Nucleotide positions of restriction endonuclease sites are indicated. Only selected sites are depicted for each plasmid. The narX transcription initiation point (V. Stewart and P. J. Bledsoe, unpublished data) is indicated by the arrow.
Plasmid pVJS2474 served as template for introducing additional silent restriction endonuclease sites into the narX gene. For example, plasmid pVJS3353 (Fig. 2) contains an NdeI site at the initiation codon (TAC ATG changed to CAT ATG), an NcoI site spanning codons Pro-181 and Trp-182 at HAMP linker positions AS-1.1 and AS-1.2 (CCG TGG changed to CCA TGG), a SacI site spanning codons Glu-222 and Leu-223 at HAMP linker positions AS-2.14 and AS-2.15 (GAA CTG changed to GAG CTC), and a SacI site spanning codons Glu-397 and Leu-398 near the amino-terminal coding region of the transmitter module (GAA CTG changed to GAG CTC). The silent SacI site at position 2355 of plasmid pVJS2474 (Fig. 2) was removed by converting it into a silent FspI site (CGA GCT CAA changed to CGT GCG CAA). Plasmid pVJS3353 or its precursors provided material for constructing a variety of narX-narQ hybrid genes, including several whose properties will be reported elsewhere (V. Stewart, L.-L. Chen, and H.-C. Wu, submitted for publication).
Four additional narX restriction sites were constructed for this study: a BssHII site spanning codons Pro-181 and Trp-182 (CCG TGG changed to CCG CGG), which changes position AS-1.2 to Ala, the residue at the corresponding position in the CpxA protein; an MscI site spanning codons His-194 through Asp-196 (CAT CGC GAT changed to CAT GGC CAG), which changes the third and fourth residues of the connector to Gly and Gln, respectively, the residues at the corresponding positions in the NarQ protein; a SacI site spanning codons Glu-208 and Met-209 (GAA ATG changed to GAG CTC), which changes position AS-2.1 to Leu, the residue at the corresponding position in the NarQ protein; and a silent SpeI site spanning codons Leu-166 through Val-168 in TM-2 (TTA CTG GTG changed to TTA CTA GTG) to facilitate recloning of small DNA fragments following mutagenesis and sequence determination. Additionally, the native PstI site in the narX gene, which spans codons Ser-220 through Glu-222 at HAMP linker positions AS-2.12 through AS-2.14 (TCT GCA GAA), was employed for construction of the NarX-CpxA-5 hybrid.
The starting point for constructions involving the narQ gene was plasmid pVJS1231 (Fig. 2) (52). This plasmid was modified in two steps to place narQ gene expression under the control of the narX transcription and translation initiation sites (see plasmid pVJS3343 in Fig. 2). First, an NdeI site was placed at the initiation codon (CGT GTG changed to CAT ATG). Second, the NdeI-AflII fragment containing the narQ gene was cloned into the superlinker plasmid pSL1180 (7), thereby placing the AflII site immediately adjacent to a HindIII site. The NdeI-HindIII fragment containing the narQ gene was then cloned in place of the corresponding narX fragment of plasmid pVJS3353 to form plasmid pVJS3343 (Fig. 2).
Plasmid pVJS3343 served as template for introducing additional silent restriction endonuclease sites into the narQ gene. For example, plasmid pVJS3354 (Fig. 2) contains a SacI site spanning codons Glu-221 and Leu-222 at HAMP linker positions AS-2.14 and AS-2.15 (GAG CTG changed to GAG CTC) and a SacI site spanning codons Glu-368 and Leu-369 near the amino-terminal coding region of the transmitter module (GAA TTG changed to GAG CTC). We also constructed an NcoI site spanning codons Pro-179 and Leu-180 at HAMP linker positions AS-1.1 and AS-1.2 (CCG CTG changed to CCA TGG), which changes codon 180 from Leu to Trp. However, the NarQ protein did not tolerate Trp at position 180 (see Discussion), so hybrid genes constructed at this junction were subjected to a further round of site-specific mutagenesis to make codon 180 revert to Leu. Plasmid pVJS3354 or its precursors provided material for constructing a variety of narX-narQ hybrid genes, including several whose properties will be reported elsewhere (Stewart et al., submitted).
Three additional narQ restriction sites were constructed for this study: a silent MscI site spanning codons His-192 through Gln-194 in the connector (CAT CGC GAT changed to CAT GGC CAT), a silent SacI site spanning codons Glu-207 and Leu-208 at HAMP linker position AS-2.1 (GAG CTT changed to GAG CTC), and a silent SpeI site spanning codons Thr-162 through Val-164 in TM-2 (ACG CTG GTC changed to ACA CTA GTC) to facilitate recloning of small DNA fragments following mutagenesis and sequence determination.
The starting point for constructions involving the cpxA gene was plasmid pOK101 (36), which carries cpxA on a 1.5-kb BamHI-EcoRI insert in vector pINIII (29). High-fidelity PCR with mutagenic oligonucleotides was used to generate double-stranded DNA fragments of the cpxA gene with the desired sites near each end. Restriction endonuclease digestion generated DNA fragments with BssHII, NcoI, MscI, or PstI cohesive ends at positions coding for AS-1.1; AS-1.2, the second residue in the connector; AS-2.1; and AS-2.14, respectively. Each fragment also had a HindIII site immediately downstream of the cpxA gene. These fragments were subcloned into restriction-digested plasmids carrying the appropriate narX allele. For all constructs, the DNA sequence of the entire subcloned region, including the flanking junctions, was determined to ensure that no spurious sequence changes had occurred.
Media, culture conditions, and β-galactosidase assays.
Two constructs expressing hybrids of the periplasmic and transmembrane domains of NarX and the cytoplasmic domain of CpxA were found to be toxic to cpxA-null strains in the absence of 40 mM nitrate. Therefore, all experiments employing plasmid-borne derivatives of cpxA were carried out by transforming the reporter strain VJS7220 with the desired plasmid and plating the transformants on nutrient agar (27) supplemented with ampicillin (200 μg/ml) and NaNO3 (40 mM). After growth overnight at 37°C, individual colonies were picked and grown aerobically in 1 ml of tryptone-yeast extract (27) supplemented with ampicillin (200 μg/ml) and NaNO3 (40 mM) for 2 h; this culture was then used to inoculate cultures for β-galactosidase assays.
