Abstract
S5 is a small subunit ribosomal protein (r-protein) linked to the functional center of the 30S ribosomal subunit. In this study we have identified a unique amino acid mutation in Escherichia coli S5 that produces spectinomycin-resistance and cold sensitivity. This mutation significantly alters cell growth, folding of 16S ribosomal RNA, and translational fidelity. While translation initiation is not affected, both +1 and −1 frameshifting and nonsense suppression are greatly enhanced in the mutant strain. Interestingly, this S5 ribosome ambiguity-like mutation is spatially remote from previously identified S5 ribosome ambiguity (ram) mutations. This suggests that the mechanism responsible for ram phenotypes in the novel mutant strain is possibly distinct from those proposed for other known S5 (and S4) ram mutants. This study highlights the importance of S5 in ribosome function and cell physiology, and suggests that translational fidelity can be regulated in multiple ways.
Keywords: E. coli, 30S ribosome, S5 protein, ram mutant, spectinomycin resistance
INTRODUCTION
Ribosomes are macromolecular machines responsible for one of the most important cellular events: translation of messenger RNA (mRNA) into protein. Ribosomes of all organisms are comprised of two asymmetric subunits, each of which is composed of many ribosomal proteins (r-proteins) and at least one ribosomal RNA (rRNA). In prokaryotes, the two functionally distinct subunits that form the 70S ribosome are the small (30S) and large (50S) subunits. It is well established that each of the main structural domains, the body, platform, and head, of the 30S subunit are involved in binding mRNA and selecting transfer RNA (tRNA). Thus, it is critical that the individual domains of the 30S subunits are accurately assembled and appropriately aligned for proper decoding and maintenance of reading frame during translation (Ramakrishnan 2002).
Only a limited number of r-proteins, S4, S5, and S12, have been genetically (for review, see Kurland et al. 1996) and structurally (Wimberly et al. 2000; Brodersen et al. 2002) linked to the functional center of the 30S subunit. Of these three r-proteins, a role for S12 in decoding is the most obvious, since it is the only small subunit r-protein proximal to the decoding center and located on the interface side of the subunit shoulder (Wimberly et al. 2000; Brodersen et al. 2002). Ribosomes containing specific S12 mutations result in hyperaccurate (restrictive) phenotypes (for review, see Kurland et al. 1996) demonstrating the important role of this protein in decoding. Additionally, r-protein S12 makes direct contacts with 16S rRNA residues that are implicated in mRNA binding and tRNA selection (Wimberly et al. 2000; Brodersen et al. 2002; Ogle et al. 2002). In contrast with S12 mutations, ribosomes containing specific mutations in S4 are less accurate than wild-type ribosomes and represent a loss of function (Donner and Kurland 1972; Funatsu et al. 1972a; Van Acken 1975; Schnier et al. 1985). Thus, S12 and S4 mutations have opposite effects on selection of cognate tRNA. r-Protein S4 forms part of the shoulder on the solvent surface of the body of the 30S subunit. Although S4 is somewhat removed from the decoding center (Wimberly et al. 2000), structural studies have begun to explain how S4 participates in decoding (Ogle et al. 2002, 2003). r-Protein S5 interacts directly with S4, and is involved in an extended interface between the shoulder and platform of the 30S subunit (Wimberly et al. 2000; Brodersen et al. 2002). S5 is located at the top of the body near the head of the 30S subunit, and is on the solvent surface of this subunit (Wimberly et al. 2000). Therefore, S5 is also quite remote from the decoding center. Nevertheless, as with S4, certain mutations in S5 can result in error-prone ribosomes, suggesting that S5 influences decoding from a distance (Itoh and Wittmann 1973; Piepersberg et al. 1975a). Recently, a model showing how S12, S4, and S5 influence tRNA selection and translational fidelity has been proposed (Ogle et al. 2002, 2003). In this model, the 30S subunit undergoes a conformational rearrangement, or domain closure, during tRNA selection, which involves movement of the 30S subunit shoulder toward the functional interface of the subunit (Ogle et al. 2002). Upon domain closure, the contacts between S4 and S5 are disrupted to accommodate movement of the 30S subunit shoulder, including S12, toward the functional center (Ogle et al. 2002). The known mutations in both S4 and S5 that result in increased miscoding (ribosome ambiguity mutants [ram]) are localized to the interface between these two r-proteins. These mutations weaken the interactions between these r-proteins, thereby allowing the closed conformation to form more readily. Thus, this model suggests that S5 is linked to the decoding center via a network of defined interactions within the 30S subunit.
Many studies also suggest a central role for r-protein S5 during formation of functional sites within the 30S subunit. Assembly of S5 onto 16S rRNA-containing ribonucleoprotein particle (RNP) facilitates the binding of two additional r-proteins, S3 and S21 (Mizushima and Nomura 1970; Held et al. 1974). It has been suggested that these proteins aid in forming the tRNA-binding sites during assembly of 30S subunits (Vladimirov et al. 1985; Ramakrishnan et al. 1986). Consistent with these observations, in vitro reconstitution of 30S subunits in the absence of S5 results in the formation of particles that have diminished ability to bind tRNA and sediment at 28S (Nomura et al. 1969). Thus, changes in S5 may have far-reaching consequences on the function of the ribosome by virtue of its facultative role in forming and maintaining functional 30S subunit architecture.
Genetic analysis has often been used to gain an understanding of the functional significance of r-proteins, and S5 is no exception. Specific amino acid changes in S5 that result in fidelity defects (ram) (Itoh and Wittmann 1973; Piepersberg et al. 1975a) and in spectinomycin resistance (spcr) (Bollen et al. 1969; Funatsu et al. 1972b; De Wilde and Wittmann-Liebold 1973; Piepersberg et al. 1975b; Wittmann-Liebold and Greuer 1978) have been identified. Additionally, mutations in S5 that yield spectinomycin resistance in combination with cold sensitivity have been reported (Guthrie et al. 1969; Nomura 1987), but the specific amino acid change(s) responsible for these phenotypes have not. Analysis of such strains would offer a unique opportunity to understand S5 function as the combination of these two phenotypes have not been reported in other S5 mutant strains. Regrettably, the original cold-sensitive and spectinomycin-resistant strain is no longer viable (M. Nomura, pers. comm.). Thus, to further expand our understanding of the role of S5 in ribosome function, Escherichia coli strains with similar phenotypes to those previously described (cold-sensitivity and spcr) were selected (Guthrie et al. 1969). In these strains we have identified a single change of a highly conserved glycine at position 28 to an aspartate in S5 (S5[G28D]) that results in multiple in vivo and in vitro effects. This mutation in S5 results in defects in ribosome biogenesis, tRNA binding, and translational fidelity. These findings are of particular interest, since the position of this mutation is quite remote from the clustering of the other S5 (and S4) ram mutations and suggest that a more complex multifaceted model for regulation of translational fidelity must be invoked. Thus, a single point mutation in the rpsE gene allows for a better understanding of the role of S5 in organizing functional conformations within the 30S subunit.
