Abstract
The study of genetic variation in mice offers a powerful experimental platform for understanding gene function. Complex trait analysis, gene-targeting and gene-trapping technologies, as well as insertional and chemical mutagenesis approaches are becoming increasingly sophisticated and provide a variety of options for cataloguing gene activities and interactions. In this review we discuss fundamental and practical concepts related to chemical mutagenesis and we highlight the growing list of strategies for performing mutagenesis screens in mice. Gene-driven and diverse types of phenotype-driven screens provide several options for the recovery of the invaluable variety of alleles generated by chemical mutagenesis. The unique advantages offered using chemical mutagenesis compare favourably to and complement the spectrum of approaches available for functional annotation of the mammalian genome.
Mutagenesis as a tool to study biology and disease
A long-standing goal of genetics has been to identify genes and determine their roles as components of integrated pathways that affect phenotypic traits. Extending genetic studies to improve human health relies upon associating mutations or allelic variants of genes with disease and disability. As a genetic model organism, the mouse offers a remarkable experimental history, elegant approaches and extensive resources to go along with its biological similarity for understanding human development and disease.
In model organisms from plants to animals the identification of spontaneous, heritable mutations has served as the starting point for their genetic characterization. In the mouse spontaneous mutations leading to defects in visible features such as tail length, coat colour or activity have been collected over the centuries. To the geneticist these are more than just ‘fancy mice’ (i.e. mice traded as pets in the 19th century in Europe and earlier in Asia, for their elaborate morphological, e.g. coat colour, or behavioural, e.g. ‘waltzing’, characteristics; see Keeler, 1931). It is impressive to consider what can be learned from the analysis of a collection of biologically related mutants (Fig. 1). For instance, a set of mice exhibiting a variety of coat colours represents an opportunity to dissect the complexities of pigmentation. Coat colour mutants have provided insights into the developmental pathways guiding neural crest cell migration, the specification of pigment-producing melanocytes, the function of melanosomes, and the biochemical control of melanin biosynthesis (Barsh, 1996). The identification of genes critical for these processes has provided a framework for ordering steps along these pathways and revealed complex genetic interactions among the various pathway components. Notably, these mutations have uncovered genes with diverse functions that extend beyond pigment biology. This has lead to an appreciation of common mechanisms in the genetic control of processes from the expansion of progenitor cell populations, to the function of lysosomal-type subcellular organelles, to the intricacy of body weight homeostasis (Swank et al. 1998; Voisey & van Daal, 2002). In addition, these studies have resulted in the identification of specific human disease genes and importantly, they have yielded a broader understanding of the complex genetic basis of a number of diseases, from Hirschsprung's and Waardenburg syndromes to bleeding disorders and glaucoma (Barsh, 1996; Swank et al. 1998; Anderson et al. 2002). Such extensive knowledge, whereby structural proteins and regulators leading to disease endpoints or leading to normalcy are related to one another in networks, is key for designing therapies and envisioning cures. This knowledge is directly attributed to the depth and scope afforded by an allelic series of mutations in a set of genes involved in a defined area of biology.
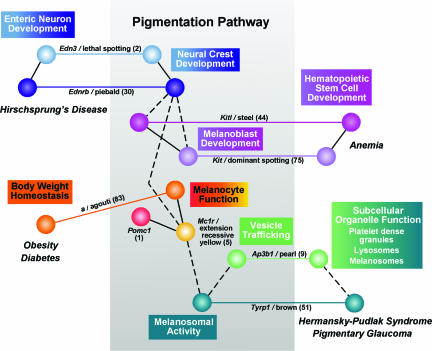
Figure 1. Connecting genes with pathways, biology and disease.
A simple network diagram built around a small subset of coat colour loci illustrates the value of a collection of mutations to model relationships that underlie genetic complexity. This framework demonstrates the progression of the pigmentation pathway (from Barsh, 1996), the comparable activities for genes in multiple biological processes, and similarities in the genetic and cellular basis of disease. Direct relationships are indicated by a continuous line, while indirect interactions are indicated by a dashed line. The number of phenotypic alleles for each locus is indicated in parentheses next to each gene name (from http://www.informatics.jax.org/). Multiple alleles at each locus were invaluable for appreciating this set of associations.
