Abstract
The DnaQ-H family exonuclease Snipper (Snp) is a 33-kDa Drosophila melanogaster homolog of 3′hExo and ERI-1, exoribonucleases implicated in the degradation of histone mRNA in mammals and in the negative regulation of RNA interference (RNAi) in Caenorhabditis elegans, respectively. In metazoans, Snp, Exod1, 3′hExo, ERI-1, and the prpip nucleases define a new subclass of structure-specific 3′-5′ exonucleases that bind and degrade double-stranded RNA and/or DNA substrates with 3′ overhangs of 2–5 nucleotides (nt) in the presence of Mg2+ with no apparent sequence specificity. These nucleases are also capable of degrading linear substrates. Snp efficiently degrades structured RNA and DNA substrates as long as there exists a minimum 3′ overhang of 2 nt to initiate degradation. We identified a Snp mutant and used it to test whether Snp plays a role in regulating histone mRNA degradation or RNAi in vivo. Snp mutant flies are viable, and display no obvious developmental abnormalities. The expression pattern and level of histone H3 mRNA in Snp mutant embryos and third instar imaginal eye discs was indistinguishable from wild type, suggesting that Snp does not play a significant role in the turnover of histone mRNA at the end of the S phase. The loss of Snp was also unable to enhance the silencing capability of two different RNAi transgenes targeting the white and yellow genes, suggesting that Snp does not negatively modulate RNAi. Therefore, Snp is a nonessential exonuclease that is not a functional ortholog of either 3′hExo or ERI-1.
Keywords: Histone mRNA, ERI-1, RNAi, exonuclease, Snipper
INTRODUCTION
Replication-dependent histone biosynthesis is tightly coupled to DNA replication during the S phase to ensure the rapid deposition of newly synthesized histones at the replication fork (Marzluff and Duronio 2002). This coupling is required for proper chromosome duplication, and reduces cell growth and viability when disrupted (Han et al. 1987; Nelson et al. 2002; Gunjan and Verreault 2003). The regulated destruction of histone mRNA is an important aspect of the coupling of histone mRNA abundance to both the cell cycle and the rate of DNA synthesis. In mammalian cells the steady-state level of histone mRNA increases ∼35-fold as cells progress from G1 to S. This results from an increase in both the synthesis and half-life of histone mRNA (Harris et al. 1991). As cells exit the S-phase, histone mRNA synthesis terminates and existing mRNAs are rapidly degraded. Histone mRNAs are also rapidly destroyed after treating cells with inhibitors of DNA replication such as hydroxyurea (Sittman et al. 1983).
While the precise mechanism of histone mRNA degradation is unknown, the 3′ end of histone mRNA is the cis element responsible for regulating histone mRNA degradation (Pandey and Marzluff 1987). Metazoan replication-dependent histone mRNAs are the only mRNAs that do not end in a polyA tail, but instead end in a 26 nucleotide (nt) stem–loop structure (Dominski and Marzluff 1999). The stem–loop binds stem–loop binding protein (SLBP), and the SLBP–histone mRNA complex plays an important role in the coordinate regulation of histone mRNA processing, translation, and stability. Ongoing translation is required for the regulated destruction of histone mRNA (Graves et al. 1987; Kaygun and Marzluff 2005a, b). SLBP may also participate in the destruction of histone mRNAs in response to replication inhibitors by recruiting the nonsense-mediated decay factor UPF1 in response to ATR checkpoint kinase activation (Kaygun and Marzluff 2005a).
Recently, an exoribonuclease termed 3′hExo was identified as a candidate regulator of histone mRNA degradation in mammalian cells. 3′hExo binds the histone mRNA stem–loop, and removes nucleotides in the 3′ flanking region of histone mRNA (Dominski et al. 2003). The 3′hExo is also capable of forming a ternary complex with the stem–loop histone mRNA and human SLBP. However, as yet there is no evidence that 3′hExo participates in histone mRNA destruction in vivo. Intriguingly, the closest Caenorhabditis elegans homolog of 3′hExo, ERI-1, has been implicated as a negative regulator of RNAi (Kennedy et al. 2004), and loss of ERI-1 leads to enhancement of gene silencing by exogenous dsRNAs. However, the precise role of ERI-1 in the RNAi pathway (Duchaine et al. 2006) or whether it has a role in histone mRNA metabolism is not clear.
To gain insight into the mechanism of histone mRNA metabolism, and to ask whether a 3′hExo/ERI-1 family member plays a role in RNAi or degradation of histone mRNAs in vivo, we biochemically and genetically characterized the only homolog of 3′hExo in Drosophila melanogaster, which we named Snipper (Snp). Comparison of the biochemical activity of Snp to 3′hExo showed that Snp is a highly active and promiscuous 3′→5′ nuclease, degrading structured nucleic acid substrates, such as the histone mRNA hairpin more efficiently than 3′hExo. Snp also has broad substrate specificity in that it is capable of degrading double-stranded as well as single-stranded DNA and RNA substrates. Similar to 3′hExo, Snp shows no apparent sequence specificity toward a particular nucleic acid sequence, and binds best to double-stranded DNA and RNA substrates with a minimum 2–5 nt 3′ flank. However, unlike 3′hExo, Snp does not require a 2′OH for substrate recognition, catalysis, or product release, making it both an efficient RNase and DNase. Our biochemical studies have helped define the properties of a new subfamily of the DnaQ-H 3′→5′exonucleases in eukaryotes. Given the broad substrate specificity of Snp in vitro, we determined whether Snp plays a role in histone mRNA degradation, RNAi, or apoptosis in vivo. We show that expression of Snp is not required for the regulated destruction of histone mRNA as cells progress from the S phase to G2. The loss of Snp expression also does not lead to an enhancement of gene silencing by exogenous dsRNAs or affect the degradation of DNA during apoptosis.
RESULTS
3′hExo/ERI-1 and Drosophila Snp define a family of closely related metazoan exonucleases
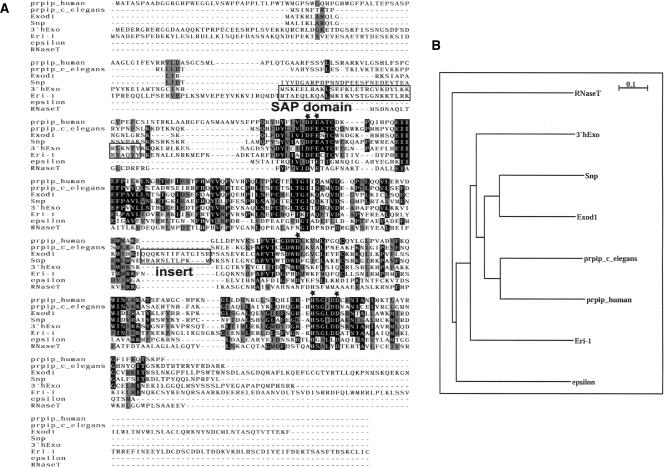
Nucleases have traditionally been classified based on the substrate hydrolyzed (DNA versus RNA), the mechanism of nucleolytic attack (endo versus exo), the hydrolytic products formed (oligonucleotides terminating in a 3′ or 5′ phosphate), and the nature of the bond hydrolyzed. Based on these criteria, 3′hExo and ERI-1 belong to the DnaQ-H family (Viswanathan and Lovett 1999) of 3′ to 5′ exoribonucleases that produce hydrolytic products releasing a nucleotide 5′ monophosphate and leaving a 3′ OH on the penultimate nucleotide. 3′hExo and ERI-1 share 38% sequence identity and 60% sequence similarity over all residues (and the same degree of identity and similarity between the respective nuclease domains). There are no other proteins that are homologous to ERI-1 and that show a similar tandem arrangement of an N-terminal SAP domain followed by an ExoIII domain in either the C. elegans or the human genome. The Drosophila genome contains only one exonuclease in the DnaQ-H superfamily homologous with ERI-1 and 3′hExo. This protein (Flybase ID CG6393) shares 31% sequence identity with 3′hExo as well as ERI-1 (Fig. 1A,B). We named CG6393 “Snipper (Snp)” based on its potent exonucleolytic activity in vitro (see below). We also identified several other putative exonucleases in C. elegans and humans (Exod1 and prpip; see “Alignments and Sequences” in Materials and Methods) that are closely related to 3′hExo, ERI-1, and Snp. These proteins comprise a subfamily of DNaQ-H exonucleases, which we call the 3′hExo/ERI-1 family.
FIGURE 1.
(A) A structure-based alignment of the 3′hExo/ERI-1 family of Exonucleases was generated using the programs CE and T-Coffee. Identical conserved residues are shown in black box shade, similar conserved residues in gray shade, and nonidentical residues are left with a white background. The location of the SAP domain (present only in 3′hExo and ERI-1) and the 13-residue insert (present only in Snp and Exod1) are boxed. The conserved DEDDh residues that coordinate the divalent metal ion cluster are highlighted with black stars. There are several residues that are conserved in Snp, Exod1, prpip, 3′hExo, and ERI-1 but not in epsilon or RnaseT. (B) Rooted Phylogenetic tree produced from ClustalW and NJPlot, showing that Snp is closely related to the uncharacterized human protein, Exod1 (AAH10503), followed by prpip nucleases and then 3′hExo and ERI-1.