All cultures for β-galactosidase assays were grown in a mixture of defined minimal medium and specific complex medium. The defined medium in this mixture was glucose-supplemented MOPS (3-[N-morpholino]propanesulfonic acid)-buffered minimal medium (46). The specific complex medium in this mixture was tryptone-yeast extract; the mixture was supplemented with ampicillin (200 μg/ml). This mixture was found to produce a favorable combination of rapid growth and robust response to nitrate by the NarX protein (3). Cultures were grown anaerobically in the presence or absence of 40 mM NaNO3− at 37°C in screw-cap tubes as previously described (46). Culture density was monitored with a Klett-Summerson photoelectric colorimeter (Klett Manufacturing Co., New York, N.Y.) equipped with a number 66 (red) filter. Cultures of strain VJS5054 [ΔnarX narQ::Tn10 φ(narG-lacZ)] derivatives were placed on ice upon reaching mid-exponential-phase growth (between 30 and 40 Klett units); cultures of strain VJS7220 [ΔcpxA φ(cpxP-lacZ)] derivatives were placed on ice after 2 h of growth.
β-Galactosidase activity was measured in CHCl3-sodium dodecyl sulfate-permeabilized cells as described by Miller (30), except that the reactions took place at room temperature (approximately 22°C). Slight variation in the absolute level of β-galactosidase activity from individual strains was seen from day to day, particularly with strains carrying derivatives of cpxA that expressed high levels of β-galactosidase; however, the relative β-galactosidase activities between different strains were quite similar. Each culture was assayed in duplicate, and the reported values are averaged from at least two independent experiments.
RESULTS
The HAMP linker in sensor kinases.
Figure 1 shows an alignment of the aminoacyl sequences of the three HAMP linkers used in this study and illustrates their salient features. These sequences show the pattern of phased hydrophobic residues characteristic of the HAMP linker amphipathic α-helices AS-1 and AS-2. These predicted α-helices are joined by an unstructured connector. Most HAMP linkers have a Pro residue within 9 aminoacyl residues of the presumed end of TM-2 at the cytoplasmic membrane and a Glu residue just before AS-2 (53). In order to facilitate comparisons between HAMP linkers, we refer here to positions in the HAMP linker by coordinates relative to these conserved residues. Positions in AS-1 are numbered starting at the conserved Pro, from residue AS-1.1 to AS-1.12 (Fig. 1). Residues between the conserved Pro and the predicted endpoint of TM-2 are assigned negative numbers. Positions in AS-2 are numbered starting at the first residue after the conserved Glu (AS-2.1) through AS-2.15. Note that the precise endpoints for these α-helices are unknown and therefore that these designations are arbitrary.
The HAMP linker is essential for intramolecular signal transduction in the E. coli nitrate-responsive sensor kinase NarX (3, 10, 22). NarX and its cognate response regulator NarL control the response to nitrate availability for anaerobic respiration. In the absence of nitrate, the NarX protein acts primarily as a phospho-NarL phosphatase (47). When nitrate is present, NarX acts primarily as a NarL kinase. Phospho-NarL controls the expression of several operons, activating transcription of some (e.g., narGHJI and fdnGHI) and repressing transcription of others (e.g., frdABCD) (12, 47). Strain VJS5054 carries a chromosomal Φ(narG-lacZ) operon fusion in a ΔnarX background, as well as the pcnB1 allele (26), which reduces the copy number of pMB9 replicon plasmids to approximately one. Expression of the Φ(narG-lacZ) operon fusion in this strain is induced about 100-fold by nitrate when the narX+ gene is present on a plasmid, providing a simple and sensitive assay of NarX protein activity. We have previously used this assay to isolate and characterize several NarX HAMP linker mutants (3).
The sensor kinase CpxA is also predicted to use a HAMP linker for intramolecular signal transduction (53). The ligand for CpxA is thought to be a periplasmic protein, CpxP, and under nonstressing conditions CpxP is thought to be bound by CpxA (38). However, the CpxA protein differs from the NarX protein in having the opposite response to ligand. When ligand is bound, CpxA acts predominantly as a phosphatase toward the phosphorylated form of the cognate response regulator, CpxR. Conditions of stress such as alkaline pH, high temperatures, and overexpression of periplasmic proteins (or spheroplast formation) are hypothesized to titrate the CpxP protein away from the CpxA periplasmic domain (37-39). This shifts the predominant activity of CpxA from phospho-CpxR phosphatase to CpxR kinase. Phospho-CpxR activates the transcription of several genes, including cpxP, degP, dsbA, and ppiA, whose products are involved in stabilizing periplasmic proteins (39). Transcription of a Φ(cpxP-lacZ) operon fusion is activated up to fivefold by overproduction of the outer membrane protein NlpE and 50-fold by stimuli such as alkaline pH, providing a simple and sensitive assay for CpxA kinase activity (11).
A NarX-CpxA hybrid sensor kinase.
We wished to determine whether sequences identified as HAMP linkers function similarly in NarX and CpxA, proteins that react to ligand binding with opposite responses. Therefore, we generated a series of hybrid proteins utilizing the sensory module of NarX and the transmitter module of CpxA (Fig. 1). The NarX and CpxA transmitter domains are classified in HPK subfamily 7 and 2b, respectively, as defined by Grebe and Stock (17). Note that NarX (and its ortholog NarQ) contains a unique central module of unknown function (see Fig. 5) (44). One of these hybrids, NarX-CpxA-1, comprised the amino-terminal cytoplasmic domain, first transmembrane α-helix, periplasmic nitrate sensing domain, and second transmembrane α-helix of NarX, fused to the HAMP linker and transmitter module of CpxA (Fig. 3; additional hybrids are discussed below). Expression of the hybrid narX-cpxA-1 gene is under the control of the narX regulatory sequences. In the absence of nitrate, a plasmid carrying this construct was toxic to cpxA-null strains even when present at a very low copy number in a pcnB background; however, essentially normal growth was observed in the presence of 40 mM nitrate or in the presence of cpxA+ on the chromosome. (Many mutant cpxA alleles are lethal or deleterious in a cpxA-null background, perhaps due to overexpression of stress response genes [T. Raivio, personal communication].)
FIG. 5.