Collections of mutant mice that encompass all areas of biology would clearly facilitate the functional annotation of the genome and link genetic networks to developmental and physiological systems. Mutagenesis screens using chemical agents such as ethylmethanesulphonate (EMS) or N-ethyl-N-nitrosourea (ENU) are an efficient approach for generating such a resource. Chemical mutagenesis accelerates forward genetics, i.e. moving from trait to identified gene, and generates valuable alleles for uncovering the diversity of gene function. EMS and ENU each induce point mutations in DNA, leading to a variety of genetic lesions that are expressed as complete loss of function, partial loss of function, or gain of function alleles. Studying genes represented by null (loss of function), hypomorphic (reduced function), hypermorphic (increased function) or neomorphic (altered function) alleles will reveal a more complete range of gene activity than knockouts. For example, null mutations are often associated with embryonic lethal phenotypes and preclude the study of gene activity at subsequent stages. In contrast, hypomorphic alleles are often more compatible with development and reveal critical gene functions at later developmental or adult stages together with dose-sensitive aspects of gene activity. Furthermore, EMS- or ENU-induced nucleotide substitutions frequently result in missense mutations changing amino acid identity. This class of mutation can be used to define functional domains required for protein interactions or biochemical activity. Connections between protein domains and their biological importance will become increasingly valuable as efforts progress toward the classification of the complete coding gene list and determining functional interactions that configure assemblies and pathways.
From mice to embryonic stem cells, expanding the mutational spectrum
ENU is used primarily as an in vivo mutagen of the male germline. Effective conditions for ENU mutagenesis are well established in mice (Justice et al. 1999). Although rates can vary substantially for a given gene, routine protocols generally yield a per locus mutation frequency of ∼1 × 10−3 for the average locus, at least a 100-fold increase over the estimated spontaneous mutation rate of ∼(5–10) × 10−6. More recently, ENU and EMS have been used to effectively mutagenize mouse embryonic stem (ES) cells, which may then be made into mice (Chen et al. 2000b; Munroe et al. 2000). Mouse ES cells can be mutagenized at rates equal to or higher than ENU-treated mice while still retaining their ability to colonize the germline. Chemical mutagenesis of ES cells offers several advantages over in vivo mutagenesis of spermatogonia. These include (i) the ability to generate and archive a bank of ES cells with a known mutational load measured at the drug-selectable Hprt locus, (ii) a choice of mutagens including ENU, EMS or other classes, and (iii) greater flexibility in the design of breeding strategies that reduce the number of generations needed to render a mutation homozygous (Chen et al. 2000a). However, a potential limitation is the production of chimeric mice, which requires the technical expertise of blastocyst microinjection when compared with the relative ease and low cost of generating a panel of ENU-treated male mice, which entails mere intraperitoneal injection and conventional matings. Another disadvantage is that successful in vitro mutagenesis has so far been limited to the most commonly available ES cell lines which represent mainly two mouse strain backgrounds, 129 and (129 × C57BL/6J)F1 hybrid, whereas in vivo ENU mutagenesis has been performed successfully on many inbred strains and hybrids (Justice et al. 2000). Specific inbred strains or hybrid combinations could play an important role in the recovery of mutants presenting a particular class of phenotypes, just as strain background has been observed to modify the viability of the Egfr knockout mouse or predispose a strain to have lower seizure thresholds (Threadgill et al. 1995; Frankel et al. 2001). Taken together, the option of generating mutations using mice or ES cell-based protocols extends the overall potential of combining chemical mutagenesis with a variety of screening strategies to build a collection of alleles to study gene function (Williams et al. 2003; Table 1).
Table 1.