A structure-based sequence alignment of members of the DnaQ-H superfamily of 3′-5′ exonucleases (Viswanathan and Lovett 1999) shows that Snp has a characteristic DEDDh motif that is essential for the catalytic activity of all these DnaQ-H family members. This family includes replicative proofreading DNases such as Exo1 (Breyer and Matthews 2000), the epsilon subunit of Pol III (Hamdan et al. 2002), and Klenow (Ollis et al. 1985), RNases such as oligoribonuclease (Yu and Deutscher 1995), 3′hExo (Dominski et al. 2003), and ERI-1 (Kennedy et al. 2004), and dual specificity nucleases such as RNase T (Deutscher 2006). In addition, the 3′hExo/ERI-1 family of proteins shares extensive sequence similarity and conservation of active site residues that are unique to this metazoan group of exonucleases (Supplemental Fig. 1; http://www.bio.unc.edu/faculty/duronio/lab/publications.htm). A similar active site suggests that Snp, 3′hExo, as well as other members of this family, could share a similar catalytic mechanism, and may recognize similar nucleotide substrates.
Recombinant Snp is a promiscuous 3′-5′ exonuclease and degrades histone stem–loop RNA substrates more efficiently than 3′hExo
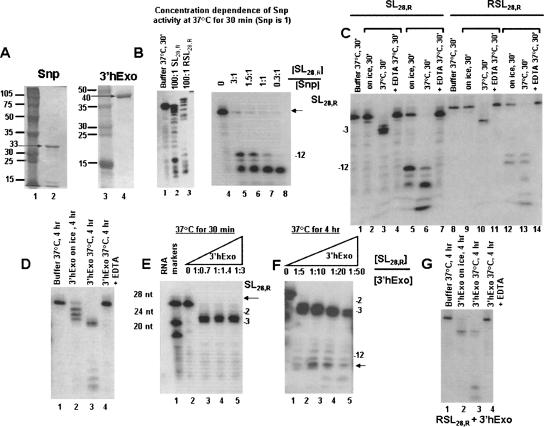
Before we undertook the biological characterization of Snp, we sought to determine (1) whether Snp was indeed a 3′→5′ exonuclease as predicted from its amino acid sequence, and (2) what substrate(s) Snp prefers. For these analyses we compared the ability of Snp to degrade structured and linear RNA and DNA substrates relative to 3′hExo under the same reaction conditions in vitro. Recombinant Snp and 3′hExo were expressed in bacteria and purified to homogeneity (Fig. 2A). The activity of recombinant Snp was tested in nuclease assays against a number of double- and single-stranded DNA and RNA substrates (Table 1). A buffer control was run under similar conditions for all reactions, and no degradation was observed in the presence of buffer alone. The observed activity was robust at a 3:1 ratio of [SL28,R]:[Snp] at 37°C and 30-min incubation (Fig. 2B, lane 1) and good even at ratios of 50:1([SL28,R]:[Snp] (data not shown). At higher enzyme concentrations, almost all the probe was degraded to –12 and −16 oligonucleotide fragments (Fig. 2B, lanes 2–5; Fig. 2C, lanes 6,13) in the presence of magnesium, and this activity was completely abolished by the addition of EDTA (Fig. 2C, lanes 4,7,11,14). This suggests that cleavage may proceed via the two metal–ion catalyzed mechanism proposed initially by Steitz and coworkers (Derbyshire et al. 1988; Beese and Steitz 1991) and later confirmed for a number of DEDDh nucleases.
FIGURE 2.
Snp and 3′hExo activity toward histone mRNA stem–loop substrates. (A) Purification of Snp and 3′hExo. Purification of Snp and 3′hExo was monitored by 12% SDS-PAGE, the gels were stained with Coomassie Brilliant Blue. Lane 1, protein markers whose apparent molecular weights are indicated; lane 2, Snp after elution from the HiTrap Ni2+ column; lane 3, protein markers; lane 4, 3′hExo after elution from the HiTrap Ni2+ column. In B, Snp activity as a function of substrate concentration is shown at 37°C and 30-min incubation. To evaluate Snp behavior at steady state, we varied the concentration of substrate from a 3:1 molar ratio of [SL28,R] relative to [Snp] in the presence of a slight excess of substrate (Snp is always 1) to a 0.3:1 ratio in the presence of excess enzyme. Panel C shows a comparison of 3′hExo and Snp nuclease activity toward RNA stem–loop and reverse stem–loop at 0°C and 37°C. Both 3′hExo and Snp were at 0. 02 pmol, and the substrate was at 0.5 pmol in a total reaction volume of 10 μL. (D–G) The activity of 3′hExo toward histone mRNA SL28,R and RSL28,R substrates as a function of time, temperature, and concentration. In D, the relative rates of degradation of stem–loop RNA by 3′hExo for a sample kept on ice at room temperature (expected temperature range 0°C –10°C) and 37°C are shown. 0.05 pmol of 3′hExo was incubated with a stem–loop RNA probe (0.5 pmol) in a reaction volume of 10 μL and cleavage analyzed after 4 h at 37°C. Two major cleavage products are observed at the −2 and −3 positions, and at the 4-h point a significant amount of the −12 cleavage product accumulates. In E and F, the activity of 3′hExo as a function of time and concentration is depicted. In G, the activity of 3′hExo toward the reverse stem–loop substrate is compared to the stem–loop under the same conditions as shown in D.
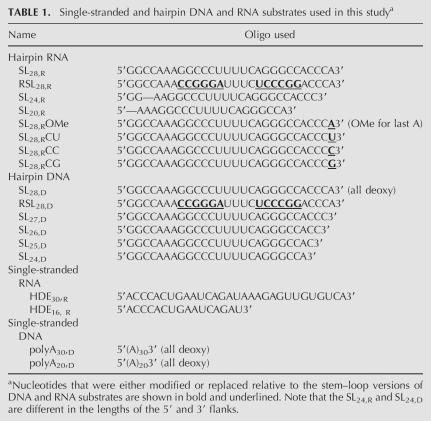
TABLE 1.
Single-stranded and hairpin DNA and RNA substrates used in this studya
The relative nuclease activities of Snp and 3′hExo were compared at different [Enzyme]:[substrate] ratios and under various time and temperature conditions using the SL28,R and RSL28,R substrates, which were previously used to characterize the nuclease activity of 3′hExo expressed in baculovirus (Fig. 2C; Dominski et al. 2003). Intriguingly, Snp is a much better nuclease than 3′hExo toward histone stem–loop RNA. Snp can readily cleave the stem beyond the –12 (UUU) position in the loop to produce −14 (Fig. 2B, lanes 4–5) and then −16 oligonucleotide fragments for both the stem–loop and the reverse stem–loop (Fig. 2C, lanes 6,13). These degradation products are observed within 30 min of incubation at 37°C in the presence of substoichiometric amounts of Snp, unlike 3′hExo, where a 4-h incubation period was required as well as a 10–50-fold excess of enzyme at the same temperature (Fig. 2F, lanes 3–5). Therefore, Snp is capable of efficiently cleaving both the single-stranded 3′flank as well as the double-stranded stem portion of histone stem–loop RNA, with faster kinetics compared to 3′hExo. As we show below for DNA substrates (Fig. 3F), a ladder of degradation products is observed for a 5′−end labeled DNA hairpin, suggesting that degradation occurs processively from the 3′ end.
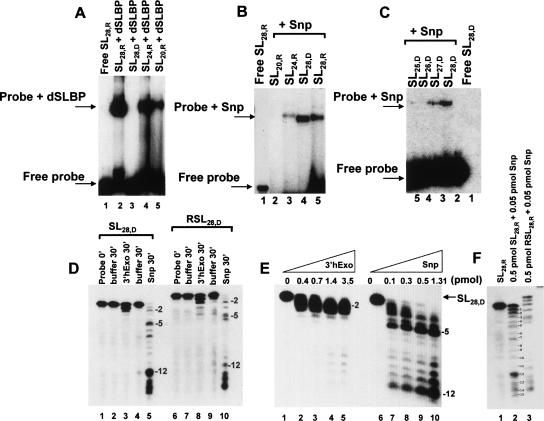
FIGURE 3.
Snp and 3′hExo activity toward DNA hairpin substrates. (A) Binding of Drosophila SLBP toward SL28,R (lane 2), SL28,D (lane 3), SL24,R (lane 4), and SL20,R (lane 5). The free probe is shown in lane 1. dSLBP binds best to SL28,R in comparison to the shorter RNA fragments as previously reported (Thapar et al. 2004). In B, the relative binding of Snp toward stem–loop RNA and DNA substrates was tested in EMSA assays in the presence of EDTA. An equivalent concentration of substrate and protein was used in all lanes as described in Materials and Methods. In C, the ability of DNA stem–loops (that differ in the number of nucleotides in the 3′ flank) to bind Snp was tested in mobility shift assays. The free probe is shown in lane 1; lane 2 shows the shift observed for SL28,D, which has five unpaired nucleotides in the 3′ flank; lane 3 shows that a weaker shift occurs upon deletion of 1 nt in SL27,D. No detectable binding is observed for SL26,D and SL25,D substrates that have three and two unpaired nucleotides, respectively. (D) The relative activities of 3′hExo and Snp (both at 0.02 pmol) toward SL28,D and RSL28,D, (both at 0.5 pmol) in the presence of excess substrate. Total reaction volume is 10 μL. The RSL28,D probe migrates a little slower than the SL28,D. Lanes 1 and 6 shows the free probe at 0 min, lanes 2, 4, 7, and 9 show the respective probes incubated in buffer for 30 min, lanes 3 and 8 show the respective probes incubated with 3′hExo, and lanes 5 and 10 show the probes incubated with Snp. (E) The concentration dependence of the activity of 3′hExo and Snp toward 0.5 pmol of SL28,D. Under limiting enzyme conditions, Snp is more active toward DNA substrates and efficiently cleaves through the stem portion of the DNA hairpin. A progressive increase in the populations of shorter nucleotide fragments is observed for Snp as the concentration is increased, at constant [substrate] (which is in excess). An appearance of fragments is observed as a function of time and constant [substrate]:[Snp] consistent with processive activity toward the stem–loop. In F, the degradation of 0.5 pmol SL28,D and RSL28,D is shown in the presence of 0.05 pmol of Snp, i.e., at the lowest [Snp] concentration tested. Under these conditions, a ladder of nucleotide products is observed for both SL28,D and RSL28,D due to processive cleavage from the 3′ end showing that Snp is a 3′→5′ exonuclease.