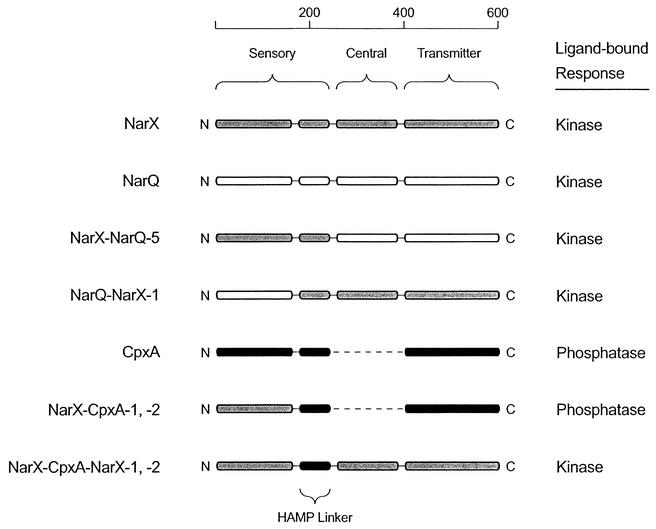
The HAMP linker in NarX, NarQ, CpxA, and hybrid sensor kinases. Sensor kinases which responded to nitrate in this study are depicted in cartoon form. The scale is in aminoacyl residues, and amino (N) and carboxy (C) termini are indicated. Modules or elements from NarX, NarQ, and CpxA are represented by gray, white, and black rounded rectangles, respectively. The absence of the central module in CpxA and the NarX-Cpx-1 and -2 hybrids is denoted by the dashed line. The primary structures of NarX-CpxA-1 and -2 differ by one amino acid, as do those of NarX-CpxA-NarX-1 and -2; the phenotypes of the two variants of each hybrid differ quantitatively but not qualitatively. The activity listed under “ligand bound” indicates the dominant activity of the sensor kinase in the presence of the ligand, nitrate. The conclusion is that the ligand-bound activity of these sensor kinases is determined not by the HAMP linker but by the transmitter module and/or the central module.
FIG. 3.
Hybrid sensor kinases with NarX sensor modules and CpxA output modules. The single-letter amino acid sequences of the HAMP linkers of hybrid sensor kinases utilizing the NarX sensor module and the CpxA output module are shown. Sequences from NarX are in plain type; sequences from CpxA are on a gray background; aminoacyl residues not native to either NarX or CpxA used for building the hybrids are boxed. The (arbitrarily assigned) endpoints of AS-1 and AS-2 are denoted by dashed lines, and the coordinates of positions within AS-1 and AS-2 are indicated as in Fig. 1. β-Galactosidase specific activity of the ΔcpxA Φ(cpxP-lacZ) strain VJS7220 expressing the indicated hybrid from a plasmid in the presence (+ NO3−) or absence (− NO3−) of 40 mM NaNO3 is indicated. Approximately the same β-galactosidase activity was seen for strain VJS7220 carrying an empty plasmid vector as with a plasmid expressing narX (data not shown). For details of culture growth during assays, see Materials and Methods.
We examined the behavior of the NarX-CpxA-1 hybrid protein by monitoring Φ(cpxP-lacZ) expression in the cpxA-null reporter strain VJS7220. Because of the toxicity of this construct in the absence of nitrate, we used cultures of cells freshly transformed with plasmid carrying the narX-cpxA-1 gene and plated on nitrate-containing medium. We assayed the β-galactosidase activity of these cultures after 2 h of anaerobic growth in liquid medium in the presence or absence of 40 mM nitrate [growth of these cultures and their Φ(cpxP-lacZ) expression in the absence of nitrate was highly variable after longer periods of time]. The narX-cpxA-1 construct mediated an approximately 10-fold reduction in Φ(cpxP-lacZ) expression in response to nitrate (Fig. 3). In the absence of nitrate, Φ(cpxP-lacZ) expression was comparable to that mediated by wild-type cpxA+, whereas nitrate reduced Φ(cpxP-lacZ) expression to the level characteristic of cpxA-null strains (Fig. 3). Therefore, the NarX-CpxA-1 hybrid sensor kinase responded to nitrate, as does the NarX sensor kinase, but reduced its kinase activity in response to ligand, as does the CpxA sensor kinase.
Hybrids among NarX, NarQ, and CpxA.
The prediction, based upon sequence analysis, that different HAMP linkers possess similar secondary structures raised the possibility that sensor kinases utilizing hybrid HAMP linkers comprising AS-1 of one HAMP linker and AS-2 of another might function normally. To test this possibility, we made a series of hybrid sensor kinases with hybrid junctions at the beginning or end of AS-1 or at the beginning or end of AS-2 (Fig. 3 and 4); one such hybrid (NarX-CpxA-1) is described above. We used NarX, CpxA, and NarQ in these constructs. NarQ is a paralog of NarX and responds similarly to the presence of nitrate in its regulation of an overlapping set of genes (47). Like the NarX protein, it phosphorylates the NarL protein and mediates full induction of the Φ(narG-lacZ) operon fusion in response to nitrate in the strain VJS5054 (35). However, the amino acid sequence of the NarQ HAMP linker is not strikingly similar to that of NarX (Fig. 1).
FIG. 4.
Hybrid sensor kinases containing elements of NarX, NarQ, and CpxA. The single-letter amino acid sequences of the HAMP linkers of hybrid sensor kinases utilizing the NarX or NarQ sensor module and the NarQ or NarX output module are shown. Sequences from NarX are in plain type, sequences from NarQ are on a gray background, and sequences from CpxA are in white on a black background. The (arbitrarily assigned) endpoints of AS-1 and AS-2 are denoted by dashed lines, and the coordinates of positions within AS-1 and AS-2 are indicated. β-Galactosidase specific activity of the ΔnarX narQ Φ(narG-lacZ) strain VJS5054 expressing the indicated hybrid from a plasmid in the presence (+ NO3−) or absence (− NO3−) of 40 mM NaNO3 is indicated. The specific β-galactosidase activity of VJS5054 carrying an empty plasmid vector was <10 Miller units (data not shown). For details of culture growth during assays, see Materials and Methods.
All of the hybrid sensor kinases that we constructed had nitrate-responsive sensory modules. The nitrate response of hybrids with NarX or NarQ transmitter modules was assayed in the Φ(narG-lacZ) strain VJS5054 [note that the uninduced, basal level of Φ(narG-lacZ) expression in VJS5054 expressing narQ (Table 2) was somewhat higher than previously observed (35, 52); this is an effect of the complex medium used in these experiments]. The nitrate response of hybrids with the CpxA transmitter module was assayed in the Φ(cpxP-lacZ) strain VJS7220.