Comparison of strategies used in chemical mutagenesis screens
| Screening strategy | Applications | Advantages | Limitations |
|---|---|---|---|
| Genome-wide dominant | Generate collection of mutants that serve as entry points for genetic characterization of defined biological processes | Surveys entire genome, single generation breeding | Mutations detected less readily when heterozygous, genetic mapping, mutation identification |
| Genome-wide recessive | Surveys entire genome, mutations detected more readily when homozygous | Multiple generation breeding, genetic mapping, mutation identification | |
| Regionally directed | Functional annotation of defined genomic interval, isolation of allelic series | Partially mapped, screen variations mark genotypic classes and offer shortened breeding schemes | Limited to genes in region, mutation identification |
| Enhancer and suppressor | Phenotype-driven dissection of genetic networks and protein complexes | Sensitized survey of entire genome | Genetic mapping, mutation identification |
| Sequence-based | Gene-driven generation of allelic series | Gene and mutation known | Phenotype unknown, limited to genes surveyed |
The evolution of mutagenesis screens in model organisms
The availability of mutagenesis approaches that significantly boost the spontaneous mutation rate has allowed genetic screens to be designed to recover more than just viable, visible phenotypes. The importance of a mutagenesis screen directed at a defined process is powerfully illustrated in the Nobel prize-winning work to build a set of mutations that disrupt the patterning of the Drosophila embryo (Nusslein-Volhard & Wieschaus, 1980). This screen uncovered key genes and provided the framework and resources for understanding evolutionarily conserved genetic pathways that control the establishment of the body plan in organisms from invertebrates to mammals. Even with their tremendous impact, a mutagenesis screen that defines an initial set of mutants can be thought of as a first-order dissection of genetic network complexity, a set of nodes around which to integrate additional components. In order to delve more deeply into a biological process, increasingly inventive screens have been applied in Drosophila. Modifier screens and region-based screens using visible markers and chromosomal deficiencies or balancing inversions have been used to fill in the gaps, making Drosophila one of the most extensively connected genetic models (Saint Johnston, 2002).
In organisms such as Drosophila and C. elegans a short generation time and ease of maintenance makes it possible to routinely screen thousands of mutagenized genomes for a relatively modest investment. By comparison, a large-scale mutagenesis screen in vertebrate models is a much more extensive undertaking. Nevertheless, their potential to advance genetic studies motivated the large-scale Boston and Tübingen mutagenesis screens in the zebrafish (Driever et al. 1996; Haffter et al. 1996). These two screens surveyed over 6000 genomes for recessive ENU-induced mutations that disrupt embryogenesis. As a result ∼2000 developmental mutants were recovered. Although an average of ∼2 alleles per locus for numerous developmental genes were identified, these screens did not achieve saturation. As in Drosophila, additional focused screens using innovative genetic schemes and phenotype detection strategies are being used to add to the inventory of zebrafish mutants (Patton & Zon, 2001).
Mutagenesis screens in the mouse, taking advantage of a regional approach
The mouse is an indispensable genetic model for understanding human biology. However, expensive husbandry requirements and a ∼10 week generation time present serious challenges for the application of mutagenesis screens in the mouse. To address these limitations, early mutagenesis screens in the mouse turned to the efficient genetic schemes commonly employed in Drosophila. Taking advantage of markers to visibly ‘genotype’ classes of mutagenized offspring together with chromosomal deletions, a defined genomic interval can be screened for recessive mutations in two generations (Rinchik, 2000). Combining ES cell mutagenesis with deletions it is possible to uncover recessive phenotypes in a single generation screen (Chen et al. 2000a). Such marked, regionally directed screens reduce the associated labour and number of mice permitting the targeted, more cost-efficient recovery of phenotypes ranging from embryonic lethality to late onset diseases. Regional screens have been applied using chromosomal deletions surrounding a number of coat colour loci. An excellent example is the albino region screen directed at a ∼6–11 cm segment of chromosome 7 that surveyed over 4500 genomes to recover 31 mutations at 10 different loci involved in fitness and survival. This screen demonstrated the capacity of a regionally directed screen in the mouse to isolate a series of ENU-induced alleles for several loci. The project also highlighted several practical considerations that serve as a guideline for conducting future regional screens (Rinchik & Carpenter, 1999).