RESULTS
In vivo genetics and selection of mutants
To isolate strains similar to those previously reported (Guthrie et al. 1969) a dual selection strategy was employed. E. coli strains that were resistant to spectinomycin (60 μg/mL) were first selected, followed by a second selection for cold sensitivity. Five independent isolates were identified that possessed both of these phenotypes, which were verified through multiple rounds of testing. Overexpression of wild-type S5 (rpsE) was shown to suppress the phenotypes associated with all five isolates, indicating that for each of these independent strains the chromosomal mutation likely occurred in the rpsE gene (data not shown) and not in any of the 16S rDNA genes or at other loci (Moazed and Noller 1987; Brink et al. 1994; Carter et al. 2000). The genomic rpsE genes were sequenced from the five mutants to identify the site(s) of mutation. All five independent isolates contained the same point mutation, suggesting that the selection was saturated. Four of the five mutants contained a single nucleotide change, resulting in an amino acid change of a highly conserved glycine to an aspartate at position 28 (G28D) (see Fig. 1; numbering corresponds to S5 sequence of E. coli). The fifth mutant contained this same change but also had a second mutation, which resulted in an aspartate replacing glycine at position 27. The phenotypes of the single and the double mutants were quite similar (data not shown); thus, only the single amino acid change was further characterized. To confirm that this change in S5 was solely responsible for the observed phenotypes, a P1 lysate from the S5(G28D) mutant strain was used to infect a wild-type strain. Given that spcr should be linked with our mutation, spcr transductants were selected. The selected strains were also cold sensitive, and sequencing of the rpsE gene from one of the strains revealed that G28D mutation was indeed present. This result effectively eliminated the possibility that additional genomic changes contribute to the observed phenotypes, and confirmed that these were due to a change of the highly conserved amino acid (glycine 28 to aspartate). Interestingly, the site of this mutation is proximal to, yet distinct from, amino acid changes in S5 that were previously reported to yield spcr (Fig. 1B; Bollen et al. 1969; Funatsu et al. 1972b; De Wilde and Wittmann-Liebold 1973; Wittmann-Liebold and Greuer 1978), and this mutation is positioned near the convergence of the major domains of the 30S subunit (see Fig. 1C; Wimberly et al. 2000. Thus, the dual selection for spcr in combination with cold sensitivity appears to have resulted in isolation of a strain carrying a novel mutation. This mutation appears to have unique properties, since this glycine 28 to aspartate was the only change isolated and it was found in multiple independent strains.
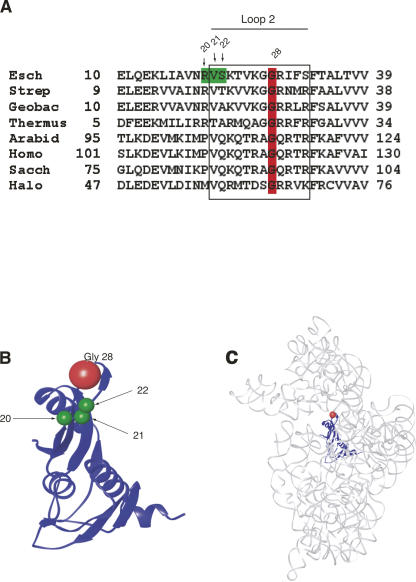
FIGURE 1.
Glycine 28 of S5 is highly conserved throughout phylogeny and located in loop 2 of the protein. (A) Sequence comparisons of loop 2 and its flanking regions of r-protein S5 (or its equivalent) from various species. A partial region of the S5 amino acid sequence is shown. The numbering of E. coli S5 is used throughout the manuscript. Conserved Gly 28 is highlighted in red and loop 2 is boxed. Positions 20, 21, and 22, highlighted in green in the E. coli sequence, are amino acids whose mutations were previously reported to result in spectinomycin resistance (Itoh and Wittmann 1973; Wittmann-Liebold and Greuer 1978). The S5 ribosomal sequences used in sequence alignment and their accession numbers in parenthesis are as follows: Esch: Escherichia coli (AAA58100), Strep: Streptococcus thermophilus (YP_142245), Geobac: Geobacillus stearothermophilus (1PKP), Ther: Thermus thermophilus (P27152), Arabid: Arabidopsis thaliana (P49688), Homo: Homo sapiens (NP_002943), Sacch: Saccharomyces cerevisiae (P25443), and Halo: Haloarcula marismortui (P26815). (B) Three-dimensional structure of r-protein S5 from T. thermophilus 30S subunits (Carter et al. 2000). Conserved Gly 28 is shown in red. Amino acids corresponding to positions 20, 21, and 22 in E. coli S5, which give spectinomycin resistance, are shown in green (Itoh and Wittmann 1973; Piepersberg et al. 1975b; Wittmann-Liebold and Greuer 1978). Ribbons (Carson 1997) were used to generate this and all structural figures within in the manuscript. (C) Three-dimensional structure of 16S rRNA with r-protein S5 from T. thermophilus 30S subunits solved in the presence of antibiotics, streptomycin, spectinomycin, and paramomycin (Carter et al. 2000) (1FJG.pdb). 16S rRNA is shown in gray, S5 is shown in blue, and glycine 28 is shown in red; all other r-proteins are omitted for clarity.
Growth properties of the S5(G28D) strain
While the cold-sensitive phenotype had been grossly established, careful examination of doubling times revealed a growth defect of the S5(G28D) mutant strain relative to the parental strain at all temperatures. Even at the permissive temperature (37°C), the S5(G28D) mutant strain had an exaggerated doubling time (43 min) compared to wild type (25 min), suggesting that while tolerated, this mutation is deleterious. As expected, at reduced temperature (20°C), growth is undetectable for the mutant strain (doubling time of >300 min versus 54 min for wild type). Similar growth rates were observed both in the presence and absence of spectinomycin in liquid and on solid media (data not shown), indicating that spectinomycin does not have an apparent effect on growth of the mutant strain. These results suggest that this mutant allele of S5 results in a strain, which is not only cold sensitive but also compromised for growth at all temperatures.
Analysis of ribosomes isolated from the S5(G28D) strain
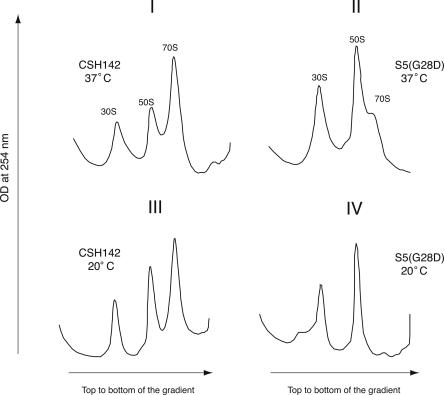
To begin to address the consequence of this mutation and to possibly identify the underlying cause of the growth defects, ribosomes isolated from the S5(G28D) strain were examined. When the mutant strain was grown at 37°C, the 30S and 50S subunit peaks were comparatively and consistently larger than those observed in wild type (Fig. 2, Panels I,II). Concomitantly, reduced amounts of 70S ribosomes and polysomes were routinely observed in profiles from the mutant strain (Fig. 2, Panel II), and the relative amounts of 70S ribosomes and polysomes in these profiles were somewhat variable. Figure 2 shows panels of representative profiles; polysomes were not observed due to an elongated run time that facilitated analysis of the free subunits and monosomes (see Materials and Methods). After a shift to the nonpermissive temperature, the ribosome profiles for the S5(G28D) strain were even more dramatically altered. Very few, if any, 70S ribosomes are evident at the nonpermissive temperature in the mutant strain (Fig. 2, Panel IV). Additionally, a distinct peak that sediments in a region consistent with pre-30S particles (Fig. 2, Panel IV) was observed in profiles from the mutant strain. This profile is consistent with ribosome biogenesis defects observed in other cold-sensitive strains (Guthrie et al. 1969; Nashimoto et al. 1971; Dammel and Noller 1995), and 16S rRNA is a component of this new peak (N. Kirthi, B. Raichoudhary, and G. Culver, unpubl.). The growth defects and abnormal ribosome profiles observed in this S5(G28D) mutant strain suggest that this mutation alters the pool of functional ribosomes in vivo.
FIGURE 2.
Altered ribosome profiles are observed in the S5(G28D) mutant strain. Sucrose gradient sedimentation analysis of ribosomes isolated from the wild-type/parental CSH142 and S5(G28D) mutant strains. Positions of the 30S, 50S, and 70S subunits are indicated. The gradients were pumped from the top to the bottom and the elution of the subunits was detected by monitoring optical density (OD) at 254 nm. Panels I and II correspond to polysome analysis of CSH142 and S5(G28D) grown at 37°C. Panels III and IV correspond to polysome profiles of CSH142 and S5(G28D) grown at 20°C. Polysomes are not resolved due to sedimentation conditions (see Materials and Methods).