TABLE 2.
Effects of characteristic deletions in the NarX, NarQ, and NarX-CpxA-1 HAMP linkers
| Deletionb | β-Galactosidase sp act for sensor kinasea:
|
|||||
|---|---|---|---|---|---|---|
| NarX
|
NarQ
|
NarX-CpxA-1
|
||||
| − NO3− | + NO3− | − NO3− | + NO3− | − NO3− | + NO3− | |
| WT | 10 | 1,990 | 320 | 1,800 | 3,550 | 350 |
| ΔAS-1.1-7 | 1,550 | 1,300 | 90 | 600 | 50 | 40 |
| ΔAS-1.5-11 | 1,660 | 1,550 | 1,980 | 1,140 | NDc | ND |
| ΔAS-1.8-con2 | 1,800 | 1,420 | 2,740 | 1,950 | 30 | 30 |
| ΔAS-2.1-7 | 1,330 | 210 | 20 | 20 | 270 | 550 |
| ΔAS-2.4-10 | 510 | 50 | 430 | 20 | 760 | 950 |
β-Galactosidase specific activity was measured as described in Materials and Methods and is expressed in Miller units. Mutant narX and narQ alleles were expressed from derivatives of plasmid pHG165 transformed into strain VJS5054 Φ(narG-lacZ) narX narPQ pcnB. Mutant narX-cpxA-1 alleles were expressed from derivatives of plasmid pHG165 transformed into strain VJS7220 Φ(cpxP-lacZ) cpxA. Cultures were grown anaerobically in the absence or presence of 40 mM NaNO3.
Deletion names indicate the positions in the HAMP linker which have been deleted; see Fig. 1 for graphic representation. WT, wild type.
ND, not determined.
Four hybrid sensor kinases responded to nitrate: NarX-CpxA-1, NarX-CpxA-2 (Fig. 3), NarX-NarQ-5, and NarQ-NarX-1 (Fig. 4). Nitrate-responsive hybrids with NarX or NarQ transmitter modules responded to ligand by increasing Φ(narG-lacZ) expression, whereas nitrate-responsive hybrids with the CpxA transmitter module responded to ligand by decreasing Φ(cpxP-lacZ) expression. Three of these have hybrid junctions at an end of the HAMP linker, and the fourth (NarX-CpxA-2) has a hybrid junction one residue removed from the end of the HAMP linker (Fig. 3). In three of these hybrids (NarX-CpxA-1, NarX-CpxA-2, and NarQ-NarX-1; Fig. 3 and 4), the HAMP linker is joined to a heterologous sensory module, whereas in one (NarX-NarQ-5; Fig. 4) the HAMP linker is joined to a heterologous transmitter module. Both the NarX-CpxA-1 and NarX-CpxA-2 constructs were toxic to the cpxA-null strain VJS7220.
Two additional hybrids were constructed in which the sensory and the central and transmitter modules of NarX were joined by the HAMP linker of CpxA (Fig. 4). The NarX-CpxA-NarX-1 hybrid mediated an approximately 40-fold increase in Φ(narG-lacZ) expression in response to nitrate, while the NarX-CpxA-NarX-2 hybrid mediated an approximately 15-fold increase in Φ(narG-lacZ) expression in response to nitrate (Fig. 4). In both of these hybrids, the HAMP linker is joined to heterologous input and transmitter modules. Although the magnitude of the response of the hybrid protein to ligand is somewhat reduced compared with wild-type protein, it is of the same sense, i.e., it mediates an increase in Φ(narG-lacZ) expression in response to nitrate.
Nine of the 15 hybrid sensor kinases that we constructed did not respond to nitrate (Fig. 3 and 4). This group includes all hybrids which have fusion junctions within the HAMP linker and, strikingly, three hybrids which have intact HAMP linkers. All nonresponsive hybrids with NarX or NarQ output modules conferred high levels of Φ(narG-lacZ) expression in the presence or absence of nitrate (Fig. 4). Two nonresponsive hybrids with the CpxA output module conferred Φ(cpxP-lacZ) expression that was significantly less than that seen with an empty plasmid vector, suggesting that the encoded protein had high phosphatase activity (Fig. 3). The output activity of all of these nonresponsive hybrid sensor kinases mimicked the ligand-bound state; only one nonresponsive hybrid (NarX-CpxA-3) deviated from this pattern, causing moderate Φ(cpxP-lacZ) expression regardless of nitrate availability (Fig. 3). None of these NarX-CpxA hybrids was toxic to strain VJS7220. We previously observed that the majority of disruptions to the HAMP linker of NarX result in a ligand-bound phenotype (3); most of these nonresponsive hybrid sensor kinases result in a similar ligand-bound phenotype.
Effects of characteristic deletions in AS-1 and AS-2 of the NarX HAMP linker.
Analysis of mutant NarX proteins with deletions in AS-1 or AS-2 indicates that these elements have markedly different roles in signal transduction (3). Deletions which shorten AS-1 of the NarX protein by 7 aminoacyl residues (approximately two α-helical turns; Fig. 1) confer constitutive Φ(narG-lacZ) expression (Table 2). Two analogous 7-residue deletions in AS-2 (Fig. 1) result in a reversed-response phenotype, with up to 10-fold-higher Φ(narG-lacZ) expression in the absence of nitrate than in its presence (3) (Table 2). The phenotypes caused by these deletions are distinctive enough that we felt that they might provide a functional assay for whether a given sequence acted as a HAMP linker similar to that in the NarX protein. Therefore, we studied the effects of these deletions in the HAMP linkers of two other sensor kinases, NarQ and the NarX-CpxA-1 hybrid. We will refer to deletions by their coordinates in the HAMP linker to facilitate comparisons of analogous deletions in different proteins (Fig. 1).
Effects of characteristic deletions in AS-1 and AS-2 of the NarQ HAMP linker.
We began by making deletions in narQ analogous to those made in narX. We assayed the nitrate response of narQ deletion mutants by using the Φ(narG-lacZ) reporter strain VJS5054. Two deletions shortening AS-1 of NarQ (ΔAS-1.5-11 and ΔAS-1.8-con2; Table 2), resulted in high, constitutive Φ(narG-lacZ) expression. The phenotypes caused by these mutations were very similar to those caused by analogous mutations in the NarX protein (Table 2).