One drawback is that deletions spanning some genomic regions are not well tolerated in mice. For example, hemizygosity for a large deletion in a gene-rich segment may even result in lethality. In these instances it may be possible to use a smaller, viable deletion that still removes a number of genes that serve as a valued set of targets. Alternatively, regional screens can be conducted using chromosomal inversions (Rinchik, 2000). Although inversions do not create hemizygosity to trap recessive mutations in a region of the genome, they are used towards the same end by virtue of the fact that they suppress local recombination and, when an appropriate genetic marker is used, they allow for a mating strategy that will readily identify ‘test-class’ mice, i.e. those that are recessive for induced mutations. As with deletions, this marked class can be used to reveal lethality as a missing class or define the mutation carrying mice for high-investment phenotyping protocols. A recessive screen using a chromosomal inversion typically involves three generations. However, mice heterozygous for large chromosomal inversions typically do not present haploinsufficiency-related phenotypes. Thus, mice carrying chromosomal inversions can be constructed for all regions of the genome.
With recently developed ES cell-based methods for introducing large-scale genomic alterations, it is possible to generate chromosomal aberration resources at any location in the mouse genome. Homologous recombination-based chromosome engineering permits the construction of Mb-scale marked inversions or deletions (Zheng et al. 1999; Mills & Bradley, 2001). An alternative strategy uses radiation mutagenesis to induce deletion complexes surrounding an integrated drug selection cassette (You et al. 1997; Thomas et al. 1998). Furthermore, the K-14-agouti transgene or enhanced green fluorescent protein (EGFP) based transgenic strategies can serve as visible markers to couple with the chromosomal aberration resources (Zheng et al. 1999; Frank et al. 2003). Thus, marked, regionally directed mutagenesis screens – once restricted to deletion complexes surrounding classic coat colour and dysmorphology loci – can now be applied across the genome. Regional genomic screens are valuable for the functional characterization of a defined chromosomal interval and traditionally have been viewed as a practical, reagent-driven approach. However, genome-wide transcription maps are beginning to suggest a clustered organization for coordinately expressed genes (Caron et al. 2001; Ramalho-Santos et al. 2002). In addition, quantitative trait loci (QTL) mapping studies are identifying regions exhibiting a complex association with a phenotypic trait, whether by evolutionary design or chance, reflecting a multigenic basis or clusters of genes that underlie single QTL (Legare et al. 2000; de Haan et al. 2002). For these reasons, the regional mutagenesis screen may offer an essential approach to further dissect the functional content of a defined interval and to explore possible relationships between biological processes and genome organization.
Extending mutagenesis in the mouse, from genome-wide to large-scale screens
A shortcoming of a regional screen is that it only surveys a limited portion of the genome. In genome-wide screens dominant mutations are recovered in a single generation permitting a high-throughput approach. A now classical example of its power is in the screens used to isolate multiple ENU-induced mutations causing eye disorders (Favour & Neuhauser-Klaus, 2000). However, dominant mutations are uncovered less frequently than recessive mutations. The price of a genome-wide recessive screen is that it involves three generations of breeding and typically 10–20 third-generation mice all need to be tested to have a chance to detect the recessive phenotype. Despite this hurdle, genome-wide recessive screens have been applied successfully in the mouse. For example, in screens designed to identify genes essential for development focusing on dysmorphology at midgestation, investigators uncovered a mutation in ∼1 out of every 20 genomes surveyed (Kasarskis et al. 1998), while focusing on defects at birth yielded a mutation in ∼1 out of every 4 genomes screened (Herron et al. 2002). Although development offers readily detectable phenotypes in a broad biological domain, these projects demonstrated that a genome-wide recessive approach can offer worthwhile returns for the investment.
Stemming from the success of the various screens in the mouse and other models, together with the prospect of the complete genomic sequence, a number of large-scale mutagenesis projects are ongoing in the mouse genetics community (Justice, 2000). Towards the functional annotation of the mouse genome, several centres have worked to establish collaborative efforts to develop high-throughput phenotyping strategies that will permit thousands of mice to be surveyed to identify models of human disease (Nadeau et al. 2001). In addition to detecting developmental defects and visible dysmorphology, the screens have been designed to identify genes that contribute to neurological and behavioural disorders, metabolic diseases and abnormalities in the immune response. These latter phenotypic classes play to the unique strength of the mouse as a biological model to define genetic networks that underlie human cardiovascular disease, diabetes, mental illness and other major disabilities.