In vitro characterization of 30S subunits and ribosomes containing S5(G28D)
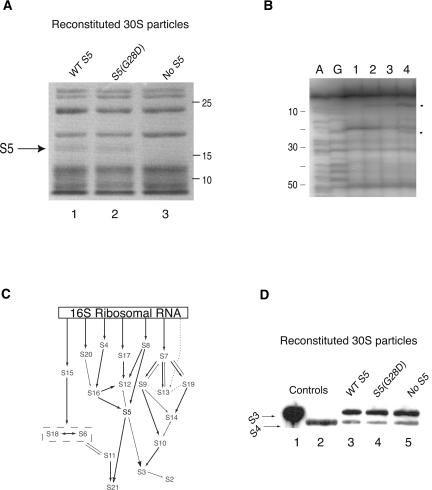
To allow further investigation of the role of S5, and particularly this specific mutation, 30S subunits containing S5(G28D) were reconstituted in vitro. The S5(G28D) protein was purified as previously described for the recombinant wild-type protein (see Materials and Methods). Reconstitution of 30S subunits using natural 16S rRNA and a complete set of recombinant small subunit r-proteins including the S5(G28D) mutant protein ensures that all other components are wild type and mature. Additionally, in vitro reconstitution allows particles formed in the absence of S5 (the gene encoding S5 is essential in vivo; M. Bububenko and D. Court, pers. comm.) to be formed as a control and compared with earlier work (Nomura et al. 1969). Particles that sediment near, but not coincident with, wild-type 30S subunits were observed when reconstitution was performed using S5(G28D) or in the absence of S5 (Fig. 3A). The altered sedimentation profile in the absence of S5 is consistent with previous work (Nomura et al. 1969). These results suggest that particles formed without S5 or with S5(G28D) are not as compact as wild-type 30S subunits.
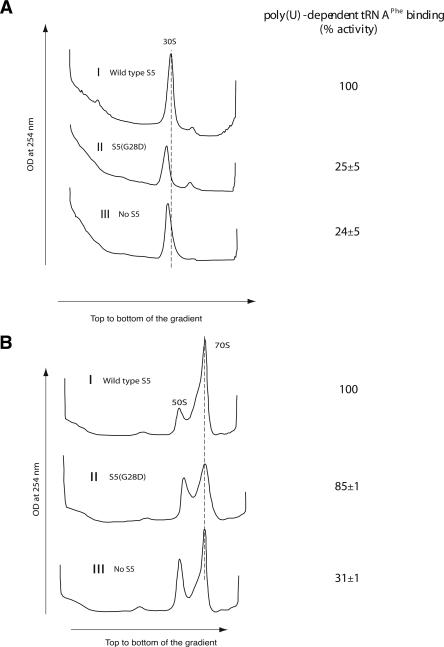
FIGURE 3.
Analysis of in vitro reconstituted particles containing S5(G28D) mutant protein. (A) Sucrose gradient sedimentation analysis of in vitro reconstituted 30S particles. Position of the 30S subunit is indicated. The gradients were pumped from the top to the bottom and the elution of the subunits was detected by monitoring OD at 254 nm. Panel I shows the 30S peak for the in vitro reconstituted 30S particle with wild-type S5. Panels II and III show the 30S-like peak for the in vitro reconstituted 30S with S5(G28D) and no S5, respectively. (B) Sedimentation analysis of ribosomal subunit association using in vitro reconstituted 30S particles and natural 50S subunits. Positions of 50S and 70S subunits are indicated. Subunit association of natural 50S subunits (40 pmol) with 30S particles reconstituted with wild-type S5 (40 pmol) (Panel I), S5(G28D) (Panel II), and no S5 (Panel III) are shown. Sedimentation is from left to right, and the absorbance was monitored at 254 nm. Poly-U-dependent tRNA binding activity of the particles is given in the right-hand column.
To begin to assess the functional consequence of the mutation of this highly conserved glycine residue, in vitro reconstituted 30S particles were assayed for their ability to associate with natural 50S subunits to form 70S ribosomes (Fig. 3B). In all cases (wild-type S5, S5[G28D], and no S5), the majority of the reconstituted particles were competent for association with 50S subunits, as indicated by the appearance of 70S peaks (Fig. 3B) and the disappearance of 30S peaks (Fig. 3, cf. A and B). The mutation of glycine to aspartate in S5 does appear to produce broader 70S peaks but is clearly not inhibitory. Thus, our in vitro results correlate well with our in vivo studies and suggest that the S5(G28D) mutation is not inhibitory for all 30S subunit functions.
In vitro reconstituted 30S subunits and 70S ribosomes containing wild-type S5, S5(G28D), and no S5 were assessed for their ability to bind tRNA to further evaluate the functional consequences of S5(G28D) mutation. The 30S-like subunits reconstituted with S5(G28D) or without S5 showed significantly reduced levels of tRNA binding (∼25%) (Fig. 3A) when compared to the 30S particles reconstituted with wild-type S5. Interestingly, in the context of 70S ribosomes, the S5(G28D) mutation did not have such a dramatic effect on tRNA binding and showed ∼85% binding compared to wild type (Fig. 3B). This “rescue” of tRNA binding in 70S ribosomes requires the presence of S5, as the subunits lacking S5 did not show the same enhanced activity in 70S ribosomes (see Fig. 3). This difference between the mutant protein and no S5 indicates that the S5(G28D) protein is incorporated into 30S subunits, and this contention is further validated by additional data (see below). These results are consistent with a role for S5 in tRNA binding and 30S subunit functions, and suggest that the specific mutation of the highly conserved glycine at position 28 to aspartate has dramatic effects on these roles.
In vivo functional analysis of ribosomes containing S5(G28D)
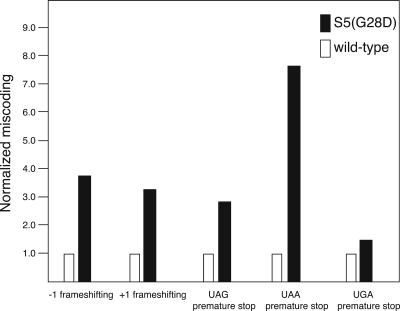
Although our isolated mutation in S5 maps outside of the region previously identified to have effects on translational fidelity (see Fig. 7; Piepersberg et al. 1975a), preliminary in vitro studies suggested that, in addition to an effect on cognate tRNA binding (Fig. 3), this glycine 28 to aspartate mutation of S5 could favor binding of near-cognate tRNA (data not shown). Therefore, the accuracy of ribosomes containing S5(G28D) was examined in vivo using a lac− strain containing the S5(G28D) chromosomal mutation (see Materials and Methods). Plasmids carrying either the wild-type lacZ gene or the lacZ gene with mutations in the 5′-coding region enable miscoding to be assessed by monitoring β-galactosidase activity in the parental and mutant strains. A moderate to significant affect on nonsense suppression was observed in the S5(G28D) mutant strain compared to the parental (wild-type) strain (Fig. 4). In addition, a dramatic increase in both +1 and −1 frameshifting was also observed in strains bearing the S5(G28D) mutation (Fig. 4). However, this S5 mutation did not significantly affect the fidelity of translation initiation in vivo (data not shown). These results clearly illustrate that the mutation of glycine 28 to aspartate in S5 yields a loss of function phenotype, very similar to those observed in the previously characterized S5 ram strains (Itoh and Wittmann 1973; Piepersberg et al. 1975a). These results in combination with the exacerbated phenotype in the presence of streptomycin (data not shown) suggest the mutation of S5 glycine 28 to aspartate produces a novel ram strain. Since this mutation site is structurally remote from the other ram sites (Itoh and Wittmann 1973; Piepersberg et al. 1975a), it is possible that translational fidelity could be modulated in multiple ways.