A third deletion in AS-1 of NarQ (ΔAS-1.1-7; Table 2) resulted in a phenotype different from that caused by the analogous deletion in NarX. This deletion conferred approximately sixfold induction by nitrate, although its induced Φ(narG-lacZ) expression was approximately one-half that of the wild-type (Table 2). The NarQ protein has nine aminoacyl residues upstream of AS-1.1, in comparison with six in NarX (Fig. 1) Thus, AS-1 of this shortened protein could comprise four amphipathic α-helical turns, unlike the shortened AS-1 of NarX. Further, this truncated protein moves an Ala residue into a position normally occupied by Pro. A Pro-to-Ala substitution confers the wild-type phenotype in NarX (3). Together, these characteristics of NarQ may permit this deletion mutant to retain rudimentary HAMP linker function.
We made two deletions in AS-2 of the NarQ HAMP linker which are analogous to two of those made in NarX. One of these deletions (ΔAS-2.4-10; Table 2) resulted in a reversed-response phenotype nearly identical to that caused by the corresponding deletion in narX, with approximately 20-fold-higher Φ(narG-lacZ) expression in the absence of nitrate than in its presence. The other deletion (ΔAS-2.1-7; Table 2) resulted in very low, uninducible Φ(narG-lacZ) expression, different from that caused by the corresponding deletion in narX. A similar ligand-unbound phenotype is caused by a different deletion in AS-2 of NarX, from position 8 to 15 (3).
Effects of characteristic deletions in AS-1 and AS-2 of the CpxA HAMP linker.
The response of the NarX-CpxA-1 hybrid to ligand is opposite from that of the NarX and NarQ proteins; therefore, it was of interest to see how this protein would respond to characteristic deletions in the HAMP linker. The phenotypes conferred by deletion mutations in narX-cpxA-1 were measured in the Φ(cpxP-lacZ) strain VJS7220. Unlike wild-type narX-cpxA-1, the genes for these deletion mutants caused no obvious growth defect in VJS7220. Two deletions in AS-1 of NarX-CpxA-1 resulted in very low Φ(cpxP-lacZ) expression (ΔAS-1.1-7 and ΔAS-1.5-11; Table 2). Indeed, the Φ(cpxP-lacZ) activity of these mutants was lower than that seen with an empty plasmid vector (approximately 300 Miller units). This suggests that these mutant proteins had very high phosphatase activity. This behavior was the opposite of that seen in the NarX (and NarQ) proteins, in which deletions in AS-1 generally resulted in very high kinase activity. However, the phenotypes of almost all the deletions in AS-1 of the NarX, NarQ, and NarX-CpxA-1 proteins were identical in that they mimicked the ligand-bound state.
We made two deletions in AS-2 of the NarX-CpxA-1 HAMP linker which were analogous to those made in NarX. Both of these deletions resulted in generally elevated Φ(cpxP-lacZ) expression, with somewhat higher (twofold or less) Φ(cpxP-lacZ) expression in the presence of nitrate than in its absence (Table 2). This phenotype is clearly different from that caused by deletions in AS-1 and may represent a weak reversed-response phenotype or a ligand-unbound phenotype. Deletion of the first seven aminoacyl residues of AS-2 in the CpxA HAMP linker resulted in a reversed-response phenotype when this sequence was in the context of the NarX-CpxA-NarX-1 hybrid. VJS5054 expressing narX-CpxA-narX-1 ΔAS-2.1-7 reduced Φ(narG-lacZ) expression from 630 to 190 Miller units in response to 40 mM NaNO3−.
DISCUSSION
We constructed hybrid sensor kinases by using the nitrate-responsive sensory modules of NarX and NarQ and the HAMP linkers and transmitter modules of NarX, NarQ, and CpxA. We used lacZ operon fusions to monitor the responses of these sensor kinases to nitrate. Expression of these operon fusions depends on the levels of phosphorylated response regulators, which in turn are dependent upon the presence of the cognate sensor kinases and the balance of their kinase and phosphatase activities. Therefore, we regard the resulting β-galactosidase activities as an indicator of the behavior of the sensor kinases. Hence, increased Φ(narG-lacZ) expression is attributable to and correlates with NarX or NarQ kinase activity. Likewise, increased Φ(cpxP-lacZ) expression is attributable to and correlates with CpxA kinase activity; furthermore, because of the relatively high basal level of Φ(cpxP-lacZ) expression, CpxA phospho-CpxR phosphatase activity can also be monitored.
The HAMP linker: a commonly used signal-transducing element.
We have generated hybrid sensor kinases in which different HAMP linkers communicate between nitrate-responsive sensory modules and heterologous transmitter modules (Fig. 5). In the NarX-NarQ-5 hybrid, the HAMP linker from NarX communicates with a heterologous transmitter module. In the NarQ-NarX-1 and NarX-CpxA-1 hybrids, HAMP linkers from NarX and CpxA communicate with heterologous sensory modules. In the NarX-CpxA-NarX hybrids, the CpxA HAMP linker communicates with heterologous sensory and transmitter modules. All of these hybrids responded to ligand with changes in transmitter module activity that are comparable to those seen in their parent proteins (Fig. 3 and 4). Also, the sense of the response to ligand—i.e., increased or decreased kinase activity—was determined not by the HAMP linker but rather by the transmitter module and/or the central module (Fig. 5).
These new hybrids can be added to the list of previously reported functional hybrids between sensor kinases and MCPs, many of which use a hybrid fusion point at position 11 in AS-2 and which therefore retain most of the HAMP linker from the sensory module. These include hybrids between the aspartate-sensing MCP Tar and the serine-sensing MCP Tsr (24), between the dipeptide-binding protein-sensing MCPs Tap and Tar (51), and between the ribose-galactose-sensing MCPs Trg and Tsr (16). Functional reciprocal hybrids between the B. subtilis MCPs McpB and McpC have been made with a fusion junction at position AS-2.8 (25). An additional hybrid between Aer and Tsr, with use of the entire Aer HAMP linker, responds appropriately to regulatory cues, although its ligand is not precisely defined (40). These and the present results indicate that the HAMP linker is capable of communicating with both heterologous sensory domains and heterologous cytoplasmic output domains, depending upon context. This supports the prediction from sequence analysis that the HAMP linker is a signal-transducing module used by many sensory proteins. Furthermore, functional hybrids in which HAMP linkers communicate with heterologous sensory domains suggest that HAMP linkers receive similar physical inputs that indicate ligand binding.