The various mouse mutagenesis programmes have adopted a variety of genetic strategies, including regionally directed and genome-wide dominant and recessive screens. However, it is important to note that even on a large scale, practical realities associated with mouse husbandry, generation time and phenotyping limit the number of genomes that can be screened. For example, although the Neuroscience Mutagenesis Facility at the Jackson Laboratory (http://nmf.jax.org/index.html) is equipped to assay ∼8000 third-generation mice each year for several disorders, this represents a survey of only ∼400 genomes because it is necessary to screen 10–20 individuals per pedigree to have a chance of detecting a recessive mutation within a given family. In a regional screen using an engineered inversion as a marked, balancer chromosome, the Baylor College of Medicine programme has surveyed 735 genomes and recovered 88 mutations mapping within a 40 cm segment of mouse chromosome 11 (Kile et al. 2003). The yield of mouse mutants establishes that these mutagenesis programmes are making significant advances. Conservatively, over 500 heritable mutations have been recovered across the various centres and hundreds of phenodeviant mice are undergoing heritability testing. Although these are relatively modest numbers compared with the size of screens performed in organisms such as Drosophila, these new mutants are an invaluable addition to the collection of resources needed to meet the challenge of genetically characterizing a model as complex as the mouse with an estimated 30 000 genes.
Phenotyping is the key
A critical factor in the potential of a mutagenesis screen to build the mouse mutant resource is phenotyping – both expertise and capacity. Phenotyping is also the greatest challenge, especially when attempting to screen for models of human disease. Using neurological traits as an example, it is almost trivial to detect motor abnormalities in mice by virtue of their abnormal gait. It is somewhat less trivial to detect models that are relatively sensitive or resistant to epilepsy (Yang et al. 2003). However, to detect mice that model more behaviourally complex disorders such as autism or depression is even more difficult because their manifestations are not so obvious in mice (Tarantino et al. 2000; Bucan & Abel, 2002). In addition, while higher-throughput phenotype screens are the most compatible with ENU mutagenesis, tests that are the best validated for these complex behaviours are relatively labour intensive. Nevertheless, as more is learned from cottage industry-style research about the manifestations of such traits in mice, it is likely that more appropriate and streamlined assays will be developed for the most complex of behaviours in high-throughput phenotype screens.
Putting it all together
The next step in large-scale mutagenesis – the ‘meta-analysis’ of mutant collections, including phenotypic classification, grouping of mutations and alleles, mapping genes and identification of the underlying molecular defects – is amongst the most rewarding but also represents a tremendous challenge. An excellent example of the promise of such meta-analysis is the characterization of a novel class of pigment mutants resulting in dark skin (Fitch et al. 2003). A group of 10 dominant mutations generated during the large-scale screen at the GSF-Munich (http://www.gsf.de/ieg/groups/enu-mouse.html) were determined to define a set of loci not previously known to be involved in pigmentation and define a unique developmental pathway. This recent study underscores the value of generating and collectively studying a group of mutations in a focused area of biology. But part of the challenge is to develop a strategy for efficiently determining which new mutations are truly unique, versus which occur in the same loci. This is because (i) many classes of mutation cannot be distinguished by phenotype alone, without a great deal of follow-up and sometimes detailed phenotype testing, and (ii) an increasing number of genes display phenotypic variability when mutated, such that it is not possible to predict allelism based on phenotype alone. For these reasons, genetic mapping is an important corollary to mutant detection. Although it requires more effort to determine map position in genome-wide mutagenesis than in regional screens which utilize marked deletions or inversions, it is well worth the effort. Often genetic mapping – even to modest chromosomal resolution (e.g. 10 cm) – leads to possible gene candidacy and important phenotyping experiments that can serve as a basis to screen for mutations in a particular gene. Genome sequence data and improvements in methods for mutation detection continue to reduce the time and costs associated with candidate gene analysis.