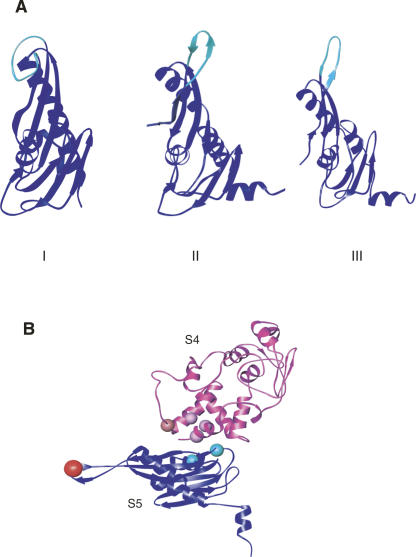
FIGURE 7.
Position of structural and functional elements in r-protein S5 (A) Three-dimensional structures of r-protein S5 reveal different conformations of loop 2. S5 is colored in blue and loop 2 of S5 is colored in cyan. (Panel I) Three-dimensional structure of free r-protein S5 from Geobacillus stearothermophilus (Ramakrishnan & White 1992) (1PKP.pdb). (Panel II) Three -dimensional structure of S5 from 30S ribosomal subunit of T. thermophilus (Wimberly et al. 2000) (1J5E.pdb). (Panel III) Three -dimensional structure of S5 from 70S ribosome of E. coli (Schuwirth et al. 2005) (2AW7.pdb). (B) Positions of functional mutations in S4 and S5 shown on the three-dimensional structure of r-proteins S4 and S5 from the 30S subunit of T. thermophilus (Wimberly et al. 2000) (1J5E.pdb). r-Protein S4 is shown in magenta and the balls represent the sites of ram mutations on r-protein S4 (for review, see Kurland et al. 1996). r-Protein S5 is shown in blue and the sites of ram mutations in S5 (Itoh and Wittmann 1973) are represented as cyan balls. Conserved glycine 28, which results in a ram mutant (this study), is shown in red. 16S rRNA and all the other r-proteins are omitted for clarity.
FIGURE 4.
Translational fidelity is affected in the S5(G28D) mutant strain. Nonsense suppression and frameshifting in β-galactosidase activity by S5(G28D). Effect of S5(G28D) mutation stop codon read-through and frameshifting is expressed as normalized values. Wild-type (white) and ΔlacZ S5(G28D) (black) strains were transformed with plasmids (see Materials and Methods) encoding constitutively expressed β-galactosidase with N-terminal nonsense codons or frame shifts or the wild-type sequence. The values are an average of at least three independent experiments.
Probing 30S subunits and ribosomes containing S5(G28D)
A possibility existed that the observed tRNA binding and translational fidelity defects could be pleotrophic and not due directly to the S5(G28D) mutation. Thus, the composition and structure of subunits containing S5(G28D) was assessed. While our tRNA-binding data indirectly suggested that the mutant protein was incorporated into subunits (see Fig. 3), analysis of the protein composition of in vitro reconstituted particles directly indicates that, like wild-type S5, S5(G28D) is associated with the subunits (Fig. 5A, lane 2), and this band is clearly absent from proteins isolated from reconstitutions lacking S5 (Fig. 5A, lane 3). Additionally, chemical probing and primer extension analysis of in vitro reconstituted particles indicates that S5(G28D) yields similar footprints (Fig. 5B, lane 3) to those reported (Stern et al. 1988a, b, c) and observed for wild-type S5 (Fig. 5A, lane 3). In contrast, footprints are clearly lacking in particles reconstituted in the absence of S5 (Fig. 5B, cf lanes 2,3 and 4).
FIGURE 5.
Analysis of r-proteins associated with 30S subunits reconstituted with S5(G28D). (A) SDS-PAGE analysis of in vitro reconstituted 30S subunits. Positions of S5 and approximate molecular weights are indicated. Lane 1, 2, and 3 correspond to in vitro reconstituted 30S particles with wild-type S5, S5(G28D), and no S5, respectively. (B) Primer extension analysis of in vitro reconstituted 30S subunits containing S5(G28D) protein. A and G correspond to dideoxy sequencing lanes. Lane 1: unmodified 30S subunits reconstituted with wild-type S5. All other lanes have been modified with kethoxal. Lanes 2, 3, and 4 correspond to reconstituted 30S particles with wild-type S5, S5(G28D), and no S5, respectively. Circles denote the sites of protection in the presence of r-protein S5. Primer 161 was used in this extension. (C) Modified in vitro assembly map for 30S subunits (Mizushima and Nomura 1970; Held et al. 1974; Grondek and Culver 2004). 16S rRNA is represented by a rectangle and arrows indicate dependency between components for association with 16S rRNA. (D) Western blot analysis of in vitro reconstituted 30S particles. Anti-S3 and -S4 antibodies were used to detect these r-proteins. Lanes 1 and 2 are purified recombinant S3 and S4 proteins, which are control proteins. Lanes 3, 4, and 5 have 40 pmol of sucrose gradient purified, in vitro reconstituted 30S particles with wild-type S5, S5(G28D), and no S5, respectively.
Although the overall r-protein patterns appear similar between reconstituted particles containing wild-type or mutant S5 (Fig. 5A, lanes 1,2), the significant functional differences between ribosomes containing these proteins necessitated more detailed analysis. Although S4 is a primary binding protein and not dependent on S5 for assembly (Fig. 5C; Held et al. 1974), defects similar to those observed for S5(G28D) have also been reported in S4 studies (Nomura et al. 1969; Donner and Kurland 1972; Funatsu et al. 1972a; Van Acken 1975; Schnier et al. 1985). Thus, the association of S4 with the subunits containing S5(G28D) was analyzed. Immunoblotting reveals that S4 is associated with 30S subunits assembled with S5(G28D) (Fig. 5D, lane 4) and in the absence of S5 (Fig. 5D, lane 5), suggesting that the functional defects are not due to the absence of S4. To further ascertain if the S5 mutant protein disrupted assembly events, the presence of ribosomal protein S3 in the reconstituted particles was analyzed. The assembly of S3 in the context of the S5 mutant is significant for two main reasons. S3 is partially dependent on S5 for assembly (Fig. 5C; Held et al. 1974), and its absence during assembly of 30S subunits has been linked to defects in tRNA binding (Nomura et al. 1969; Ramakrishnan et al. 1986). Although S3 is partially dependent on S5 for assembly (see Fig. 5C; Held et al. 1974), it is present when particles are reconstituted with S5(G28D) and with no S5 (Fig. 5D, lanes 4,5). Additionally, analysis of in vivo particles and in vitro reconstituted 30S subunits by mass spectrometry revealed that all of the small subunit r-proteins, including S21, which is partially dependent on S5 for assembly (Fig. 5C), were present when subunits were formed with either wild type or G28D S5 (data not shown). Thus, it is unlikely that the defects observed in tRNA binding and translational fidelity in subunits containing S5(G28D) result from the lack of other functionally relevant r-proteins.
While a model for how previously reported mutations in S5 (and S4) result in ram phenotypes has been put forth (Ogle et al. 2002), this model does not obviously explain the results observed for S5(G28D). Position 28 of S5 is quite remote from the S4/S5 interface (see Fig. 7B), and thus does not readily fit the current model for how the ram mutations alter tRNA selection (Ogle et al. 2002). To gain a better understanding of the effect of S5(G28D) on ribosome conformation, which could account for the functional defects described above, chemical probing and primer extension analysis were performed on 30S and 70S particles isolated from either wild-type or S5(G28D) strains.