Conserved action of the HAMP linker in signal transduction.
Our previous analysis of deletion mutants indicates that AS-1 and AS-2 of the NarX HAMP linker have different roles in signal transduction. Deletions which truncate AS-1 of NarX result in constitutive kinase activity, whereas those which truncate AS-2 result in a reversed response to ligand (3). The generally similar consequences of analogous deletions in NarQ and the NarX-CpxA-1 and NarX-CpxA-NarX-1 hybrids suggest that AS-1 and AS-2 have conserved roles in signal transduction. However, these results do not establish completely characteristic phenotypes for deletions AS-1 and AS-2 of the HAMP linker. For instance, five deletions in AS-1 of NarX (Table 2) (3), two similar deletions in AS-1 of NarQ, and two similar deletions in NarX-CpxA-1 (Table 2) resulted in a strong ligand-bound phenotype. However, one deletion in AS-1 of NarQ resulted in a protein that retains a sixfold increase in kinase activity in response to ligand (deletion AS-1.1-7; Table 2). The phenotypes of deletions in AS-2 were also variable. Two deletions in AS-2 of NarX and one similar deletion in NarQ resulted in a reversed-response phenotype (Table 2). One deletion in AS-2 of NarX and a different deletion in AS-2 of NarQ resulted in a ligand-unbound phenotype (NarX AS-2.8-15 and NarQ AS-2.1-7) (3) (Table 2). Furthermore, two deletions in AS-2 of NarX-CpxA-1 resulted in a phenotype which is intermediate between the reversed-response and ligand-unbound phenotypes (Table 2). One of these deletions resulted in a reversed-response phenotype in NarX-CpxA-NarX-1, suggesting a significant role for the output module in interpreting HAMP linker signals and determining the phenotype of a mutation. Furthermore, some variability in these deletion phenotypes may be attributable to the high degree of sequence variability between different HAMP linkers. Nonetheless, the bulk of evidence suggests conserved functions for AS-1 and AS-2 in signal transduction.
A previous investigation of NarX revealed that most mutations which disrupt HAMP linker function result in a ligand-bound phenotype (3). Most of the defective mutant or hybrid sensor kinases produced in this study also resulted in a ligand-bound phenotype. Thus, all nonresponsive hybrids with NarX or NarQ transmitter domains exhibited constitutive kinase activity (Fig. 3), whereas two of the three nonresponsive hybrids with the CpxA transmitter domain exhibited low, uninducible kinase activity (Fig. 2). An additional NarQ mutant with a Leu-to-Trp change at position AS-1.2 (generated to introduce an NcoI restriction site) similarly resulted in a ligand-bound phenotype (data not shown). Taken together, the phenotypes caused by deletions and missense mutations in AS-1 and AS-2 may provide an operational definition of the HAMP linker to complement and buttress a definition based upon primary structure.
Criteria for functional hybrid sensors.
Despite an apparently conserved structure and mode of action, the HAMP linker cannot be viewed as a freely interchangeable module. Generation of functional hybrids between sensors by using junctions in the HAMP linker is complicated by the considerable primary sequence variability of HAMP linkers and a lack of precise structural information about AS-1 and AS-2.
Some hybrid sensor kinases with junctions in the HAMP linker are functional, but their responses are difficult to interpret. Two hybrids have been made between the sensory modules of MCPs Tar and Trg and the transmitter module of the sensor kinase EnvZ, both by using the fusion point at position AS-2.11 (5, 49). The ligands for Tar include aspartate and maltose-bound maltose binding protein (MBP). The Tar-EnvZ hybrid Taz1 does not respond to MBP, and while it responds to aspartate, its sensitivity to this ligand is reduced by approximately 1,000-fold (49). Although the reciprocal hybrid between EnvZ and Tar was constructed (49), its properties have not been reported. Similarly, although the Trg-EnvZ hybrid Trz responds to ribose with increased kinase activity, its sensitivity to ribose is approximately 100-fold lower than that of Trg (5). A further hybrid has been constructed between the sensory domain of NarX and the methylation domain of Tar with a fusion point at AS-2.11 (50). This protein responds to ligand (nitrate) as a repellent, while Tar responds to its ligands Asp and MBP-maltose as attractants (13).
Many hybrid sensor kinases do not alter output activity in response to ligand or have attenuated responses to ligand. These include some hybrids in which both AS-1 and AS-2 were from the same protein (e.g., NarX-CpxA-5 [Fig. 2] and NarQ-NarX-1 and NarX-NarQ-4 [Fig. 3]) and some hybrids with a fusion point at AS-2.11 (e.g., a Tsr-Trg hybrid [16]). Significantly, no hybrids with AS-1 from one protein and AS-2 from another protein altered transmitter activity in response to ligand (e.g., NarX-CpxA-4 [Fig. 2] and NarX-NarQ-3 and NarQ-NarX-2 [Fig. 3]). To date, only hybrids in which AS-1 and the bulk of AS-2 were from the same protein function as sensors. Furthermore, the NarX-CpxA-NarX-1 protein, in which the entire NarX HAMP linker is replaced with that of CpxA, responds strongly to nitrate. Overall, these results suggest that the likelihood of a hybrid sensor being functional is increased but by no means guaranteed if it includes a complete HAMP linker, either from the sensor module or from the output module. This evidence supports the hypothesis that specific interactions within the HAMP linker, rather than specific interactions between the HAMP linker and the output module, are of paramount importance for HAMP linker function.
Mechanics of signal transduction by the HAMP linker.
Crystal structures of the MCP Tar periplasmic domain with and without ligand, cross-linking analysis of Tar transmembrane α-helices in the presence and absence of ligand, and computational models of Tar support a model in which periplasmic ligand binding displaces TM-2 of one monomer towards the cytoplasm by more than 1 Å (31, 33). This suggests that the ligand-bound signal received by HAMP linkers is a displacement of the amino-terminal end of AS-1 towards the cytoplasm. It is not clear how the HAMP linker responds to this physical input. Cross-linking studies suggest that the AS-2 elements of Tar remain close to one another in both the presence and the absence of ligand (8). The behavior of the hybrid proteins presented here suggests that specific interactions occur within the HAMP linker during signal transduction. Whatever the mechanism, the HAMP linker must be able to transmit information from periplasmic sensory modules (and perhaps a cytoplasmic PAS domain, in the case of the MCP Aer [6]) to a variety of output modules in proteins or protein complexes (4, 53).