Fine-tuned genetic dissection using sensitized screens
Once a critical set of genes involved in a particular biological process has been identified, mutagenesis screens that take advantage of existing genetic variants provide a powerful method of uncovering additional pathway components. Loss-of-function mutations may not always present a strong phenotype but the loss of activity can sensitize the genetic background such that an additional mutation will sufficiently disrupt a pathway to reveal a phenotype. Conversely, some mutations may suppress a phenotype, for example by disrupting negative feedback and permitting a reduced signalling dose to elicit a relatively normal cellular response. Modifier screens to uncover enhancer or suppressor mutations can be used to dissect functional genetic interactions. Enhancer and suppressor screens have been effectively applied in Drosophila, for example, to find components of the receptor tyrosine kinase signalling pathway by detecting alterations in eye phenotype (Therrien et al. 2000). A strong advantage of these screens is that they can easily be designed to recover dominant modifiers in a single generation breeding scheme. There are certainly several examples in the mouse of genetic interactions modifying a trait and of mutations that exhibit non-allelic non-complementation. However, mutagenesis screens to uncover modifiers have not been extensively applied and this approach is relatively unproven in the mouse. The Toronto Centre for Modelling Human Disease highlights a screen to recover mutations that interact with the Gli3 transcription factor (http://www.cmhd.ca/sub/mutagenesis.htm). Interactions between the critical developmental regulators Gli3 and Shh illustrate key aspects of the genetic hierarchy controlling limb formation (Litingtung et al. 2002; te Welscher et al. 2002). Sensitized screens that have the potential to uncover such functional relationships would greatly enhance the collection of mutations in the mouse.
Mutagenesis versus natural variation
Another method to identify collections of biologically related genes is QTL analysis. The QTL approach relies on the interplay between spontaneous mutations that have arisen in the distant past and been differentially fixed in the various inbred strains of laboratory mice (Lander & Botstein, 1989). These inbred strains are derived from a limited founder population and are thought to carry a mix of chromosomal segments mostly from two ancestral wild mouse species, M. m. domesticus and M. m. musculus (Wade et al. 2002). Careful measurements can reveal a wide range of phenotypic variation between mouse inbred strains and breeding strategies can be used to identify genomic regions and genes associated with the control of behavioural, physiological, and disease processes, and responses to the environment. These phenotypes are often inherited as complex traits, with multiple genetic loci being involved and both additive and non-additive (epistatic) interactions between loci. The Mouse Phenome Project represents an effort to proselytize this potential by defining the baseline for a battery of phenotypes displayed by over 50 different strains of mice (Paigen & Eppig, 2000).
The major challenge of the QTL approach is in managing the genetic complexity that makes it appealing in the first place. Although QTL strategies can be used to readily uncover phenotypic differences, the genetic diversity also presents difficulties for tracking contributing loci, high resolution mapping, candidate gene evaluation and identification of the molecular lesion (Nadeau & Frankel, 2000). The evidence for candidate mutations in QTL studies must be considered in the context of a higher rate of polymorphism between strains. For example, comparisons of the genomic sequence data available for a handful of inbred strains shows regional variation in the single nucleotide polymorphism (SNP) rates, ranging from 1 in every 250 bp to 1 in every 20 kb (Wade et al. 2002). Even in the low SNP density regions this rate is much higher than the base pair substitution rates reported for chemically induced mutations which range from 1 per ∼0.4 Mb to 1 per 1–2 Mb (Coghill et al. 2002; Vivian et al. 2002). Thus, gene and mutation identification tend to be more straightforward for variants recovered in a mutagenesis screen. However, strategies to relate phenotypes with haplotype comparisons across several inbred strains and plans for the construction of a 1000 line, multiparental recombinant inbred (RI) panel hold some promise of simplifying the path from phenotype to gene detection in complex trait analysis (Threadgill et al. 2002; Wade et al. 2002).
Although the goal of both mutagenesis and QTL analysis is to use tractable genetic differences to uncover gene functions, the strategies take a fundamentally different approach. QTL studies focus on the collective analysis of existing variation that has accumulated in mouse strains whereas mutagenesis screens represent a more stepwise analysis of new variation that is layered onto the genome. Both provide advantages. For example, the multiparental RI panel noted above offers the reproducibility of assessing complex phenotypes in multiple, genetically identical individuals. In chemical mutagenesis each uniquely mutagenized genome offers novel alleles for gaining insight into a protein's function. These are largely complementary approaches that will provide important resources and insights for dissecting relationships that define genetic networks.