Previous studies indicate that in the presence of a large excess of 3′hExo enzyme ([stem–loop RNA ]:[3′hExo] of 1:500) 3′hExo removes nucleotides from the 3′ end of histone stem–loop RNA, producing three major products corresponding to positions −3, −5, and −12 from the 3′ end of the histone RNA hairpin substrate (SL28,R). At a lower molar ratio of [stem–loop RNA ]:[3′hExo] of 1:5, cleavage of only 3 nt occurred from the 3′ end of the stem–loop. In the presence of at least 20-fold excess of stem–loop RNA substrate relative to 3′hExo, cleavage products of −2 and −3 nt in the 3′ flanking region are generated after 30 min, and only one product is observed at the −3 position after 60-min incubation. No degradation of the stem occurred by 3′hExo when the enzyme was limiting (Dominski et al. 2003).
A similar pattern was obtained in our studies with bacterially expressed 3′hExo (Fig. 2D,E). Even after a 4-h incubation period at 37°C, and in the presence of a 10:1 molar ratio of [SL28,R]:[3′hExo], the major products generated correspond to cleavage of 2 and 3 nt from the 3′ flank (Fig. 2E). However, at higher enzyme concentrations between 1:10 and 1:50 molar ratios of [SL28,R]:[3′hExo] and a 4-h incubation at 37°C, degradation to the −12 position is observed (Fig. 2F). Intriguingly, the cleavage of up to 3 nt in the 3′ flank occurs even for a sample kept on ice (temperature range 0–10°C) after 4 h and the −1, −2, and −3 products are clearly resolved in the gel (Fig. 2D, lane 2). This temperature and concentration dependence of nuclease activity suggests that the rate of cleavage of the first 3 nt by 3′hExo is quite fast at 37°C. Therefore, 3′hExo shows biphasic kinetics such that there is rapid hydrolysis of the last three unpaired nucleotides in the 3′ flanking sequence followed by much slower cleavage through the stem that occurs over a longer incubation period on the order of hours. No degradation of the probe is observed after 4 h at 37°C in the buffer control, or in a reaction where 3′hExo is incubated with EDTA (Fig. 2D, lane 4). Our results suggest that the biologically relevant product generated by 3′hExo is cleavage of 2–3 nt from the 3′ flank. Any further cleavage through the stem occurs only in the presence of a large excess of enzyme relative to substrate (at least a 50-fold excess) or over a longer incubation period of 4 h at 37°C at lower [3′hExo], and is not likely to be biologically relevant.
Therefore, Snp is capable of cleaving both the single-stranded 3′ flank as well as the double-stranded stem portion of histone stem–loop RNA with faster kinetics compared to 3′hExo. One reason for the increased activity of Snp relative to 3′hExo may be the absence of the SAP domain in Snp. The SAP domain is important for high affinity recognition of the histone mRNA stem–loop (Cheng and Patel 2004); D. Bernardo and R. Thapar, unpubl.). Since Snp lacks the SAP domain and is able to efficiently cleave through the stem, our data suggest that the SAP domain in 3′hExo may bind the stem of the histone hairpin, thereby protecting it from degradation.
An interesting feature of both Snp and 3′hExo is that both consistently stall in the –12(UUU) sequence of the loop. 3′hExo is not able to degrade the substrate any further, suggesting that either it requires >16 nt to efficiently bind its substrate or that a string of uridines is unfavorable for binding. Snp can remove 2–4 nt from the −12 substrate (Fig. 2B,C). As we show below (Fig. 4), 3′hExo and Snp can efficiently degrade the 16-nt single-stranded HDE16,R substrate; therefore, the length of an unstructured substrate appears not to be as critical as having a string of 2–3 uridines.
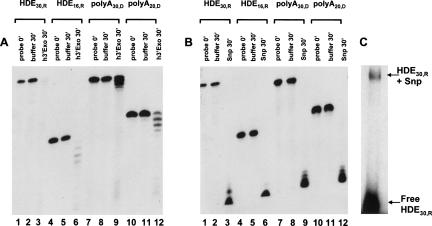
FIGURE 4.
Snp activity toward single-stranded RNA and DNA substrates. (A) The activity of 3′hExo toward single-stranded RNA (HDE30,R, HDE16,R) and DNA (polyA30,D, polyA20,D) substrates. The amount of 3′hExo used was 3.5 pmol, and that of substrate was 0.5 pmol in a 10 μL reaction volume. (B) The activity of Snp toward single-stranded RNA (HDE30,R, HDE16,R) and DNA (polyA30,D, polyA20,D) substrates. The amount of Snp was 1.3 pmol, and that of all substrates 0.5 pmol. Snp is active toward all substrates at lower enzyme concentrations compared to 3′hExo. Lanes 1, 4, 7, and 10 show the free probes at 0 min; lanes 2, 5, 8, and 11 show the free probes incubated in the reaction buffer at 37°C for 30 min; and lanes 3, 6, 9, and 12 show the probe incubated with the respective nuclease at 37°C for 30 min. (C) The ability of Snp to shift a linear RNA substrate HDE30,R was measured in an EMSA assay. At least 10-fold higher concentrations of Snp are required to observe a shift against a linear probe (compared to the RNA or DNA hairpin) and the shifted band is more diffuse, suggesting a faster off-rate for a linear substrate.
These data suggest that Snp is more active toward the histone mRNA stem–loop than 3′hExo, and easily degrades structured RNA substrates in vitro. Therefore, it is a good candidate nuclease to test for histone mRNA degradation in vivo.
Snp is also a DNase, whereas 3′hExo is more RNA-specific
In order to compare the DNase activity of Snp and 3′hExo to their ribonuclease activity described above, we tested the ability of Snp and 3′hExo to bind and degrade a deoxy-ribose version (SL28,D) of the same stem–loop sequence as was used for the histone RNA (SL28,R) under similar [Enz]:[substrate] ratios of 1:5 at 37°C for 30 min (Fig. 3A). The histone SLBP, which binds the histone stem–loop RNA with nanomolar affinity, does not bind the DNA hairpin (SL28,D) in EMSA assays (Fig. 3A), suggesting that the DNA hairpin SL28,D, although related in sequence to SL28,R, should present a structurally different substrate for recognition by Snp. The relative binding of Snp toward SL28,R and SL28,D substrates was tested in EMSA assays in the presence of EDTA. Snp binds SL28,D much better than SL28,R in mobility-shift assays (Fig. 3B, cf. lanes 4 and 5) although binding is best for structured RNA and DNA that provide a linear 3′ overhang of 5 nt (Fig. 3B,C). We find that both Snp and 3′hExo degrade SL28,D with slower kinetics as estimated from the nature of the cleavage products generated at 37°C after a 30-min incubation at similar [enzyme]:[substrate] ratios (cf. Figs. 2C and 3D). Comparison of products generated as a function of enzyme concentrations shows that Snp can cleave DNA substrates (Fig. 3E) at a much lower concentrations compared to 3′hExo, and is also able to degrade the stem portion of the DNA hairpin similar to that observed for RNA. The nuclease activity of Snp toward SL28,D and RSL28,D substrates at a 10:1 ratio of [substrate]:[Snp] shows clearly that it is a 3′→5′ exonuclease (Fig. 3F). A ladder of degradation products is observed for a probe that is labeled at the 5′ end with decreasing intensity that could result only from 3′→5′ exonucleolytic cleavage.
3′hExo cleaves only a fraction of SL28,D (Fig. 3D, lane 3) generating the −1 and −2 nucleotide products under conditions in which almost complete turnover of SL28,R occurs to a product that is cleaved to the −3 position (Fig. 2E). Similarly, a substantial population of the −5 cleavage product is observed for Snp with SL28,D under conditions in which Snp is able to completely degrade SL28,R to the −12 product as well as shorter nucleotide fragments (cf. Figs. 2B and 3E).
Previous studies on the Tetrahymena group I Ribozyme (Herschlag et al. 1993) and RNase P (Forster and Altman 1990; Smith and Pace 1993) show that a decrease in the apparent rate of cleavage observed toward DNA substrates represents contributions from both the intrinsic as well as the enzyme-catalyzed rate of hydrolysis for the phosphodiester bond in the context of a 2′deoxyribose compared to a ribose. Therefore, in order to determine the degree to which the enzymatic rates are affected for DNA versus RNA, we need to account for the contribution of the 2′deoxy to the intrinsic rate of phosphodiester bond cleavage for SL28,R and SL28,D substrates. A direct and easier method to test whether Snp and 3′hExo require a 2′OH for substrate binding or catalysis (as do most 2′,3′ phosphate cleaving RNases) involves substituting the terminal ribose 2′hydroxyl of the adenosine with a 2′O-methyl group in SL28,R and testing nuclease activity for this substrate. Most exoribonucleases show a dependence of nuclease activity on the 2′OH, through binding effects via favorable hydrogen bonding interactions with the substrate and also on the rate of catalysis via stabilization of the transition state (Forster and Altman 1990; Herschlag et al. 1993). The crystal structure of the nuclease domain of 3′hExo bound to rAMP shows that the backbone amide and carboxyl group of Ala137 hydrogen bond to the O3′ and O2′ hydroxyls of the ribose and this residue is Ala72 in Snp. Consistent with this, 3′hExo is unable to degrade the 2′O-methylated stem–loop SL28,ROMe by even 1 nt at 37°C after 30 min (Fig. 5A, lanes 13,14), suggesting that the 2′-methoxy has a large effect on the overall rate of phosphohydrolysis. The loss of a hydrogen bond and the introduction of a bulky steric group at the 2′ position is detrimental to the nuclease activity of 3′hExo, making it an RNA-specific enzyme. The complete loss of nuclease activity for 3′hExo for the 2′OMe stem–loop compared to a 2′-H (in the 2′deoxy SL28,D) suggests that in addition to loss of a hydrogen bond, the bulky methyl group could sterically exclude the catalytic water from the active site, as has been observed for RNase P (Smith and Pace 1993), thereby having a more deleterious effect on the rate of degradation. In contrast to 3′hExo, the 2′methoxy substitution does not appreciably affect nuclease activity of Snp, and almost all the substrate is degraded to the −12 and smaller degradation products. Our data suggest that Snp is a broad-specificity nuclease, capable of degrading both DNA and RNA targets, whereas 3′hExo is more RNA specific. These results also suggest that in spite of the putative similarity in their active sites, there are important mechanistic differences in substrate binding and catalysis between the two nucleases.
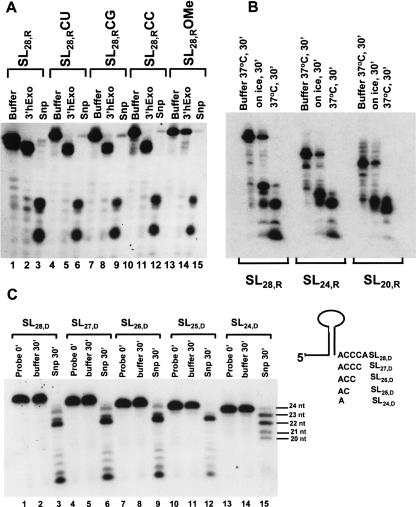
FIGURE 5.
Snp is a structure-specific nuclease. In A, the sequence specificity of Snp and 3′hExo was tested toward the 3′ nucleotides on the flanking sequence ACCCA (lanes 1–3) that is conserved in most histone mRNAs by changing the terminal adenosine to a U (lanes 4–6), G (lanes 7–9), and C (lanes 10–12), and incubating these substrates with the respective nucleases at 37°C for 30 min. The amount of 3′hExo and Snp used was 0.02 pmol and that of substrate was 0.5 pmol in a 10 μL reaction. No appreciable difference in the cleavage patterns is observed. In lanes 13–15, the effect of the 2′methoxy substitution is probed for both Snp and 3′hExo. Snp is able to cleave the terminal 2′O-methylated adenosine off the 3′ end, whereas the 3′hExo is not. In B, the nuclease activity of Snp toward SL28,R, SL24,R, and SL20,R substrates was compared at 0°C and 37°C. The amount of Snp was 0.3 pmol and that of all substrates was 0.5 pmol. Snp cleaves all RNA substrates to a similar extent under these conditions and at both temperatures. In C, the activity of Snp (1.3 pmol) toward different stem–loop DNA substrates (0.5 pmol) with decreasing lengths of the 3′ flank is shown. The data for each probe are in triplicate, with the first lane representing the probe at 0 min, the second lane representing the probe at 30 min in the presence of buffer, and the third lane representing the probe at 30 min in the presence of Snp.
Snp activity toward single-stranded RNA and DNA substrates
The ability of Snp to bind and degrade single-stranded RNA and DNA substrates was tested using HDE30,R, HDE16,R polyA30,D, and polyA20,D substrates (Fig. 4). Snp is highly active toward all these unstructured RNA and DNA substrates, and degrades them to very short nucleotide fragments, all of which run at the bottom of the gel. Under the conditions of our experiments, both deoxy and 2′ribo substrates are cleaved irrespective of length or nucleotide sequence by Snp (Fig. 4B). In contrast, 3′hExo is also able to efficiently cleave the 2′OH (ribo) single-stranded substrates HDE30,R and HDE16,R that differ only in length (Fig. 4A, lanes 1–6). However, the nuclease activity toward the single-stranded 2′deoxy substrates polyA30,D and polyA20,D is very inefficient for 3′hExo (Fig. 4A, lanes 7–12), similar to what we reported earlier for double-stranded DNA stem–loops. Snp binds the single-stranded substrate HDE30,R with lower affinity than the stem–loop substrates tested, such that higher Snp (>10-fold) concentrations are required to form a complex that can be detected in EMSA assays (Fig. 4C). The Snp-HDE30,R complex runs as a diffuse band in a native gel, suggesting a faster off-rate for linear substrates compared to RNA and DNA hairpins.
Specificity of substrate recognition by Snp and 3′hExo
Both Snp and 3′hExo degrade the histone mRNA stem–loop (SL28,R), and the reverse stem–loop (RSL28,R) (Fig. 2C,G), and Snp also degrades the DNA stem–loop (SL28,D) and reverse stem–loop (RSL28,D) (Fig. 3D), suggesting that there is no sequence specificity for the stem sequence. We tested the effect of varying the terminal nucleotide of the ACCCA 3′flank, changing it to ACCCC, ACCCG, or ACCCU (Table 1; Fig. 5A, lanes 1–12). These point mutations had no appreciable effect on nuclease activity of both proteins. Therefore, our data suggest that 3′hExo and Snp do not appear to bind or cleave double-stranded DNA or RNA in a sequence-specific manner. The specific interaction of 3′hExo with the histone message may be conferred via direct or indirect contacts with SLBP, but is not due to preference of a particular nucleotide sequence.
To test whether Snp was sensitive to the length of the 3′ flanking region, we tested binding and activity of DNA and RNA substrates with decreasing lengths of the 3′ flank toward Snp (Fig. 5B,C). Intriguingly, decreasing the length of the 3′ flanking region by 3 nt in either SL28,R (Fig. 5B) or SL28,D (Fig. 5C) did not affect nuclease activity but did affect complex formation in EMSA assays (Fig. 3C) for Snp. However, deletion of 4 or 5 nt from the 3′ end slows the kinetics of DNA degradation for Snp (Fig. 5C, lanes 13–15) as well as abolishes binding toward these substrates in EMSA assays, suggesting that a structured RNA or DNA substrate with a 1–2 nt flank is not the preferred Snp substrate. A similar effect was observed toward RNA substrates (Fig. 5A,B). The minimum length of the 3′ flank for association in a mobility shift assay between Snp and its stem–loop target for both DNA and RNA hairpins was found to be at least 2 nt. Our data suggest that Snp is a structure-specific RNA and DNA nuclease and requires two to five unpaired nucleotides in the 3′ region for efficient binding and nuclease activity.
3′hExo forms a ternary complex with human SLBP and RNA, suggesting that the two macromolecules could potentially be part of a larger macromolecular assembly that catalyzes histone mRNA degradation. We asked whether Snp could form a ternary complex with Drosophila SLBP and SL28,R in the presence of EDTA. While our recombinant 3′hExo formed a ternary complex with human SLBP and Drosophila SLBP in the presence of SL28,R, under the same conditions Snp could not form a ternary complex with Drosophila SLBP and SL28,R RNA (Supplemental Fig. 3; http://www.bio.unc.edu/faculty/duronio/lab/publications.htm). Since Snp has lower affinity toward the stem–loop RNA compared to 3′hExo, we cannot rule out the possibility that a lower affinity complex is indeed formed but is not detected in EMSA assays.
Taken together, our biochemical data indicate that Snp is a highly active nuclease with broad substrate specificity. Snp has higher affinity toward structured hairpin DNA or RNAs that have a 2–5 nt 3′ flank than for single-stranded RNAs, making it a good candidate exonuclease for histone mRNA degradation, RNAi, or DNA repair.
Characterization of the Snp gene and mutant alleles
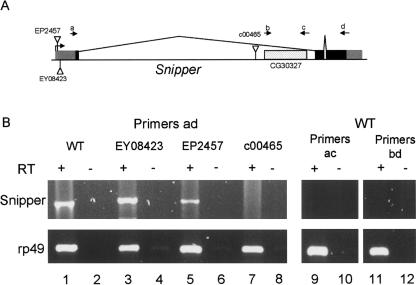
To examine whether Snp plays a role in any of these processes in vivo, we characterized a mutant allele of Snp that was present in the public collection of transposon insertion alleles. We obtained two P-element transposon insertions in the Snp 5′ UTR, EP2457 and EY08423, and one piggyBac transposon insertion in the large first intron, c00465 (Fig. 6A). All three insertions are viable and fertile as homozygotes or when placed in trans to Df(2R)Excel7170, which deletes the entire Snp locus (data not shown). RT-PCR was performed to determine if any of these transposable elements affected the expression of Snp (Fig. 6B). A Snp RT-PCR product was detected using RNA isolated from wild-type control and homozygous EP2457 and EY08423 adult female flies (Fig. 6B, lanes 1–6). In contrast, this RT-PCR product was not detected in homozygous c00465 flies (Fig. 6B, lanes 7–8). Similar data were obtained with RNA isolated from embryos or third larval instar imaginal discs (not shown). These data indicate that c00465 is a mutant allele of Snp, and that EP2457 and EY08423 affect Snp expression very little if at all. The absence of an RT-PCR product in c00465 samples suggests that this insertion may represent a null allele of Snp.
FIGURE 6.
Snp expression in transposon insertion lines. (A) Genomic locus of Snipper showing the intron and exons and the insertion sites of the three transposable elements characterized in this study. Arrows a–d indicate primers used for RT-PCR. (B) Snp RT-PCR of total RNA isolated from homozygous adult females of the indicated genotypes using the indicated primers. Reverse transcriptase was omitted from the reaction in even-numbered lanes.
The c00465 PBac insertion is located in the first intron of Snp, 614 bp upstream of an open reading frame annotated as a distinct gene called CG30327 (Fig. 6A). CG30327 is an intron-less open reading frame that has the potential to encode a protein of 407 amino acids that contains an RNA-binding RRM domain. Because of this, and because 3′hExo contains a SAP domain upstream of the exonuclease domain, we hypothesized that CG30327 may be a differentially spliced exon of Snp, even though none of the known cDNAs of Snp include the CG30327 sequence. To determine if CG30327 was an exon of Snp, RT-PCR was performed with RNA isolated from wild-type adult flies. We were unable to amplify a Snp transcript containing CG30327 from adult female RNA using two different primer pairings (Fig. 6B, lanes 9–12). Thus, it appears that CG30327 is not an exon of Snp, and that Snp does not contain an RNA-binding domain. In addition, we were unable to directly amplify a CG30327 mRNA using primers b and c, and there are no cDNA or EST sequences in current databases containing the CG30327 sequence. These data suggest that, if transcribed, CG30327 mRNA accumulates to very low abundance or in very few tissues.
Snp does not control histone mRNA degradation in vivo
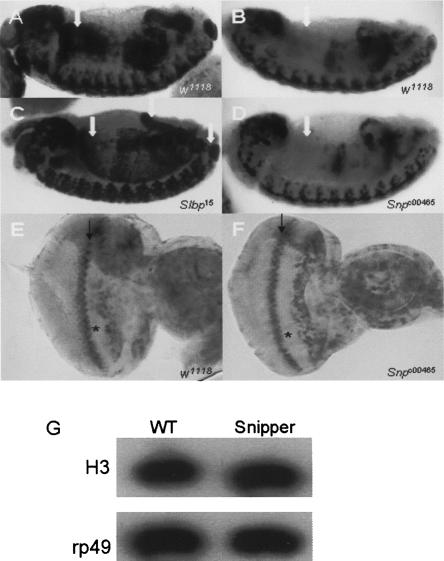
Since Snp is capable of degrading histone mRNA in vitro, we examined histone mRNA expression in Snpc00465 mutants by in situ hybridization to determine whether Snp affects histone mRNA accumulation in vivo. In endocycling cells of the embryonic midgut, histone mRNAs accumulate only during the S-phase, resulting in a dynamic but stereotypical pattern (Fig. 7A). We have previously shown that in Slbp mutants histone mRNAs are not properly processed and become polyadenylated via the use of cryptic downstream polyadenylation signals (Lanzotti et al. 2002). In the embryonic midgut, these polyadenylated histone mRNAs are not rapidly destroyed at the end of the endo S-phase and inappropriately accumulate throughout the midgut in stage 14 embryos (cf. Figs. 7BandC; Sullivan et al. 2001). This aberrant pattern provides a diagnostic for misregulation of histone mRNA destruction at the end of the S-phase. In situ hybridization of Snp mutant embryos with an H3 probe results in a wild-type pattern of expression at stage 14 (Fig. 7D), indicating that loss of Snp expression does not affect the accumulation of histone H3 mRNA in endocycling embryonic cells (Fig. 7B,D).
FIGURE 7.
Histone mRNA expression is not affected in Snp mutants. (A–B) In situ hybridization of w1118 stage 13 (A) and stage 14 (B) control embryos showing the wild-type pattern of histone H3 expression. The arrows indicate the anterior midgut, which down-regulates H3 expression by stage 14. (C) Stage 14 Slbp15 homozygous null mutant embryos with inappropriate H3 expression in the anterior midgut, hind gut, and anal pads (left to right arrows, respectively). (D) Stage 14 Snpc00465 homozygous mutant embryo obtained from homozygous mutant parents showing a normal pattern of H3 expression. Arrow as in B. (E,F) H3 in situ hybridization of w1118 and Snpc00465 third instar eye discs. The asterisks indicate the morphogenetic furrow, where all cells are arrested in G1. The arrows indicate the second mitotic wave where all cells are undergoing a synchronous round of S phase and express H3. (G) H3 Northern blot with total RNA extracted from w1118 and Snpc00465 third instar eye discs. rp49 serves as the loading control.
To examine if mutation of Snp affects histone mRNA expression in a mitotic cell population, in situ hybridizations were performed with eye imaginal discs of third instar larvae. In eye discs there is a stereotyped pattern of development in which a subset of G1-arrested retinal precursor cells undergoes a synchronous round of S phase followed by cell division before terminally differentiating (Fig. 6E). This synchronous S-phase is part of a wave of differentiation called the morphogenetic furrow that sweeps across the eye disc epithelium, and can be easily visualized as a stripe of accumulation of histone H3 mRNA (Fig. 7E). As in the embryo, the pattern of expression of H3 mRNA in Snp mutant eye discs appears identical to wild type, indicating that H3 mRNA accumulates only in replicating cells (Fig. 7F). Consistent with this, Northern blot analysis indicates that the total amount of H3 mRNA is similar between wild-type and Snp mutant eye discs (Fig. 7G). Based on the pattern of accumulation of H3 mRNA in Snp mutant embryos and eye discs, we conclude that Snp does not play a major role in the degradation of histone mRNA at the end of the S phase.
Snp does not regulate RNAi
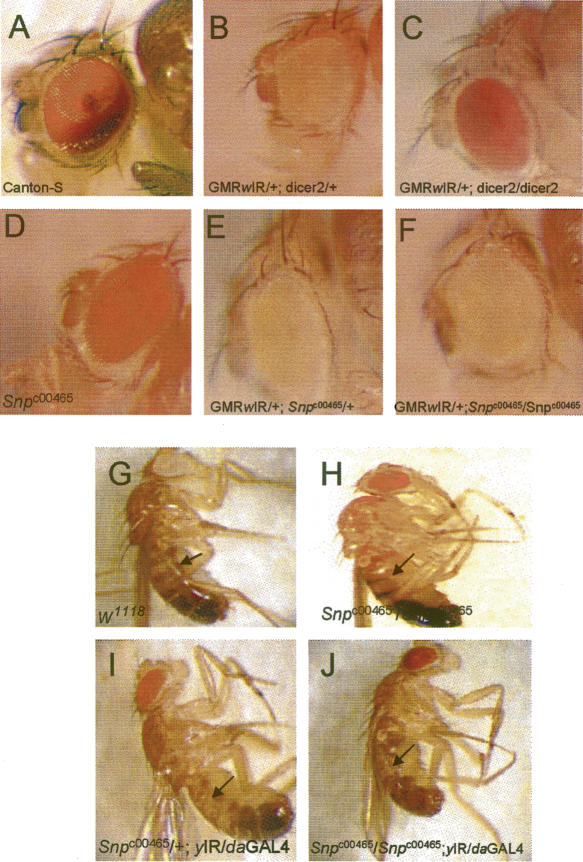
Mutations of C. elegans ERI-1 enhance the efficacy of RNAi through an unknown mechanism (Kennedy et al. 2004). To determine if Snp modulates the RNAi response to exogenous dsRNAs in Drosophila, we tested how Snpc00465 adult mutant flies responded to dsRNA producing transgenes. The first transgene, called GMRwIR, expresses a hairpin RNA of the third exon of the white gene under the control of an eye-specific promoter (Lee and Carthew 2003). One copy of GMRwIR silences white expression, resulting in a faint orange eye color (cf. Fig. 8AandB). Mutation of Dicer-2, which is required for the production of siRNAs from exogenous dsRNAs (Lee et al. 2004), suppresses the effect of GMRwIR and substantially restores eye pigmentation in GMRwIR flies (Fig. 8C). If Snp acts as a negative regulator of RNAi, as does ERI-1, then Snp mutations should enhance the ability of GMRwIR to silence white expression, resulting in a white or pale yellow eye (Lee and Carthew 2003). Homozygous Snp mutants containing GMRwIR had no further reduction in eye color when compared to siblings that are heterozygous for Snp (Fig. 8D–F). This result suggests that Snp is not a modulator of RNAi.
FIGURE 8.
Snp mutation does not affect transgene-mediated RNAi silencing. Panels A–E contain photomicrographs of an adult eye. (A) Wild-type Canton-S fly. (B) A fly carrying one copy of the GMR-wIR construct, which silences the white gene and reduces pigment deposition, and heterozygous for a mutation of Dicer-2. (C) A GMR-wIR fly homozygous mutant for Dicer-2. Note the restoration of a eye color. (D) A Snpc00465 homozygous fly. The eye color results from the mini-white in the pBac transposable element. (E) A fly carrying one copy of GMR-wIR and heterozygous for Snpc00465. (F) A fly carrying one copy of GMR-wIR and homozygous for Snpc00465. Note that silencing of the mini-white gene by GMR-wIR is not affected by loss of Snp expression. (G) A w1118 male control fly with wild-type body color. (H) A Snpc00465 homozygous male fly with wild-type body color. (I) A yIR/da-GAL4 +/Snpc00465 male fly. Note that the thorax (arrow) is lighter than the controls (G,H), resulting from silencing of the yellow gene by activation of the yIR transgene with da-Gal4. (J) A yIR/da-GAL4 Snpc00465/Snpc00465 male fly. Note that there is no obvious change in coloration when compared to the fly in panel I.
To further test this, we employed a transgene (yIR) that expresses an inverted repeat of the yellow (y) gene under the control of a UAS element (Piccin et al. 2001). Expression of yIR with a daughterless (da)-Gal4 driver phenocopies the y2 hypomorphic mutation, where the body of the fly is yellow and the bristles remain wild type in coloration (Fig. 8G–I). Other drivers (e.g., Actin-Gal4) of yIR result in a more widespread phenotype in which both the bristles and body turn yellow, as caused by a null y1 mutation (Piccin et al. 2001). We expressed the yIR transgene under the control of da-Gal4 in a Snp mutant background to determine if the loss of Snp expression is able to enhance y silencing via the yIR construct and produce yellow bristles or enhance the yellow body coloration. Snpc00465 mutant flies with yIR driven by daGal4 showed no enhancement of bristle color or yellow body color when compared to heterozygous siblings (Fig. 8I,J). Therefore, the loss of Snp failed to enhance the silencing of two genes by RNAi, indicating that Snp unlikely plays a role in negatively regulating RNAi in Drosophila.
Snp does not play a role in apoptosis
CRN-4 is a C. elegans member of the ERI-1 subfamily of exonucleases that when mutated causes a persistence of TUNEL-labeled DNA in apoptotic cells, suggesting that it plays a role in the degradation of apoptotic DNA (Parrish and Xue 2003). To determine if Snp might play a role in digesting apoptotic DNA in Drosophila, TUNEL staining was performed on Snp mutant embryos, and the amount and intensity of TUNEL staining in the Snp mutant embryos were compared to heterozygous siblings. We detected no obvious difference in the amount of TUNEL positive cells in Snp embryos compared to heterozygous siblings (Fig. 9A,B). As a control, DNaseII mutant embryos were also stained with TUNEL. DNaseII is required for degradation of apoptotic DNA in Drosophila, and apoptotic DNA persists in embryos and ovaries (Mukae et al. 2002). We detected an elevated level of TUNEL-positive apoptotic DNA in 32% (n=274) of the population collected from heterozygous parents (Fig. 9C). These embryos likely represent DNaseII mutant embryos, while the embryos with normal levels of TUNEL-positive nuclei are heterozygous siblings. This result indicates that we can score for differences in TUNEL labeling, and we therefore conclude that Snp does not play a major role in the clearance of apoptotic DNA in Drosophila.
FIGURE 9.
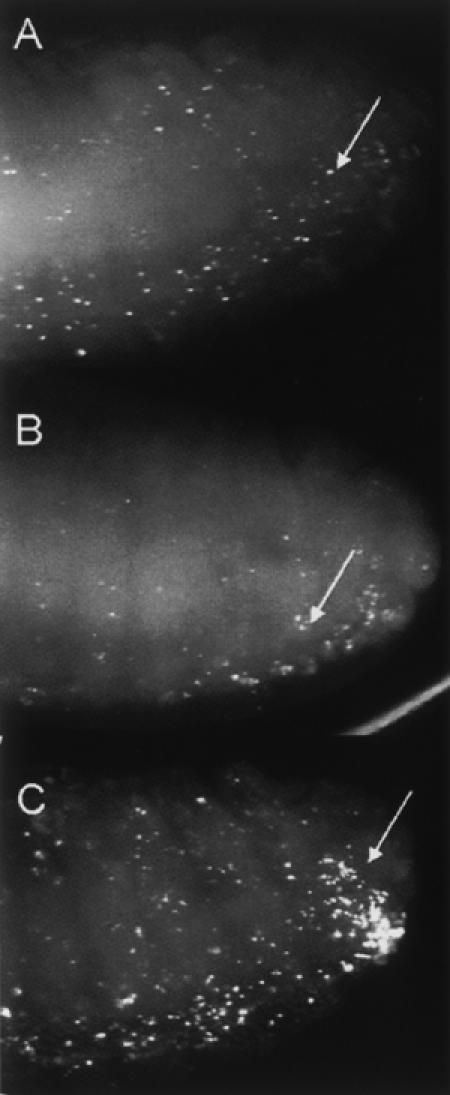
Effect of Snp mutation on digestion of apoptotic DNA in embryos. Each panel shows TUNEL-positive nuclei in the posterior portion of a stage 13 embryo (arrows). (A) Snp c00465 /CyO. (B) Snp c00465/ Snp c00465. Note the intensity and number of TUNEL-positive nuclei are similar to the heterozygous sibling shown in A. (C) DNaseII lo homozygous mutant embryo. Note the increase in the amount of TUNEL-positive nuclei, particularly in the posterior CNS (arrow).
DISCUSSION
We have presented a detailed biochemical and genetic characterization of a novel member of the DEDDh family of nucleases that we call Snipper (Snp). Our studies show that Snp is a promiscuous 3′-5′ exonuclease with broad specificity toward both RNA and DNA substrates, although it exhibits highest affinity toward double-stranded substrates with a 3′ flank. Similar to many processive 3′-5′ exonucleases such as the DNases Exo I (Breyer and Matthews 2000), RecJ (Han et al. 2006), and Exo X (Viswanathan and Lovett 1999), and the RNases PARN (Wu et al. 2005), PNPase (Symmons et al. 2002), RNase R (Cheng and Deutscher 2005), Snp is processive on hairpin DNAs and RNAs with at least three to five unpaired nucleotides in the 3′ flank and nonprocessive on single-stranded DNAs and RNAs. Snp is highly active toward histone stem–loop RNA in vitro and unlike 3′hExo (which has an N-terminal SAP domain), Snp readily digests through the stem portion of the histone RNA hairpin.
Based on Snp's sequence similarity with 3′hExo and ERI-1 and the ability to act on similar substrates, we characterized Snp's role in histone mRNA degradation and RNAi in vivo. Similar to in vivo studies on 3′hExo (Dominski et al. 2003), we have not been able to obtain evidence that Snp plays a significant role in cell cycle-regulated histone mRNA degradation in vivo. It is possible that 3′hExo and Snp act redundantly with other exonucleases, perhaps that act at the 5′ end, to destroy histone mRNA at the completion of the S phase. 3′hExo and Snp may also play orthologous roles in histone mRNA metabolism other than coupling histone mRNA degradation with the cell cycle. Alternatively, Snp and 3′hExo may not be functional orthologs, and Snp may participate in cellular functions other than histone mRNA metabolism. Our data demonstrate that any such putative functions for Snp are not required for Drosophila development.
In the absence of genetic insights, the functional role(s) of this promiscuous exonuclease remains a mystery. Our biochemical data suggest a bias toward structured DNA or RNA substrates with a 3′ flank, although it is also very active on single-stranded DNAs or RNAs. The catalytic properties of 3′hExo and Snp are remarkably similar to the prokaryotic Escherichia coli DEDDh exonucleases RNase T (Deutscher and Li 2001) and the unrelated processive 3′-5′ nuclease RNase R (Cheng and Deutscher 2005). Substrates of RNase T (like those of Snp and 3′hExo) generally consist of a double-stranded stem followed by four unpaired 3′ nucleotides. RNase T has been implicated in regulating 3′ maturation of a number of stable RNAs that include tRNAs (Deutscher et al. 1985) and 23S ribosomal RNAs (Li et al. 1999) and in the turnover of tRNAs. RNase T can suppress UV repair effects in E. coli mutants that lack DNA repair-specific exonucleases (Zuo and Deutscher 1999). A unique feature of both RNase T and 3′hExo is their ability to trim the 3′ ends of structured RNAs very close to the double-stranded stem (Zuo and Deutscher 2002). Most nucleases are unable to act near the base of a stem–loop or DNA duplex. RNase T is also a DNase and binds ssDNA more tightly compared to RNA degrading these single-stranded substrates in a nonprocessive manner (Zuo and Deutscher 2002). Like Snipper, both RNase T and RNase R are broad-specificity enzymes.
In contrast to RNase T, RNase R is very efficient in digesting single-stranded RNA and DNA substrates as well as structured RNAs as long as it has a minimum 3′ flanking sequence of 4 nt to initiate degradation (Cheng and Deutscher 2005). Substrates of RNase R include rRNA, tRNA, as well as repetitive extragenic palindromic (REP)-containing mRNA fragments that exist as stem–loop structures (Cheng and Deutscher 2005). The ability of RNase R to completely degrade structured RNA as long as it has four unpaired nucleotides in the 3′ flank to bind to is remarkably similar to that of Snp. As Cheng and Deutscher (2005) have proposed, RNase R may possess intrinsic helicase activity, or is capable of deriving energy required to unwind structured RNA/DNA from either the binding energy of association with the unpaired nucleotides, or from phosphohydrolysis of the initial nucleotides, making it an efficient nuclease for structured RNA/DNA substrates. The ability to degrade structured RNA and DNA suggests that both Snipper and RNase R have a catalytic mechanism that is distinct from other DEDDh family nucleases.
Snp and 3′hExo are predicted to be structurally distinct from both RNase T and RNase R, although they recognize similar substrates. RNase T is a dimer, and the conserved regions identified to play a role in substrate recognition in RNase T are absent in Snp and 3′hExo (Zuo and Deutscher 2002). RNase R is a 95-kDa monomer that is related to RNase II and unrelated to the DEDDh family of nucleases. However, given the similarities in their substrate specificities, it is possible that Snp may play analogous roles as RNase T or RNase R does in E. coli (Deutscher 2006). Snp may function in DNA damage repair or recombination where this structure appears during the intermediate steps of repair. Snp may also be involved in the turnover of tRNA, rRNA, or in the 3′ maturation of stable RNAs. The 3′ end of tRNA has a very similar terminal stem structure to that of histone mRNAs, and the final 4 nt of the 3′ end of tRNA and all replication-dependent histone mRNAs are similar, each ending in CCA. Therefore, tRNAs appear to be an ideal substrate for Snp, and future studies will test these hypotheses.
In conclusion, we have described the biochemical and genetic properties of two members of a new subfamily of DnaQ-H exonucleases in metazoans that we call the 3′hExo/ERI-1 subfamily. They are characterized by conservation of active site residues that are unique to this family, and the ability to degrade structured RNA/DNA substrates. Our data indicate that in metazoans nucleases that belong to this family of 3′-5′ exonucleases may show activity toward a diverse array of substrates and will play important roles in mRNA decay processes other than histone mRNA metabolism and RNAi.
MATERIALS AND METHODS
Expression and purification of recombinant Snipper and 3′hExo
The gene encoding Drosophila Snipper (CG6393) was subcloned from the vector pBS-SK into the bacterial expression vector pET28a (Novagen) between the Nco1 and Xho1 restriction sites, which produced a protein with a C-terminal His tag. The pET28a Drosophila Snipper was expressed in BL21-CodonPlus (DE3) cells (Stratagene). Cells were grown in LB medium containing 50 μg/μL kanamycin by inducing the cells with 1 mM isopropyl β-D-thiogalactopyranoside at an OD600nm of 0.4–0.6 for 10 h at 27°C. Cells were harvested and the pellet resuspended in lysis buffer, 20 mM Tris pH 8.0, 50 mM NaCl, 5 mM imidazole, 250 mM guanidine HCl, 1 mM PMSF, and protease inhibitor cocktail (Roche) and sonicated. The lysed cells were centrifuged at 18,000 rpm for 30 min. Nucleic acids were precipitated by treating the supernatant with 0.2% polyethyleneimine and stirring the solution for 20 min at 4°C followed by centrifugation at 18,000 rpm for 30 min. The clarified lysate was loaded onto a 5 mL HisTrap HP Nickel column (Amersham Biosciences), and the column was washed with 100 column volumes of buffer A (20 mM Tris pH 8.0, 300 mM NaCl, 20 mM imidazole) followed by 20 column volumes of buffer B (20 mM Tris pH 8.0, 300 mM NaCl, 40 mM imidazole) and the protein was eluted with a linear gradient of 40–300 mM imidazole in 20 mM Tris pH 8.0 and 300 mM NaCl. The Snp containing fractions were pooled, concentrated to 1 mg/mL, and immediately stored as a glycerol stock at −20°C. Snp purified in this manner (Fig. 2A) was prone to aggregation and precipitation. The protein was used immediately (within 1–2 wk) for activity assays, since some loss in activity occurred over time. The overall yield was about 5 mg/9 L of culture.
Full-length 3′hExo was cloned into the Nco1 and Xho1 restriction sites of the expression vector pET28a, and was purified using Ni+2-affinity chromatography in a similar manner as reported above for Snp. Full-length 3′hExo is quite stable (although it self-associates), and is purified with yields of 15–20 mg/L of culture.
Analytical gel filtration analysis on an S100 (Amersham Biosciences; Supplemental Fig. 2; http://www.bio.unc.edu/faculty/duronio/lab/publications.htm) shows that recombinant Snp and 3′hExo aggregate at concentrations of 1 mg/mL such that most of the proteins elute in the void volume. The net species in solution at concentrations <1 mg/mL is not a specific higher-order complex but an aggregate, as determined from sedimentation equilibrium measurements (R. Thapar, unpubl.). The amount of active monomer is estimated to be <5% of the total fraction of protein from integration of gel filtration peak volumes, and the protein concentrations or amounts reported are for the estimated monomeric species in solution.
DNA and RNA substrates
Exonuclease activity assays were performed with a number of single-stranded and stem–loop RNA and DNA substrates (summarized in Table 1). The single-stranded substrates used were HDE30,R, HDE16, R, polyA30,D, and polyA20,D, where the number of nucleotides are designated as subscripts and “D” and “R” refer to the DNA or RNA versions of the substrates, respectively. The HDE sequences correspond to the histone downstream element of histone H2A-614 pre-mRNA that binds to U7 snRNP. The stem–loop sequences used were the 2′ ribose and 2′ deoxy versions of SL28,R and SL28,D and the reverse stem–loop sequence in RSL28,R and RSL28,D, respectively. In the shorter stem–loop substrates (SL27,D, SL26,D, SL25,D, and SL24,D), the 3′ ACCCA flank in SL28,D was truncated to ACCC, ACC, AC, and A, respectively. All substrates were treated in a similar manner such that they were denatured at 95°C for 5 min and immediately put in an ice bath for 20 min so as to allow for secondary structure formation. Substrates were 5′-end labeled with T7 polynucleotide kinase and γ-[32P]ATP (specific activity 4500 Ci/mmol) for 2 h at 37°C. Free ATP was separated from the substrate by passing the labeled product over a G50 column.
Exonuclease activity assays
Nuclease reactions (10 μL each) contained 20 mTris pH 7.5, 5 mM MgCl2, 2 mM DTT, 100 mM KCl, 1 μL of a 10 mg/mL stock solution of bovine serum albumin, 5 pmol of 5′-32P probe and between 5 and 100 pmol 3′hExo or Snp or buffer for the control reaction. The reactions were incubated at 37°C for 30 min and stopped by the addition of 20 μL of stop/quench dye containing formamide to give a total volume of 30 μL. The reactions were heated at 95°C for 5 min, and 5 μL were loaded onto a prewarmed 10% denaturing polyacrylamide gel.
Electrophoretic mobility shifts assays (EMSA)
Snp and 3′hExo, with or without SLBP proteins, were incubated with a 28-nt 5′end-labeled 32P stem–loop probe (SL28,R) for 30 min on ice in binding buffer (20 mM Tris pH 7.9, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.1 μg/μL BSA) for 10 min. The total reaction volume (10 μL) was analyzed on an 8% native polyacrylamide gel in Tris-borate buffer at 125 V for 1.5 h. The gel was dried at 80°C for 2 h and then exposed to film.
Alignments and sequences
To obtain potential homologs of Snipper, the Snipper protein sequence (NP_611632) was subjected to the NCBI BLASTp program (http://www.ncbi.nlm.nih.gov/BLAST), and the following sequences were obtained from the BLAST search: ERI-1 Ce(O44406), 3′hExo (NP_699163), Crn-4(NP_508415), AAH10503, Prion Protein interacting Protein Hs (NP_076971), Prion Protein interacting Protein Ce (AAB94148), RNaseT (AAC37008). To determine the closest homologs of Snp in the DnaQ-H superfamily, a multiple sequence alignment was generated using ClustalX 1.83 (Jeanmougin et al. 1998), and a phylogenetic tree was produced using ClustalX1.83 NJ-Bootstrap Tree option with a Bootstrap of 1000; the tree was viewed and edited using the NJ Plot program (M. Gouy). Members of the DEDDh family of exonucleases have a low level of sequence identity; therefore, the reliability of the sequence alignment was tested by generating a multiple structure alignment with the combinatorial extension program (CE) (Shindyalov and Bourne 2001) using the structures of the nuclease domains of 3′hExo (PDB code 1W0H), epsilon (PDB code 1J53), and oligoribonuclease (PDB code 1J9A) as templates. This structure-based alignment was used as the input template to generate a multiple sequence alignment in the program T-coffee (Notredame et al. 2000). All approaches used produced similar dendograms, suggesting that Snp is most closely related to the hypothetical human exonuclease AAH10503, followed by members of the 3′hExo subfamily.
Fly stocks and characterization of CG6393 expression
Fly stocks EP(2)2457 and EY84235, along with da-Gal4 and DNaseIIlo flies, were obtained from the Bloomington Stock center, while P(Bac)c00465 and Df(2R)Excel7170 were obtained from the Exelixis collection of insertions at the Bloomington Stock Center. Dicer-2 stocks and GMR-wIR were kind gifts of Richard Carthew (Northwestern University), and yIR transgenes were acquired from Piccin et al. (2001), via Jeff Sekelsky (UNC Chapel Hill). First-strand synthesis was performed on duplicate samples, one containing RT(NEB), and one without RT. RT-PCR was performed on embryos, third instar eye discs, and adult females for all fly lines with an annealing temperature of 62°C for 30 cycles. RNA was isolated using TRIzol Reagent (Gibco). The following primers were used to amplify the Snp transcript: Primer A: 5′ GGGCCATGGCTGCCTTGATAAAACTA 3′, Primer B: 5′ GCGCGCTTAGCAGTAAACTTC 3′, Primer C: 5′ TCCAGCGAGCAGGGGATCCGT 3′ Primer D: 5′CAACACAAGCCTGGGATTAAG 3′. Primers for the coding region of rp49 were as follows: Forward primer: 5′ATCCGCCCAGCATACAGG 3′, Reverse primer: 5′CTCGTTCTCTTGAGAACGCAG.
Northern analysis
For Northern blots total cellular RNA was isolated from third instar eye discs with TRIzol Reagent (Gibco). For the analysis of histone mRNAs, 2 μg of RNA per lane were subjected to electrophoresis in a 1× MOPS in a 1% agarose gel containing 0.01MOPS (PH 7.0) and 6.75% Formaldehyde. Separated RNAs were transferred to a N+ nitrocellulose membrane (Amersham) using the wick method in 20× SSC. DNA containing histone H3 or rp49 coding regions were labeled with α[ 32P]-dCTP using a random primer labeling kit (Stratagene). Hybridizations were performed at 60°C using Quikhybe (Stratagene).
In situ hybridization and TUNEL labeling
Eye discs were dissected from wandering third instar larvae and were fixed in 4% formaldehyde. Embryos were collected and aged at 18°C until the appropriate stage and were dechorionated in 50% bleach and fixed in a 1:1 mixture of Heptane and 4% Formaldehyde for 15 min. In situ hybridization was performed using digoxigen-labeled riboprobes complimentary to the coding region of H3 as previously described (Lanzotti et al. 2002). For TUNEL labeling, embryos were fixed in a 1:1 mixture of paraformaldehyde and heptane for 20 min, then devitilineized by shaking in a 1:1 mixture of heptane and methanol. Embryos were then incubated in 100 mM Sodium Citrate for 30 min at 60°C for 30 min, then TUNEL labeling was performed using an In Situ Cell Death Kit (Roche).
RNAi transgenes
For RNAi analysis the following fly crosses were performed. ♀GMRwIR/GMRwIR X ♂+/Y; Snpc00465/Snpc00465. Male off spring of the following genotype ♂GMRwIR/Y; Snpc00465/+, were then crossed to ♀ Snpc00465/Snpc00465. The eye colors of the resulting progeny were analyzed and photographed with a Nikon digital camera. An identical crossing scheme was used to analyze Dicer2’s function on the GMRwIR, since Dcr2 is also on the second chromosome. To analyze Snps possible fuction in RNAi using a yIR Transgene the following crosses were performed. ♀Snpc00465/Snpc00465 X ♂+/+: yIR/yIR and ♀Snpc00465/Snpc00465 X ♂ +/+, da-GAL4/da-GAL4. The using the progeny from each class the following crosses were performed. ♀ Snpc00465/+, yIR/+ X ♂ Snpc00465/+, daGAL4/+. The resulting offspring were analyzed and photographed with a Nikon digital camera.
ACKNOWLEDGMENTS
We thank Richard Carthew for the Dicer-2 and GMRwIR flies, the Bloomington stock center for fly stocks, and Jeff Sekelsky for the yIR flies. This work was supported by NSF Grant MCB-0342847 to R.J.D. and NIH Grant 1RO1-GM076660-01 to W.F.M and R.T.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.186706.
REFERENCES
- Beese, L.S., Steitz, T.A. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: A two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyer, W.A., Matthews, B.W. Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nat. Struct. Biol. 2000;7:1125–1128. doi: 10.1038/81978. [DOI] [PubMed] [Google Scholar]
- Cheng, Z.F., Deutscher, M.P. An important role for RNase R in mRNA decay. Mol. Cell. 2005;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- Cheng, Y., Patel, D. Crystallographic structure of the nuclease domain of 3′hExo, a DEDDh family member, bound to rAMP. J. Mol. Biol. 2004;343:305–312. doi: 10.1016/j.jmb.2004.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire, V., Freemont, P.S., Sanderson, M.R., Beese, L., Friedman, J.M., Joyce, C.M., Steitz, T.A. Genetic and crystallographic studies of the 3′, 5′-exonucleolytic site of DNA polymerase I. Science. 1988;240:199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- Deutscher, M.P. Degradation of RNA in bacteria: Comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher, M.P., Li, Z. Exoribonucleases and their multiple roles in RNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:67–105. doi: 10.1016/s0079-6603(00)66027-0. [DOI] [PubMed] [Google Scholar]
- Deutscher, M.P., Marlor, C.W., Zaniewski, R. RNase T is responsible for the end-turnover of tRNA in Escherichia coli . Proc. Natl. Acad. Sci. 1985;82:6427–6430. doi: 10.1073/pnas.82.19.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., Marzluff, W.F. Formation of the 3′ end of histone mRNA. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- Dominski, Z., Yang, X.C., Kaygun, H., Dadlez, M., Marzluff, W.F. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell. 2003;12:295–305. doi: 10.1016/s1097-2765(03)00278-8. [DOI] [PubMed] [Google Scholar]
- Duchaine, T.F., Wohlschlegel, J.A., Kennedy, S., Bei, Y., Conte D., Jr, Pang, K., Brownell, D.R., Harding, S., Mitani, S., Ruvkun, G., et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Forster, A.C., Altman, S. External guide sequences for an RNA enzyme. Science. 1990;249:783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- Graves, R.A., Pandey, N.B., Chodchoy, N., Marzluff, W.F. Translation is required for regulation of histone mRNA degradation. Cell. 1987;48:615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Gunjan, A., Verreault, A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae . Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- Hamdan, S., Carr, P.D., Brown, S.E., Ollis, D.L., Dixon, N.E. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure. 2002;10:535–546. doi: 10.1016/s0969-2126(02)00738-4. [DOI] [PubMed] [Google Scholar]
- Han, E.S., Cooper, D.L., Persky, N.S., Sutera, V.A., Jr., Whitaker, R.D., Montello, M.L., Lovett, S.T. RecJ exonuclease: Substrates, products, and interaction with SSB. Nucleic Acids Res. 2006;34:1084–1091. doi: 10.1093/nar/gkj503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, M., Chang, M., Kim, U.J., Grunstein, M. Histone H2B repression causes cell-cycle-specific arrest in yeast: Effects on chromosomal segregation, replication, and transcription. Cell. 1987;48:589–597. doi: 10.1016/0092-8674(87)90237-6. [DOI] [PubMed] [Google Scholar]
- Harris, M.E., Bohni, R., Schneiderman, M.H., Ramamurthy, L., Schumperli, D., Marzluff, W.F. Regulation of histone mRNA in the unperturbed cell cycle: Evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag, D., Eckstein, F., Cech, T.R. Contributions of 2′-hydroxyl groups of the RNA substrate to binding and catalysis by the Tetrahymena ribozyme. An energetic picture of an active site composed of RNA. Biochemistry. 1993;32:8299–8311. doi: 10.1021/bi00083a034. [DOI] [PubMed] [Google Scholar]
- Jeanmougin, F., Thompson, J.D., Gouy, M., Higgins, D.G., Gibson, T.J. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Kaygun, H., Marzluff, W.F. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005a;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- Kaygun, H., Marzluff, W.F. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol. Cell. Biol. 2005b;25:6879–6888. doi: 10.1128/MCB.25.16.6879-6888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, S., Wang, D., Ruvkun, G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Lanzotti, D.J., Kaygun, H., Yang, X., Duronio, R.J., Marzluff, W.F. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ end processing in vivo. Mol. Cell. Biol. 2002;22:2267–2282. doi: 10.1128/MCB.22.7.2267-2282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.S., Carthew, R.W. Making a better RNAi vector for Drosophila: Use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Lee, Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Li, Z., Pandit, S., Deutscher, M.P. Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA. 1999;5:139–146. doi: 10.1017/s1355838299981669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff, W.F., Duronio, R.J. Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 2002;14:692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- Mukae, N., Yokoyama, H., Yokokura, T., Sakoyama, Y., Nagata, S. Activation of the innate immunity in Drosophila by endogenous chromosomal DNA that escaped apoptotic degradation. Genes & Dev. 2002;16:2662–2671. doi: 10.1101/gad.1022802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.M., Ye, X., Hall, C., Santos, H., Ma, T., Kao, G.D., Yen, T.J., Harper, J.W., Adams, P.D. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 2002;22:7459–7472. doi: 10.1128/MCB.22.21.7459-7472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D.G., Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Ollis, D.L., Kline, C., Steitz, T.A. Domain of E. coli DNA polymerase I showing sequence homology to T7 DNA polymerase. Nature. 1985;313:818–819. doi: 10.1038/313818a0. [DOI] [PubMed] [Google Scholar]
- Pandey, N.B., Marzluff, W.F. The stem–loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 1987;7:4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish, J.Z., Xue, D. Functional genomic analysis of apoptotic DNA degradation in C. elegans . Mol. Cell. 2003;11:987–996. doi: 10.1016/s1097-2765(03)00095-9. [DOI] [PubMed] [Google Scholar]
- Piccin, A., Salameh, A., Benna, C., Sandrelli, F., Mazzotta, G., Zordan, M., Rosato, E., Kyriacou, C.P., Costa, R. Efficient and heritable functional knock-out of an adult phenotype in Drosophila using a GAL4-driven hairpin RNA incorporating a heterologous spacer. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.12.e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindyalov, I.N., Bourne, P.E. A database and tools for 3-D protein structure comparison and alignment using the Combinatorial Extension (CE) algorithm. Nucleic Acids Res. 2001;29:228–229. doi: 10.1093/nar/29.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman, D.B., Graves, R.A., Marzluff, W.F. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc. Natl. Acad. Sci. 1983;80:1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D., Pace, N.R. Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry. 1993;32:5273–5281. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- Sullivan, E., Santiago, C., Parker, E.D., Dominski, Z., Yang, X., Lanzotti, D.J., Ingledue, T.C., Marzluff, W.F., Duronio, R.J. Drosophila stem–loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes & Dev. 2001;15:173–187. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons, M.F., Williams, M.G., Luisi, B.F., Jones, G.H., Carpousis, A.J. Running rings around RNA: A superfamily of phosphate-dependent RNases. Trends Biochem. Sci. 2002;27:11–18. doi: 10.1016/s0968-0004(01)01999-5. [DOI] [PubMed] [Google Scholar]
- Thapar, R., Marzluff, W.F., Redinbo, M.R. Electrostatic contribution of serine phosphorylation to the Drosophila SLBP–histone mRNA complex. Biochemistry. 2004;43:9401–9412. doi: 10.1021/bi036315j. [DOI] [PubMed] [Google Scholar]
- Viswanathan, M., Lovett, S.T. Exonuclease X of Escherichia coli. A novel 3′-5′ DNase and Dnaq superfamily member involved in DNA repair. J. Biol. Chem. 1999;274:30094–30100. doi: 10.1074/jbc.274.42.30094. [DOI] [PubMed] [Google Scholar]
- Wu, M., Reuter, M., Lilie, H., Liu, Y., Wahle, E., Song, H. Structural insight into poly(A) binding and catalytic mechanism of human PARN. EMBO J. 2005;24:4082–4093. doi: 10.1038/sj.emboj.7600869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D., Deutscher, M.P. Oligoribonuclease is distinct from the other known exoribonucleases of Escherichia coli . J. Bacteriol. 1995;177:4137–4139. doi: 10.1128/jb.177.14.4137-4139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, Y., Deutscher, M.P. The DNase activity of RNase T and its application to DNA cloning. Nucleic Acids Res. 1999;27:4077–4082. doi: 10.1093/nar/27.20.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, Y., Deutscher, M.P. Mechanism of action of RNase T.I. Identification of residues required for catalysis, substrate binding, and dimerization. J. Biol. Chem. 2002;277:50155–50159. doi: 10.1074/jbc.M207706200. [DOI] [PubMed] [Google Scholar]