Information transmitted by the HAMP linker.
What is the nature of the signal transmitted by the HAMP linker? Two extreme models provide a starting point for discussion. One possibility is that the HAMP linker receives mechanical input from the sensory module and interprets it for the output domain. Thus, the CpxA HAMP linker would receive a ligand-bound signal and assume a configuration which decreases the ratio of kinase to phosphatase activities in the transmitter, whereas the NarX HAMP linker would receive a ligand-bound signal and assume a conformation which increases that ratio. However, this model requires that HAMP linkers which appear to be very similar in predicted structure, and which apparently use the same structural elements in similar ways, nevertheless have opposite responses to the same signal.
Another possibility is that the HAMP linker simply transmits a signal from the sensory module about whether ligand is bound; the cytoplasmic portion of the protein interprets this information and adjusts the ratio of kinase to phosphatase activity (or alters the kinase/phosphatase activity ratio of an associated histidine kinase, in the case of MCPs) appropriately. Thus, the HAMP linkers of NarX or CpxA can be in one of two physical conformations, corresponding to ligand-bound or ligand-unbound input from the sensory module; the different proteins respond to these conformations differently. The signal from the HAMP linker would be interpreted by the central module, the transmitter module, or both. Ultimately, similar transmitter modules generate opposite responses to the same input from the HAMP linker.
The behavior of the NarX-CpxA-NarX hybrids is consistent with the latter model (Fig. 5). In the NarX-CpxA-NarX-1 hybrid protein, the HAMP linker from CpxA apparently transmits a signal of the same sense and comparable magnitude as that of the HAMP linker from NarX in the wild-type protein. This suggests that different HAMP linkers, even from proteins with opposite responses to ligand, transmit similar signals to indicate that ligand is bound. Opposite responses to ligand may be generated by different output modules or by the presence or absence of a central module. Our data do not allow us to distinguish between these possibilities; however, we note that other sensor kinases lacking central modules have either kinase-activating (2, 20, 21, 23, 42) or kinase-inactivating (18) responses to ligand. Specific interactions between the HAMP linker and the cytoplasmic potion of the protein may alter the magnitude of the response. Furthermore, sensors such as the MCPs manifest different responses to different ligands (e.g., Tsr, which responds to the ligand serine as an attractant [19] but responds to nickel as a repellent [28]). It is not clear whether these different sensory inputs generate different mechanical inputs to the HAMP linker and, if so, how the HAMP linker responds to them.
Structural similarities between HAMP linkers and the similarities of mutant phenotypes in different HAMP linkers, as well as the number of functional hybrid sensors that use the HAMP linker, all point to a common underlying mechanism for the action of this element in signal transduction. The HAMP linker is an active participant in signal transduction, receiving a signal about ligand binding from the sensory module and conveying that signal to the output module. How that signal is sent and interpreted and what interactions occur within the HAMP linker and between the HAMP linker and the output module remain unknown. It is clear from the properties of hybrid sensor kinases that apparently minor differences in HAMP linker sequence have significant effects on HAMP linker function. Our present lack of understanding makes it impossible to predict whether a given hybrid that uses the HAMP linker will respond properly to ligand or not, complicating the task of modeling HAMP linker function. It is to be hoped that structural data and information about interactions between the HAMP linker and the rest of the sensor protein will clarify the role of this widely used signal-transducing element.
Acknowledgments
We thank Tracy Raivio and Tom Silhavy for strains, plasmids, and helpful information about CpxA. We thank members of our laboratory for helpful advice and interest.
This study was supported by Public Health Service grant GM36877 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Ames, P., and J. S. Parkinson. 1988. Transmembrane signaling by bacterial chemoreceptors: E. coli transducers with locked signal output. Cell 55:817-826. [DOI] [PubMed] [Google Scholar]
- 2.Ansaldi, M., C. Jourlin-Castelli, M. Lepelletier, L. Theraulaz, and V. Mejean. 2001. Rapid dephosphorylation of the TorR response regulator by the TorS unorthodox sensor in Escherichia coli. J. Bacteriol. 183:2691-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appleman, J. A., and V. Stewart. 2003. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J. Bacteriol. 185:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signaling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 5.Baumgartner, J. W., C. Kim, R. E. Brissette, M. Inouye, C. Park, and G. L. Hazelbauer. 1994. Transmembrane signaling by a hybrid protein: communication from the domain of chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor EnvZ. J. Bacteriol. 176:1157-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosius, J. 1989. Superpolylinkers in cloning and expression vectors. DNA 8:759-777. [DOI] [PubMed] [Google Scholar]
- 8.Butler, S. L., and J. J. Falke. 1998. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry (Moscow) 37:10746-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadaban, M. J. 1976. Transposition and fusion of lac genes to selected promoters in Escherichia coli using bacteriophages lambda and mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 10.Collins, L. A., S. M. Egan, and V. Stewart. 1992. Mutational analysis reveals functional similarity between NarX, a nitrate sensor in Escherichia coli K-12, and the methyl-accepting chemotaxis proteins. J. Bacteriol. 174:3667-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danese, P. N., and T. Silhavy. 1998. CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darwin, A. J., J. Li, and V. Stewart. 1996. Analysis of nitrate regulatory protein NarL-binding sites in the fdnG and narG operon control regions of Escherichia coli K-12. Mol. Microbiol. 20:621-632. [DOI] [PubMed] [Google Scholar]
- 13.Falke, J. J., R. B. Bass, S. L. Butler, S. A. Chervitz, and M. A. Danielson. 1997. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 13:457-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falke, J. J., and S.-H. Kim. 2000. Structure of a conserved receptor domain that regulates kinase activity: the cytoplasmic domain of bacterial taxis receptors. Curr. Opin. Struct. Biol. 10:462-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 16.Feng, X., J. Baumgartner, and G. Hazelbauer. 1997. High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J. Bacteriol. 179:6714-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41:139-227. [DOI] [PubMed] [Google Scholar]