A gene-driven alternative: sequence-based chemical mutagenesis screens
Chemical mutagenesis approaches have traditionally been coupled with phenotype-driven screens affording a random, unbiased identification of genes with essential biological functions. However, the availability of high-throughput sequencing and mutation detection technologies makes it possible to screen directly for ENU- or EMS-induced mutations in any gene of interest. In this gene-driven extension, DNA from mutagenized mice or ES cells can be screened for mutations predicted to result in complete inactivation or altered gene function. Analysis of exons from selected genes in DNA from ENU-treated mice estimates a mutation frequency of 1 bp change per 1–2 Mb of coding sequence surveyed (Coghill et al. 2002). However, only a handful of loci have been studied and mutagen dose and sensitivity of mutation detection influence this number. In ENU-treated ES cells a rate of 1 in 464 kb has been reported. In the ES cell study a large allelic series of mutations in the Smad2 and Smad4 members of the transforming growth factor (TGF)-β signalling pathway were recovered (Vivian et al. 2002). Selected alleles in mice showed hypomorphic or dominant negative activity. Interestingly, three out of four ENU-induced missense substitutions of conserved amino acid residues in Smad4 did not appear to produce a phenotype in mice. Significantly, these studies demonstrate the capacity of high-throughput sequence-based screening to develop an allelic series at a given locus. Banks of mutagenized ES cells and sperm from mutagenized mice will dramatically extend the dimension of chemical mutagenesis approaches for genetic analysis.
Prospects
Experimental genetics has revealed the exquisite simplicity and the remarkable complexity of biological systems. Mutagenesis screens contribute an essential context for understanding systems properties by providing a genetically proven set of related components controlling a biological or disease-related process. Complete genomic sequence data and increasingly sophisticated computational approaches for integrating genomic information will yield predictions about protein interactions, developmental and physiological networks, and regulatory hierarchies. With both phenotype-based and sequence-based screening approaches, chemical mutagenesis can offer returns comparable to gene-trap, conditional and high-throughput gene knockout strategies to provide the complete complement of reagents needed to test biological hypotheses and computational predictions. However, perhaps the greatest value of the mutagenesis screen remains its potential to reveal biological relationships that escape prediction, a purely discovery-driven approach to identify novel functions for genes and expose connections that establish unappreciated genetic pathways.
Acknowledgments
We thank Drs Greg Cox and John Schimenti for comments on the manuscript and Sarah Williamson for help with figure preparation.
References
- Anderson MG, Smith RS, Hawes NL, Zabaleta A, Chang B, Wiggs JL, John SW. Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice. Nat Genet. 2002;30:81–85. doi: 10.1038/ng794. [DOI] [PubMed] [Google Scholar]
- Barsh GS. The genetics of pigmentation: from fancy genes to complex traits. Trends Genet. 1996;12:299–305. doi: 10.1016/0168-9525(96)10031-7. [DOI] [PubMed] [Google Scholar]
- Bucan M, Abel T. The mouse: genetics meets behaviour. Nat Rev Genet. 2002;3:114–123. doi: 10.1038/nrg728. [DOI] [PubMed] [Google Scholar]
- Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, Hermus MC, van Asperen R, Boon K, Voute PA, Heisterkamp S, van Kampen A, Versteeg R. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- Chen Y, Schimenti J, Magnuson T. Toward the yeastification of mouse genetics: chemical mutagenesis of embryonic stem cells. Mamm Genome. 2000a;11:598–602. doi: 10.1007/s003350010114. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yee D, Dains K, Chatterjee A, Cavalcoli J, Schneider E, Om J, Woychik RP, Magnuson T. Genotype-based screen for ENU-induced mutations in mouse embryonic stem cells. Nat Genet. 2000b;24:314–317. doi: 10.1038/73557. [DOI] [PubMed] [Google Scholar]
- Coghill EL, Hugill A, Parkinson N, Davison C, Glenister P, Clements S, Hunter J, Cox RD, Brown SD. A gene-driven approach to the identification of ENU mutants in the mouse. Nat Genet. 2002;30:255–256. doi: 10.1038/ng847. [DOI] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SCF, Malicki J, Stemple DL, Stainier DYR, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Favor J, Neuhauser-Klaus A. Saturation mutagenesis for dominant eye morphological defects in the mouse Mus musculus. Mamm Genome. 2000;11:520–525. doi: 10.1007/s003350010099. [DOI] [PubMed] [Google Scholar]
- Fitch KR, McGowan KA, van Raamsdonk CD, Fuchs H, Lee D, Puech A, Herault Y, Threadgill DW, Hrabe de Angelis M, Barsh GS. Genetics of dark skin in mice. Genes Dev. 2003;17:214–228. doi: 10.1101/gad.1023703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AC, Meyers KA, Welsh IC, O'Brien TP. Development of an EGFP & -based dual-color reporter to facilitate genetic screens for the recovery of mutations in mice. Proceedings of the Natl Acad Sci USA. 2003;100:14103–14108. doi: 10.1073/pnas.1936166100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel WN, Taylor L, Beyer B, Tempel BL, White HS. Electroconvulsive thresholds of inbred mouse strains. Genomics. 2001;74:306–312. doi: 10.1006/geno.2001.6564. [DOI] [PubMed] [Google Scholar]
- de Haan G, Bystrykh LV, Weersing E, Dontje B, Geiger H, Ivanova N, Lemischka IR, Vellenga E, Van Zant G. A genetic and genomic analysis identifies a cluster of genes associated with hematopoietic cell turnover. Blood. 2002;100:2056–2062. doi: 10.1182/blood-2002-03-0808. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nusslein-Volhard C. The identification of genes with unique and essential functions in the development of the zebrafish. Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Herron BJ, Lu W, Rao C, Liu S, Peters H, Bronson RT, Justice MJ, McDonald JD, Beier DR. Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat Genet. 2002;30:185–189. doi: 10.1038/ng812. [DOI] [PubMed] [Google Scholar]
- Justice MJ. Capitalizing on large-scale mouse mutagenesis screens. Nat Rev Genet. 2000;1:109–115. doi: 10.1038/35038549. [DOI] [PubMed] [Google Scholar]
- Justice MJ, Carpenter DA, Favor J, Neuhauser-Klaus A, Hrabe de Angelis M, Soewarto D, Moser A, Cordes S, Miller D, Chapman V, Weber JS, Rinchik EM, Hunsicker PR, Russell WL, Bode VC. Effects of ENU dosage on mouse strains. Mamm Genome. 2000;11:484–488. doi: 10.1007/s003350010094. [DOI] [PubMed] [Google Scholar]
- Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A. Mouse ENU mutagenesis. Hum Mol Genet. 1999;8:1955–1963. doi: 10.1093/hmg/8.10.1955. [DOI] [PubMed] [Google Scholar]
- Kasarskis A, Manova K, Anderson KV. A phenotype-based screen for embryonic lethal mutations in the mouse. Proc Natl Acad Sci USA. 1998;95:7485–7490. doi: 10.1073/pnas.95.13.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeler CE. The Laboratory Mouse. Its Origin, Heredity, and Culture. Cambridge: Harvard University Press; 1931. [Google Scholar]
- Kile BT, Hentges KE, Clark AT, Nakamura H, Salinger AP, Lui B, Box N, Stockton DW, Johnson RL, Behringer RR, Bradley A, Justice MJ. Functional genetic analysis of mouse chromosome 11. Nature. 2003;425:81–86. doi: 10.1038/nature01865. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legare ME, Bartlett FS, 2nd, Frankel WN. A major effect QTL determined by multiple genes in epileptic EL mice. Genome Res. 2000;10:42–48. [PMC free article] [PubMed] [Google Scholar]
- Litingtung Y, Dahn RD, Li Y, Fallon JF, Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418:979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- Mills AA, Bradley A. From mouse to man: generating megabase chromosome rearrangements. Trends Genet. 2001;17:331–339. doi: 10.1016/s0168-9525(01)02321-6. [DOI] [PubMed] [Google Scholar]
- Munroe RJ, Bergstrom RA, Zheng QY, Libby B, Smith R, John SW, Schimenti KJ, Browning VL, Schimenti JC. Mouse mutants from chemically mutagenized embryonic stem cells. Nat Genet. 2000;24:318–321. doi: 10.1038/73563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH, Balling R, Barsh G, Beier D, Brown SD, Bucan M, Camper S, Carlson G, Copeland N, Eppig J, Fletcher C, Frankel WN, Ganten D, Goldowitz D, Goodnow C, