Abstract
The peptidoglycan cell wall determines the shape and structural integrity of a bacterial cell. Class B penicillin-binding proteins (PBPs) carry a transpeptidase activity that cross-links peptidoglycan strands via their peptide side chains, and some of these proteins are directly involved in cell shape determination. No Bacillus subtilis PBP with a clear role in rod shape maintenance has been identified. However, previous studies showed that during outgrowth of pbpA mutant spores, the cells grew in an ovoid shape for several hours before they recovered and took on a normal rod shape. It was postulated that another PBP, expressed later during outgrowth, was able to compensate for the lack of the pbpA product, PBP2a, and to guide the formation of a rod shape. The B. subtilis pbpH (ykuA) gene product is predicted to be a class B PBP with greatest sequence similarity to PBP2a. We found that a pbpH-lacZ fusion was expressed at very low levels in early log phase and increased in late log phase. A pbpH null mutant was indistinguishable from the wild-type, but a pbpA pbpH double mutant was nonviable. When pbpH was placed under the control of an inducible promoter in a pbpA mutant, viability was dependent on pbpH expression. Growth of this strain in the absence of inducer resulted in conversion of the cells from rods to ovoid/round shapes and lysis. We conclude that PBP2a and PbpH play redundant roles in formation of a rod-shaped peptidoglycan cell wall.
Most bacteria have a rigid cell wall that maintains the cell shape and resists the internal turgor pressure. The major structural component of the bacterial cell wall is peptidoglycan, which is a net-like macromolecule of glycan strands cross-linked by peptides (reviewed in reference 5). Polymerization involves glycosyl transferases, which produce the glycan strands, and transpeptidases, which cross-link the glycan strands via their peptide side chains. The family of penicillin-binding proteins (PBPs) carry out these polymerization activities. Based on sequence similarities, the PBPs are classified as multimodular class A or B high-molecular-weight PBPs or monofunctional low-molecular-weight PBPs (reviewed in reference 19). The PBPs have a conserved C-terminal penicillin-binding domain that possesses d,d-carboxypeptidase or endopeptidase activity in low-molecular-weight PBPs and transpeptidase activity in most high-molecular-weight PBPs.
Most species possess multiple PBPs within each class, and multiple PBPs of the same class within a species often exhibit redundant function (14, 23, 32, 39, 47, 55). Class A PBPs are bifunctional proteins that also possess an N-terminal glycosyl transferase domain. The N-terminal domain of the class B PBPs has no clearly defined activity but may be required for the roles that these proteins play in determining cell morphology (19, 20). Analysis of the Bacillus subtilis genome sequence (25) reveals 16 PBP-encoding genes recognizable from sequence similarities (17). These include four class A, six class B, and six low-molecular-weight PBP-encoding genes. Studies of peptidoglycan synthesis and morphology throughout growth, sporulation, and germination have revealed specific roles played by a number of PBPs (reviewed in references 17 and 40).
Rod-shaped bacteria appear to have two peptidoglycan synthetic complexes, one for cell elongation and another for the formation of the new poles by septation (reviewed in reference 22). The shape of the cell is determined by alternating activation of these two systems. Each apparatus may contain multiple PBPs and probably interacts with autolytic and morphogenic system components. Class B PBPs have been found to play unique essential roles in septation and regulation of cell shape, and it has been proposed that they segregate to specific elongation and septation peptidoglycan synthetic complexes.
Among the B. subtilis class B PBPs, PBP2b (the product of pbpB) is essential for septation (54), and its sequence places it in a class of PBPs that have an analogous function in other species (19), which includes Escherichia coli PBP3 (FtsI) (50). B. subtilis PBP2a, the product of pbpA, is involved in the formation of rod-shaped cells from oval spores during spore outgrowth (36) and is most similar to E. coli PBP2, which is required for maintenance of the rod shape of the cell (50). B. subtilis PBP3, encoded by pbpC, was found to be dispensable under all growth, sporulation, and germination conditions tested (37). The product of spoVD is a class B PBP required for spore peptidoglycan synthesis (9, 10). The functions of two additional class B PBP-encoding genes, ykuA and yrrR, identified during analysis of the genome sequence have not been determined.
The studies presented here concern ykuA and its gene product. Due to extensive sequence similarity between the ykuA product and other class B PBPs, this gene was renamed pbpH. We demonstrate that PbpH plays a redundant role with PBP2a in determining the rod shape of the cell during vegetative growth and spore outgrowth.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All B. subtilis strains were derived from strain PS832, a prototrophic derivative of strain 168, and are listed in Table 1. Transformation of B. subtilis was performed as described previously (3). Transformants were selected on 2× SG (27) plates containing appropriate antibiotics: chloramphenicol (3 μg/ml), spectinomycin (100 μg/ml), kanamycin (10 μg/ml), tetracycline (10 μg/ml), and erythromycin (0.5 μg/ml) plus lincomycin (12.5 μg/ml). Vegetative growth and sporulation were done routinely in 2× SG medium at 37°C. Spores were purified by washing with water (38). Spore heat resistance and chloroform resistance were measured after sporulation for 24 h as previously described (38). Spore germination and outgrowth were analyzed in 2× YT medium (43) containing 4 mM l-alanine after a 30-min heat shock in water at 65°C (38).
TABLE 1.
B. subtilis strains used
| Strain | Genotypea | Constructionb | Source or reference |
|---|---|---|---|
| DPVB133 | ΔpbpH::Sp | pDPV113→PS832 | This work |
| DPVB168 | pbpH::pbpH-lacZ::Cm | pDPV125→PS832 | This work |
| DPVB171 | pbpC::Cm ΔpbpH::Sp | DPVB133→PS2352 | This work |
| DPVB182 | pbpA::Cm pbpC::Sp | PS2328→PS2465 | This work |
| DPVB202 | ΔpbpH::Sp amyE::xylAp-pbpH::Cm | pDPV138→DPVB133 | This work |
| DPVB203 | pbpA::Erm | pCm::Er→PS2465 | This work |
| DPVB207 | pbpA::Erm ΔpbpH::Sp amyE::xylAp-pbpH::Cm | DPVB203→DPVB202 | This work |
| DPVB213 | amyE::xylAp-bgaB::Cm | pSWEET-bgaB→PS832 | This work |
| PS832 | Wild type, Trp+ revertant of 168 | Lab stock | |
| PS2328 | pbpC::Sp | pTMM4→PS832 | 37 |
| PS2352 | pbpC::Cm | pTMM5→PS832 | 37 |
| PS2465 | pbpA::Cm | pTMA4→PS832 | 36 |
Abbreviations for antibiotic resistance genes: Cm, chloramphenicol; Erm, lincomycin and erythromycin; Kn, kanamycin; Sp, spectinomycin.
Strains were constructed by natural transformation. The designation preceding the arrow is the plasmid or source of donor chromosomal DNA, and the designation following the arrow is the recipient strain.
For induction of the xylose-regulated promoter, strains were grown at 37°C in 2× SG lacking glucose, and xylose was added to a final concentration of 0.4% (for the study of pbpH function) or 2% (for maximum expression). To resuspend an exponentially growing culture into a new medium, the culture was centrifuged at 5,000 × g for 5 min at 24°C and resuspended into warm (37°C) fresh medium. β-Galactosidase assays were performed with the substrate o-nitrophenyl-β-d-galactopyranoside (38), and activity was expressed in Miller units (34). Glucose dehydrogenase activity was assayed as previously described (38). For analysis of vegetative peptidoglycan structure, cultures were grown to an optical density of 0.5 to 1.0 at 37°C in 100 ml of 2× SG. Vegetative cell peptidoglycan (6, 33) and spore peptidoglycan (42) were purified and analyzed as previously described.
Construction of plasmids and strains.
Primers pbpH1 (5′-CGTTGAATCACTAAGAATAAGG) and pbpH2 (5′-TCATCTCTCCTTGGAGATAGCC) were used to amplify a 2,628-bp fragment containing 272 bp of upstream sequence, the coding sequence, and 302 bp of sequence downstream of pbpH from the chromosomal DNA of PS832. This fragment was cloned into pGEM-T (Promega), generating plasmid pDPV103, in which the pbpH sequence is in the same orientation as lacZ. The insert of this plasmid was sequenced and found to be identical to that in the published genome sequence (25). To construct a deletion mutation in pbpH, pDPV103 was digested with HindIII and BamHI to remove 40% of the coding sequence, including the penicillin-binding active site. The deleted region was replaced with a spectinomycin resistance gene cassette obtained from HindIII- and BamHI-digested pJL73 (26) to generate pDPV113. This plasmid was linearized by digestion at an XmnI site within the pGEM-T vector sequence. The linearized DNA was transformed into strain PS832, with selection for the spectinomycin resistance marker, to produce strain DPVB133 (ΔpbpH::Sp), in which the mutant allele was exchanged with the wild-type allele in the chromosome via double crossover.
Primer pbpHa (5′-CGGAATTCCGTTGAATCACTAAGAATAAGG) contained an added EcoRI site (underlined) to assist in cloning a fragment for constructing a lacZ fusion. Primers pbpHa and pbpH2 were used to PCR amplify the 2,628-bp insert in pDPV103. The 2,637-bp PCR product was cut with EcoRI and HindIII to produce a 682-bp fragment that contained the upstream region and the first 402 bp of the pbpH coding sequence. This fragment was inserted into EcoRI- and HindIII-digested pDPC87 (45) to generate pDPV125, in which the 682-bp fragment was placed in front of a promoterless lacZ gene. The insert of the plasmid was sequenced and confirmed to be identical to that in the published genome sequence. The plasmid was transformed into B. subtilis PS832, with selection for chloramphenicol resistance so that the lacZ fusion construct could insert into the chromosomal pbpH locus via a single crossover, producing strain DPVB168.
Primers pbpH5 (5′-GCGCTTAATTAATGATTTGAGAGGGGTA) and pbpH3 (5′-GAAGATCTTTTAGTTGTGCACCCTG) were designed with added PacI and BglII sites (underlined), respectively. They were used to PCR amplify a 2,162-bp fragment containing 80 bp upstream of the predicted start codon (25), the coding sequence, and 25 bp downstream of pbpH from chromosomal DNA of PS832. The resulting 2,176-bp PacI-BglII fragment was cloned into PacI- and BamHI-digested pSWEET-bgaB (8), which contains a xylose-regulated expression system. The sequence of the pbpH gene in the resulting plasmid, pDPV138, was found to be identical to the sequence from the genome. The plasmid was linearized at a PstI site in the vector and used to transform DPVB133 to place the xylAp-pbpH construct at the amyE locus. The resulting Amy− Cmr Spr strain, DPVB202 (ΔpbpH::Sp amyE::xylAp-pbpH::Cm), was transformed with chromosomal DNA from DPVB203, with selection for erythromycin and spectinomycin resistance (Ermr and Spr) in the presence of 0.2% xylose, to give strain DPVB207. As a control, DPVB213 was generated by transforming PS832 with the plasmid pSWEET-bgaB. DPVB203 was constructed by transforming PS2465 (pbpA::Cm) (36) with plasmid pCm::Er (51) to switch the resistance marker in the pbpA locus from chloramphenicol to erythromycin.
Primer extension analysis.
Strain PS832 was grown in 2× SG medium until approximately 30 min after the initiation of sporulation, and total RNA was purified with an RNeasy kit (Qiagen). Primer pbpH-PE (5′-CCCGAGCTTAAAAATTAATGC) was 5′-end labeled with 32P as directed in the fmol DNA cycle sequencing kit (Promega). DNA sequencing with the end-labeled primer was done as directed in the fmol DNA cycle sequencing kit, and primer extension with 40 μg of RNA was done as directed in the primer extension system-avian myeloblastosis virus reverse transcriptase kit (Promega). Primer extension products were ethanol precipitated and concentrated prior to electrophoresis on denaturing polyacrylamide gels. Radioactive signals were detected with Kodak Phosphorimager screens and a Storm 860 phosphorimager (AP Biotech).
MIC determination.
Aliquots (50 μl) of serial dilutions of antibiotics in water were placed in the wells of 96-well microtiter plates. Mid-log cultures (optical density ≈0.5) growing at 37°C in 2× YT medium (16 g of tryptone,10 g of yeast extract, and 5 g of NaCl per liter) lacking antibiotics were diluted 1,000-fold in sterile 2× YT medium, and 200 μl was introduced into each well of a microtiter plate containing antibiotics. Wells containing no antibiotic and sterile medium were included as controls. Samples of the diluted cultures were plated to determine the approximate CFU introduced into each well. For all cultures, the inoculum was between 100 and 400 cells/well. Repetitions revealed that variation within this inoculum range had no effect on the MIC. Plates were incubated at 37°C for 18 to 20 h and then scored for growth. The MIC reported is the concentration that completely inhibited visible growth.
Phase contrast and electron microscopy.
For phase contrast light microscopy, 0.5-ml culture samples were centrifuged, resuspended in 0.5 ml of fixing solution (2.4% paraformaldehyde and 0.01% glutaraldehyde in 30 mM sodium phosphate buffer [pH 7.0]) for 15 min at room temperature and 30 min on ice, washed twice with 0.5 ml of phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.76 mM KH2PO4, pH 7.4), and resuspended in 50 to 150 μl of PBS. Fixed cell suspensions were placed on slides with polylysine-coated coverslips and photographed on an Olympus Provis AX70 microscope equipped with an Olympus UPlanF1 100X/1.30 oil Ph3 objective.
For electron microscopy, 10-ml culture samples were centrifuged, resuspended gently in fixing solution (840 μl of 0.5 M sodium phosphate buffer [pH 7.0] plus 50 μl of 25% electron microscopy-grade glutaraldehyde), placed at 4°C overnight, washed four times with 1 ml of cold 0.1 M sodium phosphate buffer (pH 6.7), resuspended in 1% osmium in cold 0.1 M sodium phosphate buffer (pH 6.7), and stored at 4°C overnight. Fixed cells were then washed in 0.5 M NH4Cl, suspended in 2% warm agar, and immediately spun at 10,000 × g for 3 to 5 min while cooling to pellet the cells in agar. A standard dehydration was performed with 30%, 50%, 70%, 95%, and 100% ethanol, then with ethanol-Spurr's resin (1:1) overnight, and finally in 100% Spurr's resin overnight. The samples were embedded in Spurr's resin and cut with a diamond knife. Sections were placed on 200-mesh copper grids and stained with 1% uranyl acetate for 12 min and with lead citrate for 5 min. Samples were viewed and photographed with a Jeol 100 CX-II transmission electron microscope at an accelerating voltage of 80 kV. Cell lengths and widths were measured from electron micrographs. Photographic negatives were scanned to produce digital images and processed for publication with Adobe Photoshop software.
RESULTS
Identification of pbpH gene.
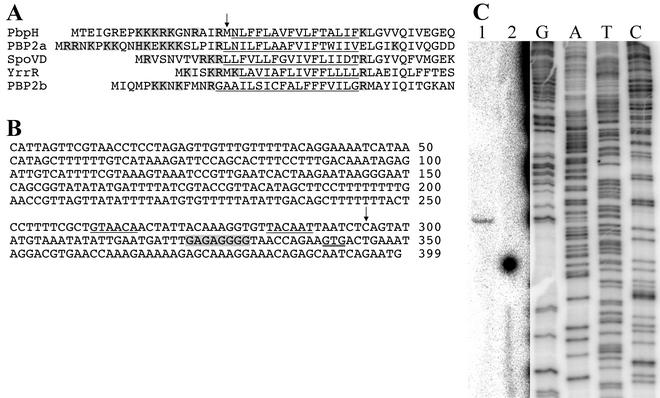
A sequence alignment of the predicted gene product of pbpH (ykuA) against the microbial protein database with the Blast software (2) revealed that the most similar protein in B. subtilis was PBP2A (42% identical and 60% similar) and in E. coli was PBP2 (22% identical and 40% similar). PBP2A is required for the normal outgrowth of spores (36), and PBP2 of E. coli is required in cell elongation and maintenance of the rod shape (50). These data suggested that pbpH encodes a class B PBP of a subclass required for maintaining the rod shape of the cell (19). Examination of the PBP alignment suggested that the previously predicted pbpH start codon (25) might be incorrect. Alignment of the signal peptide hydrophobic cores and several conserved residues immediately downstream of the signal peptides of five B. subtilis class B PBPs indicated that the predicted PbpH N-terminal methionine residue was immediately upstream of the hydrophobic core (Fig. 1A). Translation initiation 19 codons upstream would produce a protein with the expected basic residues upstream of the hydrophobic core, similar to the situation seen in the other PBPs. This upstream start codon is also preceded by a better, putative purine-rich ribosome-binding site than the originally proposed start (Fig. 1B).
FIG. 1.
Predicted pbpH transcription and translation start sites. (A) The N-terminal regions of five B. subtilis class B PBPs are aligned relative to their signal peptide hydrophobic cores (underlined) and several conserved residues immediately downstream of the signal peptides. The predicted PbpH N-terminal methionine residue (25) is indicated by an arrow. Several basic amino acids (shaded) would be incorporated upstream of the hydrophobic core if translation initiated 19 codons further upstream. (B) The DNA sequence begins with the start codon of a divergently oriented upstream gene, ykwD, and ends with the pbpH start codon predicted in the published genome sequence (25). The proposed alternative pbpH start codon is double underlined, and the putative ribosome-binding site is highlighted in gray. The apparent transcription start site at position 295 is indicated by an arrow, and potential σA recognition sequences are underlined. (C) RNA was purified from wild-type B. subtilis cells during early sporulation and used for reverse transcriptase primer extension (lane 1) with a 32P-, 5′-end-labeled primer that anneals within the pbpH sequence. Lane 2 is an identical primer extension reaction lacking RNA. DNA cycle sequencing reactions (lanes G, A, T, and C) utilized the same end-labeled primer. Due to the weak signal strength in lanes 1 and 2, the contrast of the image was adjusted independently for these lanes and for the sequencing lanes. The positions of the lanes relative to one another were not altered.
Expression of pbpH.
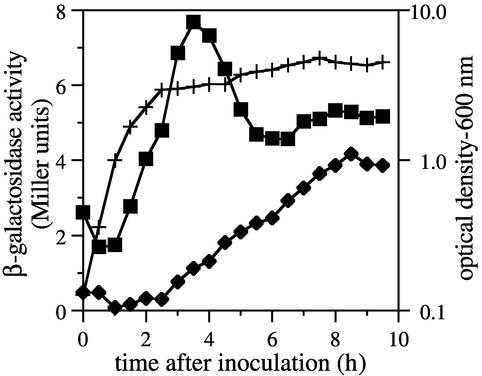
A PCR-amplified fragment containing 402 bp of the N-terminal coding region and 272 bp upstream of pbpH was used to generate a transcriptional lacZ fusion, and this construct was recombined in single copy into the chromosomal pbpH locus. Transcription of pbpH was evident only during vegetative growth, increasing progressively during log phase, peaking during early stationary phase, and then decreasing as the cells progressed into sporulation (Fig. 2). No expression was observed during the first 2 h of spore germination and outgrowth, during which outgrowing cells underwent approximately two generations (data not shown). The peak pbpH expression level was very low, even compared to those of other PBP-encoding genes (44-46) which are known to encode very low abundance proteins (16, 48).
FIG. 2.
Expression of a pbpH-lacZ fusion. β-Galactosidase activity was determined in samples of cultures throughout growth and sporulation (sporulation initiated approximately 2.5 h after inoculation). Activity in the pbpH-lacZ strain DPVB168 (▪) was compared to the background activity found in the wild-type strain PS832 (⧫). The optical densities of the two cultures were essentially identical (+).
Primer extension mapping was used to determine a pbpH transcription start site. One RNA 5′ end was detected 102 bases upstream of the originally predicted pbpH start codon, 45 bases upstream of what we now believe to be the pbpH start codon (Fig. 1C). Examination of the sequence upstream of this apparent transcription start site revealed reasonable matches to σA-dependent promoter consensus recognition sequences (Fig. 1B). Further study will be required to clarify the identity of this promoter, the true PbpH translation start site, and the mechanism of growth phase-dependent expression of pbpH.
We attempted to identify the product of pbpH (predicted to be 77 or 79 kDa, depending on the start codon utilized) with radiolabeled penicillin (24, 29, 32). We could find no difference in the PBPs visualized from membranes prepared from wild-type, ΔpbpH, and pbpH-overexpressing cells (data not shown). An identical experiment was successful in demonstrating the product of another low abundance B. subtilis class B PBP, YrrR (unpublished data).
Phenotypic properties of pbpH single mutant and multiple mutants.
To identify the function of pbpH, we constructed a strain with a pbpH deletion (DPVB133) as well as multiple mutant strains containing the pbpH deletion and deletions of two other class B PBP-coding genes, pbpC (37) and yrrR (unpublished data). The phenotypic properties of the mutant strains were compared to those of the wild-type strain PS832. There were no significant differences in the doubling times, cell morphologies, sporulation efficiencies, vegetative cell (6, 33) and spore peptidoglycan (42) structures, or germination and outgrowth rates (data not shown).
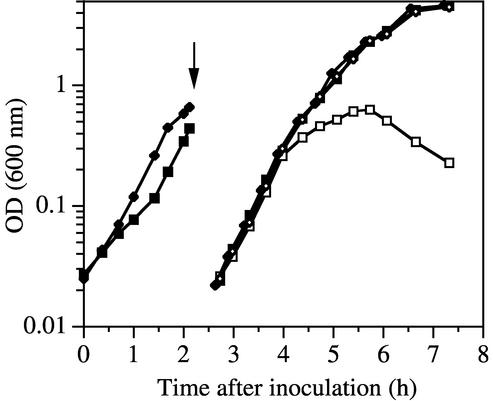
We were unable to construct a double mutant lacking both pbpA and pbpH, suggesting that such a strain was not viable. We therefore constructed a pbpH deletion strain containing a xylose-regulated pbpH gene inserted into the chromosome at the amyE locus and then introduced a pbpA deletion into it. This strain, DPVB207, had the appearance of the wild-type strain on plates containing xylose, but in the absence of xylose it produced only a few suppressor colonies, at a frequency of ≈10−7. These suppressor strains had colony phenotypes indistinguishable from that of the wild-type strain and were not characterized further. They most likely resulted from mutations inactivating the xylose repressor or from recombination events that exchanged the pbpH null and wild-type alleles between the constructs at the pbpH and amyE loci. DPVB207 and the wild-type strain, PS832, were grown to mid-log phase in liquid medium containing xylose, and then samples of the cultures were centrifuged and resuspended in 20-fold volumes of fresh medium with and without xylose. Growth of the wild type was not affected by the presence or absence of xylose (Fig. 3). In the presence of xylose, the growth of DPVB207 was similar to that of the wild type. However, when resuspended in the absence of xylose, these cells resumed growth for only three generations. This indicates that depletion of PbpH in the absence of xylose led to growth arrest and that pbpA and pbpH play redundant, essential roles in vegetative growth.
FIG. 3.
Growth is dependent on pbpH expression in the absence of pbpA. The wild-type strain PS832 (⧫ and ◊) and the pbpA pbpH amyE::xylAp-pbpH strain DPVB207 (▪ and □) were grown in the presence of xylose and then centrifuged (at the time indicated by the arrow), resuspended at a 20-fold dilution, and incubated in medium containing (solid symbols) or lacking (open symbols) xylose. OD, optical density.
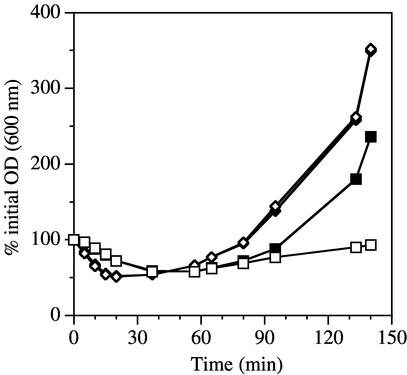
Upon entering the stationary phase in medium containing xylose, strain DPVB207 initiated sporulation and generated apparently normal spores. These spores exhibited heat resistance indistinguishable from that of spores produced by the wild-type strain in the presence and absence of xylose (data not shown). Spore germination was initiated in both the presence and absence of xylose (Fig. 4). The germination and outgrowth of wild-type spores was the same under both conditions. The initiation of germination (indicated by the decrease in optical density) of the DPVB207 spores was slightly delayed relative to that of the wild type. DPVB207 spore outgrowth was significantly impaired in the medium lacking xylose. In the presence of xylose, DPVB207 spores were slightly delayed in initiating outgrowth but eventually exhibited a rate of outgrowth similar to that of the wild-type spores. This delay was reproducibly observed in three independent spore preparations and was probably an outcome of the slow germination. Slow germination, like that seen for DPVB207 spores, was previously associated with highly cross-linked spore peptidoglycan (41), a situation that could be affected by PbpH. However, the structure of cortex peptidoglycan of DPVB207 spores was indistinguishable from that of the wild type (data not shown).
FIG. 4.
Germination and outgrowth of spores expressing and lacking pbpH. Spores were produced from the wild-type strain PS832 (⧫ and ◊) and the pbpA pbpH amyE::xylAp-pbpH strain DPVB207 (▪ and □) in the presence of xylose and germinated in the presence (solid symbols) or absence (open symbols) of xylose.
Cell morphology of pbpA pbpH double mutant.
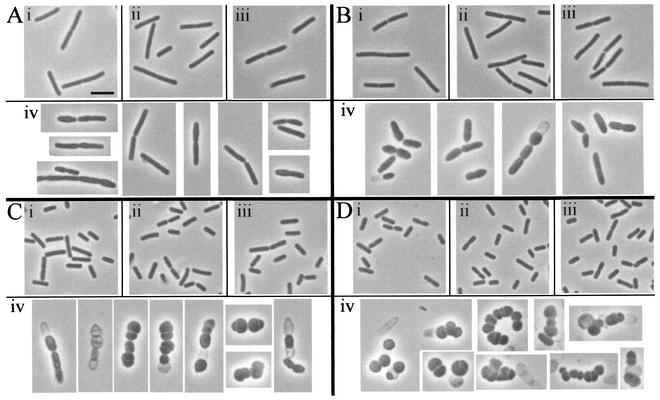
Phase contrast microscopy revealed that vegetative cells of the wild-type strain grown in the presence and absence of xylose and DPVB207 cells grown in the presence of xylose looked similar (Fig. 5). However, between 60 and 80 min after resuspension in the absence of the xylose (two to three mass doublings), DPVB207 cells started to swell (Fig. 5Aiv). After 2 h, a few cells had lysed, many cells had swelled dramatically, and some cell divisions were asymmetric (Fig. 5Biv). After 3 h, many cells had lysed, and most cells were nearly spherical (Fig. 5Civ). In cells that survived up to 4 h, morphology was extremely variable, and division septa were irregular (Fig. 5Div).
FIG. 5.
Phase contrast microscopy of vegetative cells. Cultures were grown in the presence of xylose and then resuspended in medium containing or lacking xylose, as described for Fig. 3. Culture samples were examined 80 min (A), 120 min (B), 180 min (C), and 240 min (D) after resuspension. Cells are from the wild-type strain PS832 with (i) and without (ii) xylose and the pbpA pbpH amyE::xylAp-pbpH strain DPVB207 with (iii) and without (iv) xylose. All images are at the same magnification. Scale bar in Ai, 2 μm.
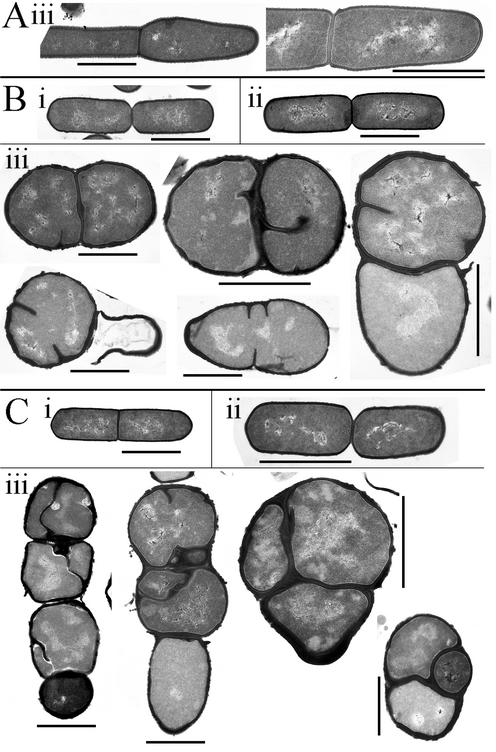
Vegetative cells were examined in greater detail by transmission electron microscopy (Fig. 6). Wild-type cells were unaffected by the presence of xylose (data not shown). DPVB207 cells incubated in the presence of xylose had the same morphology as those of the wild type. However, DPVB207 cells had a pleiomorphic spherical or distorted shape when incubated in the absence of xylose, and eventually many of the cells lysed. In these aberrant cells, septa were relatively randomly localized, although in some cases successive divisions seemed to occur in perpendicularly alternating planes. Most cell walls and septa were thicker than those seen in wild-type cells. Four hours after suspension in the absence of xylose, the average DPVB207 cell width (Table 2) and apparent volume were larger than those of wild-type and DPVB207 cells grown in xylose-containing medium.
FIG. 6.
Electron microscopy of vegetative cells. Cultures were grown in the presence of xylose and then resuspended in medium containing or lacking xylose as described for Fig. 3. Culture samples were examined 80 min (A), 180 min (B), and 240 min (C) after resuspension. Cells are from the wild-type strain PS832 with xylose (i) and the pbpA pbpH amyE::xylAp-pbpH strain DPVB207 with (ii) and without (iii) xylose. Scale bars, 1 μm.
TABLE 2.
Dimensions of wild type and pbpA pbpH cells
| Strain | Xylose in growth mediuma | No. of cells measured | Avg cell width (μm) ± SD | Avg cell length (μm) ± SD |
|---|---|---|---|---|
| PS832 | Yes | 40 | 0.57 ± 0.04 | 1.7 ± 0.5 |
| No | 21 | 0.55 ± 0.02 | 1.5 ± 0.2 | |
| DPVB207 | Yes | 10 | 0.60 ± 0.04 | 1.5 ± 0.3 |
| No | 56 | 1.4 ± 0.3 | 1.5 ± 0.4 |
Cells were fixed and prepared for electron microscopy 4 h after resuspension in the presence and absence of xylose.
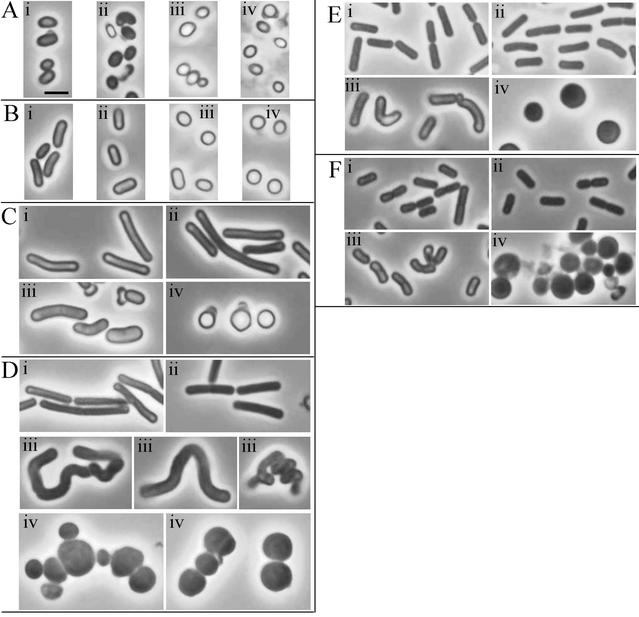
The morphologies of germinating and outgrowing wild-type spores were identical in the presence and absence of xylose (Fig. 7). The delayed germination of DPVB207 spores was evident under phase contrast microscopy (Fig. 7). The spores remained ovoid and partially refractile longer than the wild-type spores. The morphology of outgrowing DPVB207 spores was different in the presence and absence of xylose. Spores germinated without xylose were spherical, gradually enlarged, and eventually lysed. Few of these cells were able to divide, and the septa formed were generally asymmetric. At later stages (240 min), a few rare rod-shaped cells appeared. We believe that these were the result of the potential suppressor mutations described above, since plating these cultures in the absence of xylose produced rare colonies (from about ≈1 in 106 spores) with a phenotype like that of the wild-type strain. DPVB207 spores germinated in xylose-containing medium produced outgrowing cells that were bent and had a greater width than those of the wild type. Some of these cells became helical, but eventually they became shorter bent rods and grew similarly to the wild type.
FIG. 7.
Phase contrast microscopy of germinating spores. Spores were germinated in medium containing or lacking xylose. Culture samples were examined 45 min (A), 60 min (B), 90 min (C), 120 min (D), 180 min (E), and 240 min (F) after resuspension. Cells are from the wild-type strain PS832 with (i) and without (ii) xylose and the pbpA pbpH amyE::xylAp-pbpH strain DPVB207 with (iii) and without (iv) xylose. All images are at the same magnification. Scale bar in Ai, 2 μm.
Peptidoglycan structure.
To determine if peptidoglycan structural changes could be correlated with losses of class B PBPs, the muropeptides derived from vegetative cell peptidoglycan were separated and quantified by reversed-phase high-pressure liquid chromatography. Muropeptide chromatograms revealed no reproducible differences between the vegetative cell peptidoglycan of the wild-type strain and that of the pbpA and pbpH mutant strains (data not shown). In addition, when DPVB207 was grown for 2 h in the presence and absence of xylose, the muropeptide profiles still revealed no clear differences from each other or from the wild-type strain.
MICs for β-lactam antibiotics.
One possible explanation for the maintenance of redundant PBPs is that they have different sensitivities to certain antibiotics and could thus provide an advantage in the natural setting. We determined the MICs of several β-lactam antibiotics for the wild-type and class B PBP single and multiple mutant strains (Table 3). Vancomycin, a non-β-lactam antibiotic that also targets peptidoglycan synthesis, was also examined. While none of the strains exhibited altered sensitivities to vancomycin, several had increased or decreased sensitivities to a number of β-lactams. The MIC changes were small but reproducible in three independent cultures of each strain. The pbpA and pbpC mutants were both more sensitive to the cephalosporins cefoxitin, ceftriaxone, and cefuroxime, and these increased sensitivities seemed to be additive in the pbpA pbpC double mutant. This suggests that another PBP is partially redundant with the pbpA and pbpC products and is sensitive to these antibiotics. The pbpH mutation had no effect on sensitivity to these antibiotics. The pbpA mutant also showed increased sensitivity to cephalexin, amdinocillin, and piperacillin, while the pbpC mutant did not. Interestingly, the pbpH mutant was more resistant to these antibiotics, suggesting a distinction in the binding specificities of these two redundant PBPs.
TABLE 3.
MICs for class B PBP mutant strainsa
| Strain | Relevant genotype | MIC (μg/ml)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | Carbenicillin | Cefoxitin | Ceftriaxone | Cefuroxime | Cephalexin | Furazolidone | Mecillinam | Penicillin | Piperacillin | Vancomycin | ||
| PS832 | Wild type | 0.06 | 2 | 1 | 0.25 | 0.13 | 0.13 | 2 | 5 | 0.015 | 0.25 | 0.25 |
| PS2465 | pbpA | 0.06 | 1 | 0.5 | 0.06 | 0.06 | 0.06 | 2 | 1.8 | 0.03 | 0.13 | 0.25 |
| PS2328 | pbpC | 0.13 | 2 | 0.5 | 0.06 | 0.06 | 0.13 | 1 | 2.5 | 0.015 | 0.25 | 0.25 |
| DPVB133 | pbpH | 0.13 | 1 | 1 | 0.25 | 0.13 | 0.25 | 1 | 10 | 0.03 | 0.5 | 0.25 |
| DPVB182 | pbpA pbpC | 0.13 | 1 | 0.25 | 0.03 | 0.015 | 0.03 | 1 | 1.3 | 0.015 | 0.13 | 0.25 |
| DPVB171 | pbpC pbpH | 0.13 | 1 | 0.5 | 0.06 | 0.03 | 0.13 | 1 | 2.5 | 0.015 | 0.13 | 0.25 |
Three independent trials for each strain produced identical results.
DISCUSSION
The high level of sequence similarity between PbpH, B. subtilis PBP2a, and E. coli PBP2 suggested that PbpH might be involved in rod shape maintenance. Our studies revealed that PBP2a and PbpH play redundant roles in determining the rod cell shape and that the activity of one of these proteins is required for viability. It is not clear why B. subtilis maintains two PBPs to play this role. The fact that pbpH is expressed most strongly in late log phase and during the transition to stationary phase suggests that PbpH may play a unique role during this part of the life cycle, one that is important under growth conditions other than those encountered in the laboratory.
We envision four possible reasons why we could not detect PbpH with radiolabeled penicillin. PbpH is expressed at very low levels, perhaps even below the detection limit of this very sensitive method. PbpH may not remain associated with the membrane, in the cell or during cell fractionation, and thus would not be recovered in our membrane fraction. PbpH may be obscured by another PBP that runs on SDS-PAGE in the 75- to 80-kDa range. Finally, PbpH may not bind this particular β-lactam with high affinity. The fact that strains lacking PBP2a or PbpH exhibit small but different and in some cases opposite changes in MICs for several β-lactam antibiotics supports the idea that these two proteins have different binding specificities. Late log phase, when cells are expressing PbpH and are under nutrient-limiting conditions, is a time at which B. subtilis might expect to encounter antibiotics produced by competitors in the environment. The presence of redundant PBPs with different β-lactam sensitivities could allow B. subtilis to compete under a wider range of conditions.
pbpH was not expressed during the early stages of spore outgrowth. On the contrary, the expression of pbpA began to increase 30 to 40 min after the initiation of germination, increased throughout vegetative growth, and then decreased upon entry into stationary phase and sporulation (36). The expression pattern of pbpH explains the phenotype of the pbpA mutant. Spore germination does not require peptidoglycan synthesis, and thus both the pbpA and pbpA pbpH strains can initiate this process. Peptidoglycan synthesis is required for outgrowth of the germinated spores into rod-shaped vegetative cells, and while pbpA mutant spores begin outgrowth at a normal rate, they are unable to produce rod-shaped cells (36). After several hours, these cells finally recover their rod shape and then grow with a wild-type morphology. During outgrowth of the pbpA mutant spores, pbpH is expressed at an extremely low level, and thus PbpH cannot replace the function of PBP2a in production of the rod shape. Eventually, the expression of pbpH must increase to a level that allows the cells to resume normal shape and growth. In the pbpA pbpH double mutant, the outgrowing cells are never able to achieve a rod shape.
Current models of bacterial cell wall synthesis suggest that distinct wall-synthetic complexes act in alternating fashion during the life cycle to first drive cell elongation by insertion of peptidoglycan into the cylindrical wall, followed by a switch of the majority of the wall-synthetic activity to septum production (reviewed in reference 22). E. coli PBP2 and B. subtilis PBP2a and PbpH are predicted to function as transpeptidases (which cross-link the peptidoglycan strands) in complexes with peptidoglycan glycosyl transferases (which polymerize the strands) and with other proteins that direct the localization of their activities. One protein that has consistently been associated with the functions of these class B PBPs is RodA (21, 31, 52), a member of the SEDS (shape, elongation, division, and sporulation) family (21). In E. coli, the rodA and pbpA genes are in an operon (31), and their gene products are believed to interact with each other (30). Loss of the RodA integral membrane protein causes both E. coli (52) and B. subtilis (21) cells to alter their morphology in a manner similar to that observed upon loss of PBP2a and PbpH. The presence of a single rodA ortholog in the B. subtilis genome suggests that this gene product is able to function with both PBP2a and PbpH in production of the cylindrical wall.
In the absence of the cylindrical wall synthetic machinery (in the absence of RodA or both PBP2a and PbpH), the majority of peptidoglycan synthesis may be carried out by the septum synthetic machinery. This would be consistent with the appearance of round cells and with the production of aberrant septa. Previous observations that the peptidoglycan of polar caps is turned over more slowly than that of the cylindrical wall (4, 35) would be consistent with the general thickening of the wall in the absence of PBP2a and PbpH. A thickening of the wall could potentially contribute to failure in cell separation and the formation of cell chains. It might be expected that the peptidoglycan being synthesized in the absence of PBP2a and PbpH would have structural characteristics that are indicative of poles. However, we detected no clear difference between the overall muropeptide profiles of peptidoglycan derived from rod-shaped and round cells. The same result has been obtained several times with E. coli cells caused to become round or filamentous for various reasons (11, 12, 18). Only under very particular conditions was it possible to demonstrate structural differences between cylindrical wall and polar peptidoglycan, and in this case the greatest difference was in the glycan chain length distribution (49). This indicates that the different shapes of the poles and cylindrical walls are derived from differences in the three-dimensional arrangement of the glycan strands and cross-links, which is lost during muramidase digestion to produce muropeptides, rather than any chemical differences in the peptidoglycan.
In the spherical cells produced by a B. subtilis pbpA pbpH double mutant, there was still septum formation, but the septa were formed extremely irregularly, suggesting that the septation localization system is affected by the loss of the rod shape. It is possible that PbpH or PBP2a is required for complete assembly of the division machinery. This would be consistent with the observation that E. coli PBP2 can localize to division sites and can affect cell pole diameter (13). Alternatively, the greater diameter of the mutant cells itself may impair septation, possibly by disrupting the formation of the complete FtsZ rings required for normal septation (15). Polymerization of FtsZ at a division site in large-diameter cells may lead to partial rings or spirals (1, 7, 28) that are unable to direct a normal invagination of the envelope. Overexpression of FtsZ can suppress the lethal effect of inhibition of PBP2 in E. coli (53), perhaps by allowing the production of larger FtsZ rings.
It is not absolutely clear why cells lacking PBP2a and PbpH lyse. Among the relatively small number of cells observed in electron micrographs in which the site of cell rupture appeared obvious, about 50% of those sites were at or near the transition point between the cylindrical and septal walls (data not shown). Certainly a failure of either the cylinder- or septum-synthesizing machinery might be expected to lead most rapidly to a failure at this transition point. There is no way to be sure if any of these rupture sites was the primary cause of lysis or was a secondary site of wall breakdown. It is also possible that the increase in cell size leads to cell lysis. If some control mechanism, internal to the cell, is required for coordinating wall synthetic and degradative activities, then this system may not be able to function over a significantly larger surface area. Further dissection of the peptidoglycan synthetic systems of B. subtilis will further our understanding of this process in gram-positive species and will enable the more effective development of antibacterial agents active against this attractive target.
Acknowledgments
This work was supported by grant GM56695 (to D.L.P.) from the National Institutes of Health.
We thank Eric Brown for providing pSWEET-bgaB; Amanda Dean, Kathy Lowe, Jill Sible, Ann Stevens, and Nan Qin for technical assistance; and Marita Seppanen Popham for editing the manuscript.
REFERENCES
- 1.Addinall, S. G., and J. Lutkenhaus. 1996. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol. 22:231-237. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:74-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, A. J., R. S. Green, A. J. Sturman, and A. R. Archibald. 1978. Cell wall assembly in Bacillus subtilis: location of wall material incorporated during pulsed release of phosphate limitation, its accessibility to bacteriophages and concanavalin A, and its susceptibility to turnover. J. Bacteriol. 136:886-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis, and turnover, p. 381-410. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 6.Atrih, A., G. Bacher, G. Allmaier, M. P. Williamson, and S. J. Foster. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257-266. [DOI] [PubMed] [Google Scholar]
- 8.Bhavsar, A. P., X. Zhao, and E. D. Brown. 2001. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl. Environ. Microbiol. 67:403-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coote, J. G. 1972. Sporulation in Bacillus subtlis. Characterization of oligosporogenous mutants and comparison of their phenotypes with those of asporogenous mutants. J. Gen. Microbiol. 71:1-15. [DOI] [PubMed] [Google Scholar]
- 10.Daniel, R. A., S. Drake, C. E. Buchanan, R. Scholle, and J. Errington. 1994. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J. Mol. Biol. 235:209-220. [DOI] [PubMed] [Google Scholar]
- 11.de Jonge, B. L. 1990. Isogenic variants of Escherichia coli with altered morphology have peptidoglycan with identical muropeptide composition. J. Bacteriol. 172:4682-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Jonge, B. L., F. B. Wientjes, I. Jurida, F. Driehuis, J. T. Wouters, and N. Nanninga. 1989. Peptidoglycan synthesis during the cell cycle of Escherichia coli: composition and mode of insertion. J. Bacteriol. 171:5783-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Den Blaauwen, T., M. E. Aarsman, N. O. Vischer, and N. Nanninga. 2003. Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol. Microbiol. 47:539-547. [DOI] [PubMed] [Google Scholar]
- 14.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Pedro, M. A., W. D. Donachie, J. V. Holtje, and H. Schwarz. 2001. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J. Bacteriol. 183:4115-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dougherty, T. J., K. Kennedy, R. E. Kessler, and M. J. Pucci. 1996. Direct quantitation of the number of individual penicillin-binding proteins per cell in Escherichia coli. J. Bacteriol. 178:6110-6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, S. J., and D. L. Popham. 2002. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules, p. 21-41. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its close relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 18.Glauner, B., J. V. Holtje, and U. Schwarz. 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 263:10088-10095. [PubMed] [Google Scholar]
- 19.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guzman, L. M., D. S. Weiss, and J. Beckwith. 1997. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J. Bacteriol. 179:5094-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 22.Holtje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoskins, J., P. Matsushima, D. L. Mullen, J. Tang, G. Zhao, T. I. Meier, T. I. Nicas, and S. R. Jaskunas. 1999. Gene disruption studies of penicillin-binding proteins 1a, 1b, and 2a in Streptococcus pneumoniae. J. Bacteriol. 181:6552-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacoby, G. H., and K. D. Young. 1988. Unequal distribution of penicillin-binding proteins among inner membrane vesicles of Escherichia coli. J. Bacteriol. 170:3660-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessières, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S.-K. Choi, J.-J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Düsterhöft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S.-Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Hénaut, H. Hilbert, S. Holsappel, S. Hosono, M.-F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. Klaerr-Blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S.-M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauël, C. Médigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'reilly, K. Ogawa, A. Ogiwara, B. Oudega, S.-H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B.-S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H.-F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 26.LeDeaux, J. R., and A. D. Grossman. 1995. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J. Bacteriol. 177:166-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leighton, T. J., and R. H. Doi. 1971. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J. Biol. Chem. 254:3189-3195. [PubMed] [Google Scholar]
- 28.Ma, X., D. W. Ehrhardt, and W. Margolin. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by with green fluorescent protein. Proc. Natl. Acad. Sci. USA 93:12998-13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masson, J. M., and R. Labia. 1983. Synthesis of a 125I-radiolabeled penicillin for penicillin-binding proteins studies. Anal. Biochem. 128:164-168. [DOI] [PubMed] [Google Scholar]
- 30.Matsuhashi, M., M. Wachi, and F. Ishino. 1990. Machinery for cell growth and division: penicillin-binding proteins and other proteins. Res. Microbiol. 141:89-103. [DOI] [PubMed] [Google Scholar]
- 31.Matsuzawa, H., S. Asoh, K. Kunai, K. Muraiso, A. Takasuga, and T. Ohta. 1989. Nucleotide sequence of the rodA gene, responsible for the rod shape of Escherichia coli: rodA and the pbpA gene, encoding penicillin-binding protein 2, constitute the rodA operon. J. Bacteriol. 171:558-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McPherson, D. C., A. Driks, and D. L. Popham. 2001. Two class A high-molecular-weight penicillin-binding proteins of Bacillus subtilis play redundant roles in sporulation. J. Bacteriol. 183:6046-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPherson, D. C., and D. L. Popham. 2003. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J. Bacteriol. 185:1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, J. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Mobley, H. L., A. L. Koch, R. J. Doyle, and U. N. Streips. 1984. Insertion and fate of the cell wall in Bacillus subtilis. J. Bacteriol. 158:169-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray, T., D. L. Popham, and P. Setlow. 1997. Identification and characterization of pbpA encoding Bacillus subtilis penicillin-binding protein 2A. J. Bacteriol. 179:3021-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray, T., D. L. Popham, and P. Setlow. 1996. Identification and characterization of pbpC, the gene encoding Bacillus subtilis penicillin-binding protein 3. J. Bacteriol. 178:6001-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 39.Paik, J., I. Kern, R. Lurz, and R. Hakenbeck. 1999. Mutational analysis of the Streptococcus pneumoniae bimodular class A penicillin-binding proteins. J. Bacteriol. 181:3852-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popham, D. L. 2002. Specialized peptidoglycan of the bacterial endospore: the inner wall of the lockbox. Cell. Mol. Life Sci. 59:426-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Popham, D. L., M. E. Gilmore, and P. Setlow. 1999. Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties. J. Bacteriol. 181:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Analysis of the peptidoglycan structure of Bacillus subtilis endospores. J. Bacteriol. 178:6451-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc. Natl. Acad. Sci. USA 93:15405-15410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popham, D. L., and P. Setlow. 1995. Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for penicillin-binding protein (PBP) 1 and a PBP-related factor. J. Bacteriol. 177:326-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popham, D. L., and P. Setlow. 1993. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis pbpF gene, which codes for a putative class A high-molecular-weight penicillin-binding protein. J. Bacteriol. 175:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popham, D. L., and P. Setlow. 1994. Cloning, nucleotide sequence, mutagenesis, and mapping of the Bacillus subtilis pbpD gene, which codes for penicillin-binding protein 4. J. Bacteriol. 176:7197-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Popham, D. L., and P. Setlow. 1996. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin-binding proteins. J. Bacteriol. 178:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pucci, M. J., and T. J. Dougherty. 2002. Direct quantitation of the numbers of individual penicillin-binding proteins per cell in Staphylococcus aureus. J. Bacteriol. 184:588-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romeis, T., U. Kohlrausch, K. Burgdorf, and J. V. Holtje. 1991. Murein chemistry of cell division in Escherichia coli. Res. Microbiol. 142:325-332. [DOI] [PubMed] [Google Scholar]
- 50.Spratt, B. G. 1975. Distinct penicillin-binding proteins involved in the division, elongation, and shape of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 72:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinmetz, M., and R. Richter. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142:79-83. [DOI] [PubMed] [Google Scholar]
- 52.Tamaki, S., H. Matsuzawa, and M. Matsuhashi. 1980. Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J. Bacteriol. 141:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vinella, D., D. Joseleau-Petit, D. Thevenet, P. Bouloc, and R. D'Ari. 1993. Penicillin-binding protein 2 inactivation in Escherichia coli results in cell division inhibition, which is relieved by FtsZ overexpression. J. Bacteriol. 175:6704-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yanouri, A., R. A. Daniel, J. Errington, and C. E. Buchanan. 1993. Cloning and sequencing of the cell division gene pbpB, which encodes penicillin-binding protein 2B in Bacillus subtilis. J. Bacteriol. 175:7604-7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yousif, S. Y., J. K. Broome-Smith, and B. G. Spratt. 1985. Lysis of Escherichia coli by beta-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J. Gen. Microbiol. 131:2839-2845. [DOI] [PubMed] [Google Scholar]