- 18.Gunn, J. S., E. L. Hohmann, and S. I. Miller. 1996. Transcriptional regulation of Salmonella virulence: a PhoQ periplasmic domain mutation results in increased net phosphotransfer to PhoP. J. Bacteriol. 178:6369-6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hedblom, M., and J. Adler. 1980. Genetic and biochemical properties of Escherichia coli mutants with defects in serine chemotaxis. J. Bacteriol. 144:1048-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Island, M. D., and R. J. Kadner. 1993. Interplay between the membrane-associated UhpB and UhpC regulatory proteins. J. Bacteriol. 175:5028-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, T., and M. Inouye. 1993. Ligand binding to the receptor domain regulates the ratio of kinase to phosphatase activities of the signaling domain of the hybrid Escherichia coli transmembrane receptor, Taz1. J. Mol. Biol. 232:484-492. [DOI] [PubMed] [Google Scholar]
- 22.Kalman, L. V., and R. P. Gunsalus. 1990. Nitrate-independent and molybdenum-independent signal transduction mutations in narX that alter regulation of anaerobic respiratory genes in Escherichia coli. J. Bacteriol. 172:7049-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaspar, S., R. Perozzo, S. Reinelt, M. Meyer, K. Pfister, L. Scapozza, and M. Bott. 1999. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol. Microbiol. 33:858-872. [DOI] [PubMed] [Google Scholar]
- 24.Krikos, A., M. P. Conley, A. Boyd, H. C. Berg, and M. I. Simon. 1985. Chimeric chemosensory transducers of Escherichia coli. Proc. Natl. Acad. Sci. USA 82:1326-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kristich, C. J., G. D. Glekas, and G. W. Ordal. 2003. The conserved cytoplasmic module of the transmembrane chemoreceptor McpC mediates carbohydrate chemotaxis in Bacillus subtilis. Mol. Microbiol. 47:1353-1366. [DOI] [PubMed] [Google Scholar]
- 26.Lopilato, J., S. Bortner, and J. Beckwith. 1986. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol. Gen. Genet. 205:285-290. [DOI] [PubMed] [Google Scholar]
- 27.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Mao, H., P. S. Cremer, and M. D. Manson. 2003. A sensitive, versatile microfluidic assay for bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 100:5449-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masui, Y., T. Mizumo, and M. Inouye. 1984. Novel high-level expression cloning vehicles: 104-fold amplifications of Escherichia coli minor protein. Bio/Technology 2:81-85. [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Ottemann, K. M., W. Xiao, Y.-K. Shin, and D. E. Koshland, Jr. 1999. A piston model for transmembrane signaling of the aspartate receptor. Science 285:1751-1754. [DOI] [PubMed] [Google Scholar]
- 32.Park, H., and M. Inouye. 1997. Mutational analysis of the linker region of EnvZ, an osmosensor in Escherichia coli. J. Bacteriol. 179:4382-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peach, M. L., G. Hazelbauer, and T. P. Lybrand. 2002. Modeling the transmembrane domain of bacterial chemoreceptors. Protein Sci. 11:912-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabin, R., and V. Stewart. 1993. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 175:3259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabin, R., and V. Stewart. 1993. Either of two functionally redundant sensor proteins, NarX and NarQ, is sufficient for nitrate regulation in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 88:6941-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rainwater, S., and P. M. Silverman. 1990. The Cpx proteins of Escherichia coli K-12: evidence that cpxA, ecfB, ssd, and eup mutations all identify the same gene. J. Bacteriol. 172:2456-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raivio, T. L., M. W. Laird, J. C. Joly, and T. J. Silhavy. 2000. Tethering of CpxP to the inner membrane prevents spheroplast induction of the Cpx envelope stress response. Mol. Microbiol. 37:1186-1197. [DOI] [PubMed] [Google Scholar]
- 38.Raivio, T. L., D. L. Popkin, and T. J. Silhavy. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J. Bacteriol. 181:5263-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raivio, T. L., and T. J. Silhavy. 1999. The σe and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2:159-165. [DOI] [PubMed] [Google Scholar]
- 40.Repik, A., A. Rebbapragada, M. S. Johnson, J. O. Haznedar, I. B. Zhulin, and B. L. Taylor. 2000. Pas domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh, M., B. Berger, P. S. Kim, J. M. Berger, and A. G. Cochran. 1998. Computational learning reveals coiled coil-like motifs in histidine kinase linker domains. Proc. Natl. Acad. Sci. USA 95:2738-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809-821. [DOI] [PubMed] [Google Scholar]
- 43.Stewart, G. S. A. B., S. Lubinsky-Mink, C. G. Jackson, A. Kassel, and J. Kuhn. 1986. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid 15:172-181. [DOI] [PubMed] [Google Scholar]
- 44.Stewart, V. 2003. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem. Soc. Trans. 33:1-10. [DOI] [PubMed] [Google Scholar]
- 45.Stewart, V. 1982. Requirement of FNR and NarL functions for nitrate reductase expression in Escherichia coli K-12. J. Bacteriol. 151:1320-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart, V., and J. J. Parales. 1988. Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli. J. Bacteriol. 170:1589-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart, V., and R. S. Rabin. 1995. Dual sensors and dual response regulators interact to control nitrate- and nitrite-responsive gene expression in Escherichia coli, p. 233-252. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 48.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 49.Utsumi, R., R. E. Brissette, A. Rampersaud, S. A. Forst, K. Oosawa, and M. Inouye. 1989. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science 245:1246-1249. [DOI] [PubMed] [Google Scholar]
- 50.Ward, S. M., A. Delgado, R. P. Gunsalus, and M. D. Manson. 2002. A NarX-Tar chimera mediates repellent chemotaxis to nitrate and nitrite. Mol. Microbiol. 44:709-719. [DOI] [PubMed] [Google Scholar]
- 51.Weerasuriya, S., B. M. Schneider, and M. D. Manson. 1998. Chimeric chemoreceptors in Escherichia coli: signaling properties of Tar-Tap and Tap-Tar hybrids. J. Bacteriol. 180:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams, S. B., and V. Stewart. 1997. Discrimination between structurally related ligands nitrate and nitrite controls autokinase activity of the NarX transmembrane signal transducer of Escherichia coli. Mol. Microbiol. 26:911-925. [DOI] [PubMed] [Google Scholar]
- 53.Williams, S. B., and V. Stewart. 1999. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol. Microbiol. 33:1093-1102. [DOI] [PubMed] [Google Scholar]
- 54.Williams, S. B., and V. Stewart. 1997. Nitrate- and nitrite-sensing protein NarX of Escherichia coli K-12: mutational analysis of the amino-terminal tail and first transmembrane segment. J. Bacteriol. 179:721-729. [DOI] [PMC free article] [PubMed] [Google Scholar]