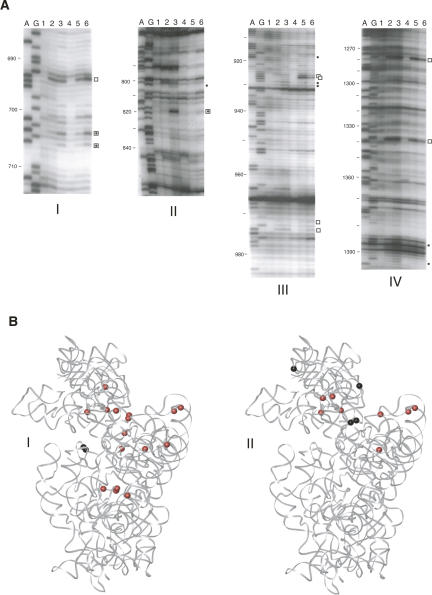
The 30S subunits containing the mutant protein were more highly modified than those containing wild-type S5 (Fig. 6A). In 30S subunits containing S5(G28D) the majority of reactivity changes are at sites implicated in mRNA and tRNA binding (Moazed and Noller 1986; Yusupova et al. 2001), intersubunit connections (Merryman et al. 1999; Yusupov et al. 2001), and functional pseudoknot formation (Powers and Noller 1991; Poot et al. 1998) and helices (Woese et al. 1980). Additionally, the majority of reactivity changes attributed to r-protein footprints are similar to those previously reported (Fig. 6A; Powers et al. 1988; Stern et al. 1988a,b). These results are highly consistent with our in vitro (Fig. 3) and in vivo (Fig. 4) data, and suggest that S5(G28D) mutant protein alters the architecture of functional sites within the 30S subunit but does not grossly alter the binding of the other r-proteins.
FIGURE 6.
Ribosomal subunits of S5(G28D) have altered 16S rRNA conformation. (A) Primer extension analysis of 30S subunits and 70S ribosomes isolated from the S5(G28D) strain. A and G correspond to dideoxy sequencing lanes. Lanes 1 and 4 are unmodified 30S subunits and 70S ribosomes isolated from the wild-type S5 strain. All other lanes have been modified with kethoxal. Lanes 2 and 5 are 30S subunits and 70S ribosomes, respectively, isolated from the wild-type S5 strain. Lanes 3 and 6 are 30S subunits and 70S ribosomes, respectively, isolated from the S5(G28D) strain. Filled circles denote the sites of altered reactivity in the S5(G28D) mutant 30S subunits, open squares denote sites of altered reactivity in the S5(G28D) mutant 70S ribosomes, and open squares with filled circles inside denote sites of altered reactivity in both mutant 30S subunits and 70S ribosomes. Panels I, II, III, and IV correspond to primer extensions done with primers 795, 1046 (Panels II,III), and 1490, respectively. (B) Kethoxal probing data mapped on the tertiary structure of E. coli 30S subunit (Schuwirth et al. 2005). Red circles denote enhancement in reactivity and black circles denote protections. Panels I and II show the guanine residues with altered reactivity in S5(G28D) 30S subunits and 70S ribosomes compared to wild-type particles, respectively. The 16S rRNA is shown in gray, and none of the r-proteins are shown.
The number of reactivity differences is greater in mutant 30S subunits than in the corresponding 70S ribosomes (Fig. 6A,B). Since the S5(G28D) mutation has some effect on 30S subunit biogenesis (see Fig. 2), it is possible that the differences arise from probing a heterogeneous population of 30S-like particles. In contrast, the 30S subunit components of the 70S ribosomes have been functionally selected via subunit association, and therefore might represent a more homogeneous population. Additionally, given our fidelity results in the S5(G28D) mutant strain (see Fig. 4), understanding differences between wild-type and these S5 mutant 70S ribosomes will yield more functionally relevant information. Many of the modifications that were observed in helical elements of the mutant 30S subunits are not present in the corresponding 70S ribosomes (data not shown; nucleotides 299, 301, 302, 928, 929, 1182, 1385, and 1392; Fig. 6A,B). Also, the changes at the intersubunit connections are less pronounced or absent in the 70S ribosomes (data not shown; nucleotides 703, 705, 803, 818, and 1094; Fig. 6A,B). These data are consistent with our in vitro results (see Fig. 3). Most importantly, the majority of reactivity differences between wild-type and S5(G28D) 70S ribosomes are found at functional sites, even more so than in the 30S subunits (Fig. 6A,B). Again, sites that have been implicated in tRNA and mRNA binding are differently modified in the mutant ribosomes (data not shown; nucleotides 693, 925, 926, 971, 973, and 1338; Fig. 6A,B; Moazed and Noller 1990; Yusupova et al. 2001). The changes are concentrated at the bottom of the head, along the neck, and in the top of the platform (Fig. 6B). These changes in accessibility to chemical probes are quite distinct from those observed when subunits containing one of the S4 ram mutants, which maps to the S4/S5 interface, were probed (Allen and Noller 1989). These S4 ram containing ribosomes showed differential reactivity at only two nucleotides, A8 and A26 (Allen and Noller 1989). Thus, it appears that the structural changes associated with this S5 mutant protein are distinct from those associated with another ram mutation. Our data suggest that the alignment of the head and platform is altered in the S5 mutant ribosomes, and this could have a significant impact on the functional capacity of the ribosomes, especially in maintenance of the reading frame.
DISCUSSION
In an attempt to gain insight into the functional role of r-protein S5, E. coli strains with spectinomycin-resistant and cold-sensitive phenotypes were selected. We identified a unique mutation in the rpsE gene of E. coli that results in a glycine 28 to aspartate (G28D) substitution in S5. This mutation is responsible for the selected phenotypes as well as the marked effect on ribosome function, namely, tRNA binding and translational fidelity. Thus, this single amino acid change, in the context of this large macromolecular complex, has a profound effect on ribosome function. This mutation is distinct from previously identified ribosome ambiguity mutations, and thus offers new insight into the role of S5 in maintenance of translational fidelity and reveals that translational fidelity can be modulated in different manners.
Spectinomycin resistance of the S5(G28D) strain
One phenotype that our mutant strain exhibits is spectinomycin resistance. Glycine 28 is located in loop 2 of S5 (Ramakrishnan and White 1992), and is within 5 Å of the spectinomycin binding site in the 30S subunit (Carter et al. 2000). Spectinomycin binding is disrupted in ribosomes containing the S5(G28D) mutation (data not shown), and this lack of binding would account for resistance. It is possible that spectinomycin, a fused ring antibiotic, cannot bind the mutant ribosomes due to introduction of a bulky charged aspartate residue in the place of the highly conserved glycine. Earlier studies have shown that mutations at other positions of loop 2 in S5 (residues 20, 21, or 22) (Itoh and Wittmann 1973; Piepersberg et al. 1975b; Wittmann-Liebold and Greuer 1978) and mutation of specific 16S rRNA residues (Sigmund et al. 1984; Makosky and Dahlberg 1987) also result in spcr strains (Fig. 1A,B); however, unlike the S5(G28D) mutation, cold sensitivity (Piepersberg et al. 1975b; Wittmann-Liebold and Greuer 1978; Sigmund et al. 1984; Makosky and Dahlberg 1987) and translational fidelity defects (Piepersberg et al. 1975a; Andersson et al. 1986) have not been associated with these mutations. Thus, our mutation appears to be distinct from those previously characterized, and was likely identified due to selection for both spcr and cold-sensitive phenotypes. The isolation of a strain bearing a single amino acid change in r-protein S5 that displays such remarkable phenotypes clearly indicates the importance of S5 for ribosome formation and function.
Conservation and position of Glycine 28 of S5
The mutation that we have isolated occurs at a highly conserved glycine residue (G28), which is located in loop 2 of S5 (Fig. 1; Ramakrishnan and White 1992). The overall identity between S5 proteins of E. coli and Homo sapiens is only about 29%; however, the glycine at position 28 is conserved in all current prokaryotic and eukaryotic S5 sequences (Fig. 1A). Additionally, the conservation does not extend to the whole of loop 2 (Fig. 1A). Of particular note are the amino acids at positions 20–22 of S5, which are in the same loop as glycine 28 (Fig. 1B; Ramakrishnan and White 1992) and can also be mutated to result in spcr (Sigmund et al. 1984; Makosky and Dahlberg 1987). However, conservation at these positions is low (Fig. 1A). While mutation of these other residues in loop 2 yield resistance to spectinomycin, changes at these other loop 2 residues have not been associated with cold sensitivity or ribosome ambiguity (Itoh and Wittmann 1973; Piepersberg et al. 1975a, b; Wittmann-Liebold and Greuer 1978). Additionally, mutation of G28 to D in S5 appears to be the only change to yield both spcr and cold sensitivity. Thus, the functional importance of glycine 28 to S5 revealed in our work is underscored by its marked level of conservation.
Structural studies also suggest that loop 2 of S5 may have functional significance. The positioning of this loop differs in the structure of free S5 (Ramakrishnan and White 1992) compared to the structure of S5 as part of the 30S subunit (Brodersen et al. 2002). In the structure of S5 alone (Fig. 7A, Panel I; Ramakrishnan and White 1992) this loop is folded back on the body of the protein, while in the T. thermophilus 30S subunit (Fig. 7A, Panel II; Wimberly et al. 2000) and E. coli 70S ribosome (Fig. 7A, Panel III; Schuwirth et al. 2005) it is an extended beta sheet. Our findings that mutation of the conserved glycine in this loop has a dramatic effect on function concur with the proposal that this loop of S5 is important for the appropriate orientation of the head and body of the 30S subunit, and therefore crucial for the function of the ribosome (Carter et al. 2000). Additionally, elements of loop 2 of S5 form part of a tunnel that defines the mRNA path through the 30S subunit (Yusupova et al. 2001). Thus, it is possible that substitution of glycine 28 for aspartate in this loop alters the formation or mobility of the loop, disrupting the appropriate positioning of this central element, and thereby affecting 30S subunit assembly, structure, and function.
Functional consequences of the S5(G28D) mutation
While finding that a mutation in ribosomal protein S5 has an effect on translational fidelity is not surprising, that such a mutation is located in loop 2 of S5 is surprising. The two previously characterized ram mutations in S5 map to the interface with S4 and are proximal to the ram mutations in S4 (see Fig. 7B; Itoh and Wittmann 1973). The positioning and the amino acid changes of these mutations led to an elegant model whereby destabilization of the S4/S5 interface promoted closure of the 30S subunit, and thus allowed compromised tRNA selection (Ogle et al. 2002). Glycine 28 in S5 is remote from the S4/S5 interface, and this glycine does not appear to interact with other r-proteins or 16S rRNA (Fig. 7B; Wimberly et al. 2000). Thus, it is unlikely that substitution of glycine 28 for aspartate in S5 manifests its effects in an identical manner as the other ram mutations. The ribosomes containing S5(G28D) show altered reactivity to chemical probes compared to wild-type ribosomes (Fig. 6), and these changes are distinct from other ram mutations (Allen and Noller 1989). These reactivity changes correlate very well with observed changes in the accuracy of decoding by S5(G28D) ribosomes. Many of the changes are observed in the neck of the 30S subunit, and therefore it is possible that the deleterious effects associated with this change result from a misalignment of the head relative to the body and platform. Again, this would be consistent with a hypothesized role for loop 2 of S5 (Carter et al. 2000). The phenotypic changes associated with this novel S5 mutation could result from an indirect effect at the S4/S5 interface due to this remote mutation. Alternatively, changes in the conformation of loop 2 due to this single amino acid change or changes associated with the dramatic changes in chemical/physical properties that occurred when aspartate is substituted for glycine could be responsible for these findings. Overall, our findings could suggest that there are different means by which changes in S5 result in ram phenotypes and dramatically alter the functional capabilities of ribosomes.
MATERIALS AND METHODS
Selection of spectinomycin-resistant, cold-sensitive strains
E. coli strain CSH142 was grown in 2× yeast extract and tryptone media (2× YT) (40 individual cultures) containing 60 μg/mL spectinomycin overnight at 37°C. Cultures were then plated on 2× YT plates containing 60 μg/mL spectinomycin (spc60). Colonies that appeared on independent plates were restreaked to spc60 plates to confirm spectinomycin resistance and to generate single colonies. Single spectinomycin-resistant colonies were then patched and tested for growth at both 37°C and 20°C. Colonies that were able to grow at 37°C but not at 20°C in the presence of spc60 were further characterized.
DNA preparation and sequencing
Competent cells were prepared and transformed essentially as described (Hanahan 1983). Transformants were plated on 2×YT plates containing 60 μg/mL spectinomycin and/or 12.5 μg/mL tetracycline. Plasmid DNA and genomic DNA were prepared by the alkaline lysis method (Sambrook et al. 1989) or commercial DNA preparation kits (Quiagen). DNA sequencing was performed at the Iowa State University DNA Sequencing and Synthesis Facility. The S5 gene was sequenced using the following primer:
S5G3: 5′-TTGACACTGCGGGTTTGAG-3′
Determination of doubling times
Overnight cultures with 60 μg/mL spectinomycin (depending on the requirement) were grown in 2× YT and were used to inoculate 50 mL 2× YT media. Cells were grown at 37°C (permissive temperature) for the length of the experiment or grown at 37°C for 30–60 min prior to being shifted to 20°C (nonpermissive temperature). Optical density at 600 nm was measured at several time points during the course of the growth (at least a total of 7 h). Growth curves were generated and used to determine the doubling times.
Polysome analysis
Polysome analysis was essentially done as described (Maki et al. 2002). The polysomes were analyzed by sucrose gradient sedimentation using 10%–40% sucrose gradients in 20 mM Tris-HCl (pH 7.8), 10 mM magnesium chloride, and 100 mM ammonium chloride in a SW41 rotor (26,000 rpm) for 17 h at 4°C. The speed and time were optimized to enable a better separation of the 30S and the pre-30S peak, therefore not allowing the polysomes to be readily observed. Additionally, peak fractions were collected from the gradients, and particles were concentrated on 100 K cutoff Centricons and then used for chemical probing.
Overexpression and purification of S5(G28D) mutant protein
Site-directed mutagenesis of the S5 gene previously cloned in pET24b (Culver and Noller 1999) was carried out using the Stratagene QuickChange Site-Directed Mutagenesis kit. The primers used for site directed mutagenesis were:
Sense primer: 5′-CCGTTAAAGGTGATCGTATTTTCTCC-3′ and
Antisense primer: 5′-GGAGAAAATACGATCACCTTTAACGG-3′
The mutant clones were authenticated by sequencing with the T7 forward primer. The S5(G28D) protein was overexpressed and purified as described for the wild-type protein (Culver and Noller 1999).
In vitro reconstitution of 30S subunits and subunit association of reconstituted 30S Subunits and natural 50S subunits
Reconstitution of 30S particles was done using a ninefold molar excess of each purified recombinant protein over 16S rRNA, following an ordered assembly protocol as described (Culver and Noller 1999). Subunit association of reconstituted 30S subunits and native 50S subunits was done as described (Culver and Noller 1999). Reconstituted 30S and 70S particles were purified on 10%–40% sucrose density gradients as described (Culver and Noller 1999). Purified and concentrated reconstituted particles were used for chemical probing experiments.
SDS-PAGE and Western blot analysis of in vitro reconstituted 30S particles with anti-S3 and S4 antibodies
In vitro reconstituted 30S particles with either wild-type S5, S5(G28D), or no S5 were purified on sucrose density gradients and concentrated on 100 K cutoff centricons. Equal amounts of the particles (∼40 pmol) were loaded onto 15% urea SDS-PAGE gel along with purified S3 and S4 proteins as controls. Gels were either stained with Coomassie blue or the proteins were transferred onto nitrocellulose membrane and subsequently probed with antibodies against S3 and S4 and visualized by autoradiography.
Transfer RNA binding
Transfer RNA binding was done as previously described (Culver and Noller 1999). The reported tRNA-binding activities are an average from three to six independent experiments. tRNA binding to 50S subunits alone was 5%, and this has been subtracted from the values reported for 70S ribosomes.
Bacterial strains and plasmids used for fidelity studies
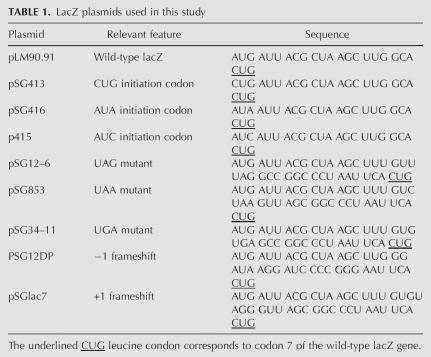
An isogenic strain carrying the S5 (G28D) mutation was constructed by transducing MC136 (Δ[lac-pro]thi-aroE::Tn10[Tetracycline]) (M. O'Connor, pers. comm.) to aromatic amino acid independence and loss of tetracycline resistance with Phage P1 grown on the S5 (G28D) mutant strain. This strain was called the ΔlacZ S5(G28D) strain. The presence of the mutation in this strain was ascertained by PCR amplification of the S5 gene and its subsequent sequencing. All the lacZ plasmids used in this study are as shown below (O'Connor et al. 1997). pSG plasmids encoding wild-type lacZ, non-AUG initiation condon containing lacZ, lacZ with premature stops at the N terminus and lacZ with N-terminal frame shift mutation (Table 1) were transformed into the Δ lacZ S5(G28D) strain and the β-galactosidase activity was measured in miller units (O'Connor et al. 1997).
TABLE 1.
LacZ plasmids used in this study
β-Galactosidase assay
Cells to be assayed for the activity of β-galactosidase were grown in luria-broth medium containing 12.5 μg/mL of tetracycline. Overnight cultures were used as precultures to inoculate fresh media containing antibiotics to generate logarithmically growing cells that were used for the assay of β-galactosidase activity as described previously (O'Connor et al. 1997). β-Galactosidase activity from the plasmid encoding wild-type lacZ in the wild-type strain, MC 256 (O'Connor et al. 1997), and the mutant strain, ΔlacZ S5(G28D) strain (from this study), were used for normalization. The level of activity observed in the wild-type strain was set to one for all plasmids.
Chemical probing and primer extension analysis
Chemical probing of the 16S rRNA in the in vitro reconstituted particles and in vivo particles was done with kethoxal, as described (Merryman and Noller 1998; Holmes and Culver 2004). RNA extraction of the modified particles was performed as described (Culver and Noller 2000) and used as a template for primer extension analysis. Primer extension analysis was done as described (Moazed et al. 1986; Culver and Noller 2000).
Examination of r-protein S5 and 16S rRNA contacts
Crystal structures (1FJG.pdb, 1J5E.pdb, and 2AW7.pdb) were analyzed using ENTANGLE (Allers and Shamoo 2001) to determine if glycine 28 of r-protein S5 (or its equivalent) interacts with 16S rRNA.
ACKNOWLEDGMENTS
We thank Dr. Rachel Green for helpful discussions and Dr. Green, Dr. Kristi Holmes, Laura Dutca, Nathan Napper, and Deepika Calidas for critical comments on the manuscript. The MC136 Lac− strain and all the lacZ plasmids used in this study were kind gifts from Dr. Michael O'Connor (University of Misssouri–Kansas City). The authors also acknowledge Jae-Hyung Lee for help in making Figure 5 and looking at the structural impact of the mutation in S5. This work was supported by NIH Grant no. GM62432 to G.M.C.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.302006.
REFERENCES
- Allen, P.N., Noller, H.F. Mutations in ribosomal proteins S4 and S12 influence the higher order structure of 16 S ribosomal RNA. J. Mol. Biol. 1989;208:457–468. doi: 10.1016/0022-2836(89)90509-3. [DOI] [PubMed] [Google Scholar]
- Allers, J., Shamoo, Y. Structure-based analysis of protein–RNA interactions using the program ENTANGLE. J. Mol. Biol. 2001;311:75–86. doi: 10.1006/jmbi.2001.4857. [DOI] [PubMed] [Google Scholar]
- Andersson, D.I., Andersson, G.E., Kurland, C.G. Functional interactions between mutated forms of ribosomal proteins S4, S5, and S12. Biochimie. 1986;68:705–713. doi: 10.1016/s0300-9084(86)80164-x. [DOI] [PubMed] [Google Scholar]
- Bollen, A., Davies, J., Ozaki, M., Mizushima, S. Ribosomal protein conferring sensitivity to the antibiotic spectinomycin in Escherichia coli . Science. 1969;165:85–89. doi: 10.1126/science.165.3888.85. [DOI] [PubMed] [Google Scholar]
- Brink, M.F., Brink, G., Verbeet, M.P., de Boer, H.A. Spectinomycin interacts specifically with the residues G1064 and C1192 in 16S rRNA, thereby potentially freezing this molecule into an inactive conformation. Nucleic Acids Res. 1994;22:325–331. doi: 10.1093/nar/22.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, D.E., Clemons, W.M.J., Carter, A.P., Wimberly, B.T., Ramakrishnan, V. Crystal structure of the 30S ribosomal subunit from Thermus thermophilus:Structure of the proteins and their interactions with 16S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- Carson, M. Ribbons. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- Carter, A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Wimberly, B.T., Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Culver, G.M., Noller, H.F. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA. 1999;5:832–843. doi: 10.1017/s1355838299990714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver, G.M., Noller, H.F. In vitro reconstitution of 30S ribosomal subunits using complete set of recombinant proteins. Methods Enzymol. 2000;318:446–460. doi: 10.1016/s0076-6879(00)18069-3. [DOI] [PubMed] [Google Scholar]
- Dammel, C.S., Noller, H.F. Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes & Dev. 1995;9:626–637. doi: 10.1101/gad.9.5.626. [DOI] [PubMed] [Google Scholar]
- De Wilde, M., Wittmann-Liebold, B. Localization of the amino-acid exchange in protein S5 from an Escherichia coli mutant resistant to spectinomycin. Mol. Gen. Genet. 1973;127:273–276. doi: 10.1007/BF00333767. [DOI] [PubMed] [Google Scholar]
- Donner, D., Kurland, C.G. Changes in the primary structure of a mutationally altered ribosomal protein S4 of Escherichia coli . Mol. Gen. Genet. 1972;115:49–53. doi: 10.1007/BF00272217. [DOI] [PubMed] [Google Scholar]
- Funatsu, G., Puls, W., Schiltz, E., Reinbolt, J., Wittmann, H.G. Ribosomal proteins. XXXI. Comparative studies on altered proteins S4 of six Escherichia coli revertants from streptomycin dependence. Mol. Gen. Genet. 1972a;115:131–139. doi: 10.1007/BF00277293. [DOI] [PubMed] [Google Scholar]
- Funatsu, G., Schiltz, E., Wittmann, H.G. Ribosomal proteins. XXVII. Localization of the amino acid exchanges in protein S5 from two Escherichia coli mutants resistant to spectinomycin. Mol. Gen. Genet. 1972b;114:106–111. doi: 10.1007/BF00332781. [DOI] [PubMed] [Google Scholar]
- Grondek, J.F., Culver, G.M. Assembly of the 30S ribosomal subunit: Positioning ribosomal protein S13 in the S7 assembly branch. RNA. 2004;10:1861–1866. doi: 10.1261/rna.7130504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., Nashimoto, H., Nomura, M. Structure and function of E. coli ribosomes. VIII. Cold-sensitive mutants defective in ribosome assembly. Proc. Natl. Acad. Sci. 1969;63:384–391. doi: 10.1073/pnas.63.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Held, W.A., Ballou, B., Mizushima, S., Nomura, M. Assembly mapping of 30S ribosomal proteins from Escherichia coli . J. Biol. Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- Holmes, K.L., Culver, G.M. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat. Struct. Mol. Biol. 2004;11:179–186. doi: 10.1038/nsmb719. [DOI] [PubMed] [Google Scholar]
- Itoh, T., Wittmann, H.G. Amino acid replacements in proteins S5 and S12 of two Escherichia coli revertants from streptomycin dependence to independence. Mol. Gen. Genet. 1973;202:446–454. doi: 10.1007/BF00267779. [DOI] [PubMed] [Google Scholar]
- Kurland, C.G., Hughes, D., Ehrenberg, M. Escherichia coli and Salmonella typhimurium: Cellular and molecular biology. American Society for Microbiology; Washington, D.C: 1996. Limitations of translational accuracy; pp. 979–1004. [Google Scholar]
- Maki, J.A., Schnobrich, D.J., Culver, G.M. The DnaK chaperone system facilitates 30S ribosomal subunit assembly. Mol. Cell. 2002;10:129–138. doi: 10.1016/s1097-2765(02)00562-2. [DOI] [PubMed] [Google Scholar]
- Makosky, P.C., Dahlberg, A.E. Spectinomycin resistance at site 1192 in 16S ribosomal RNA of E. coli: An analysis of three mutants. Biochimie. 1987;69:885–889. doi: 10.1016/0300-9084(87)90216-1. [DOI] [PubMed] [Google Scholar]
- Merryman, C., Noller, H.F. Footprinting and modification-interference analysis of binding sites on RNA. In: Smith C.W.J., editor. RNA:Protein interactions, A practical approach. Oxford University Press; New York: 1998. pp. 237–253. [Google Scholar]
- Merryman, C., Moazed, D., McWhirter, J., Noller, H.F. Nucleotides in 16S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 1999;285:97–105. doi: 10.1006/jmbi.1998.2242. [DOI] [PubMed] [Google Scholar]
- Mizushima, S., Nomura, M. Assembly mapping of 30S ribosomal proteins in E. coli . Nature. 1970;226:1214–1218. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Noller, H.F. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Noller, H.F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Noller, H.F. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16 S rRNA. J. Mol. Biol. 1990;211:135–145. doi: 10.1016/0022-2836(90)90016-F. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Stern, S., Noller, H.F. Rapid chemical probing of conformations in 16S ribosomal RNA and 30S ribosomal subunits using primer extension. J. Mol. Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Nashimoto, H., Held, W., Kaltschmidt, E., Nomura, M. Structure and function of bacterial ribosomes. XII. Accumulation of 21S particles by some cold-sensitive mutants of E. coli . J. Mol. Biol. 1971;62:121–138. doi: 10.1016/0022-2836(71)90135-5. [DOI] [PubMed] [Google Scholar]
- Nomura, M. The role of RNA and protein in ribosome function: A review of early reconstitution studies and prospects for future studies. Cold Spring Harb. Symp. Quant. Biol. 1987;52:653–663. doi: 10.1101/sqb.1987.052.01.075. [DOI] [PubMed] [Google Scholar]
- Nomura, M., Mizushima, S., Ozaki, M., Traub, P., Lowry, C.V. Structure and function of ribosomes and their molecular components. Cold Spring Harb. Symp. Quant. Biol. 1969;34:49–61. doi: 10.1101/sqb.1969.034.01.009. [DOI] [PubMed] [Google Scholar]
- O'Connor, M., Thomas, C.L., Zimmermann, R.A., Dahlberg, A.E. Decoding fidelity at the ribosomal A and P sites: Influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res. 1997;25:1185–1193. doi: 10.1093/nar/25.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle, J.M., Murphy, F.V., Tarry, M.J., Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Carter, A.P., Ramakrishnan, V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- Piepersberg, W., Böck, A., Wittmann, H.G. Effect of different mutations in ribosomal protein S5 of Escherichia coli on translational fidelity. Mol. Gen. Genet. 1975a;140:91–100. doi: 10.1007/BF00329777. [DOI] [PubMed] [Google Scholar]
- Piepersberg, W., Bock, A., Yaguchi, M., Wittmann, H.G. Genetic position and amino acid replacements of several mutations in ribosomal protein S5 from Escherichia coli . Mol. Gen. Genet. 1975b;143:43–52. doi: 10.1007/BF00269419. [DOI] [PubMed] [Google Scholar]
- Poot, R.A., van den Worm, S.H., Pleij, C.W., van Duin, J. Base complementarity in helix 2 of the central pseudoknot in 16S rRNA is essential for ribosome functioning. Nucleic Acids Res. 1998;26:549–553. doi: 10.1093/nar/26.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, T., Noller, H.F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, T., Stern, S., Changchien, L.M., Noller, H.F. Probing the assembly of the 3′ major domain of 16 S rRNA. Interactions involving ribosomal proteins S2, S3, S10, S13 and S14. J. Mol. Biol. 1988;201:697–716. doi: 10.1016/0022-2836(88)90468-8. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, V., Graziano, V., Capel, M.S. A role for proteins S3 and S14 in the 30 S ribosomal subunit. J. Biol. Chem. 1986;261:15049–15052. [PubMed] [Google Scholar]
- Ramakrishnan, V., White, S.W. The structure of ribosomal protein S5 reveals sites of interaction with 16S rRNA. Nature. 1992;358:768–771. doi: 10.1038/358768a0. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., Maniatis, T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Schnier, J., Isono, S., Cumberlidge, A.G., Isono, K. Unstable mutations caused by regional tandem multiplications in the gene for ribosomal protein S4 show thermosensitivity in Escherichia coli . Mol. Gen. Genet. 1985;199:265–270. doi: 10.1007/BF00330268. [DOI] [PubMed] [Google Scholar]
- Schuwirth, B.S., Borovinskaya, M.A., Hau, C.W., Zhang, W., Vila-Sanjurjo, A., Holton, J.M., Cate, J.H. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Sigmund, C.D., Ettayebi, M., Morgan, E.A. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli . Nucleic Acids Res. 1984;12:4653–4663. doi: 10.1093/nar/12.11.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, S., Changchien, L.-M., Craven, G.R., Noller, H.F. Interaction of proteins S16, S17 and S20 with 16S ribosomal RNA. J. Mol. Biol. 1988a;200:291–299. doi: 10.1016/0022-2836(88)90241-0. [DOI] [PubMed] [Google Scholar]
- Stern, S., Powers, T., Changchien, L.M., Noller, H.F. Interaction of ribosomal proteins S5, S6, S11, S12, S18 and S21 with 16 S rRNA. J. Mol. Biol. 1988b;201:683–695. doi: 10.1016/0022-2836(88)90467-6. [DOI] [PubMed] [Google Scholar]
- Stern, S., Weiser, B., Noller, H.F. Model for the three-dimensional folding of 16 S ribosomal RNA. J. Mol. Biol. 1988c;204:447–481. doi: 10.1016/0022-2836(88)90588-8. [DOI] [PubMed] [Google Scholar]
- Van Acken, U. Protein chemical studies on ribosomal proteins S4 and S12 from ram (ribosomal ambiguity) mutants of Escherichia coli . Mol. Gen. Genet. 1975;140:61–68. doi: 10.1007/BF00268989. [DOI] [PubMed] [Google Scholar]
- Vladimirov, S.N., Graifer, D.M., Karpova, G.G., Semenkov Yu, P., Makhno, V.I., Kirillov, S.V. The effect of GTP hydrolysis and transpeptidation on the arrangement of aminoacyl-tRNA at the A-site of Escherichia coli 70 S ribosomes. FEBS Lett. 1985;181:367–372. doi: 10.1016/0014-5793(85)80294-5. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons, W.M.J., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., Ramakrishnan, V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Wittmann-Liebold, B., Greuer, B. The primary structure of protein S5 from the small subunit of the Escherichia coli ribosome. FEBS Lett. 1978;95:91–98. doi: 10.1016/0014-5793(78)80059-3. [DOI] [PubMed] [Google Scholar]
- Woese, C.R., Magrum, L.J., Gupta, R., Siegel, R.B., Stahl, D.A., Kop, J., Crawford, N., Brosius, J., Gutell, R., Hogan, J.J., et al. Secondary structure model for bacterial 16S ribosomal RNA: Phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980;8:2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., Noller, H.F. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Yusupova, G.Z., Yusupov, M.M., Cate, J.H.D., Noller, H.F. The path of the messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]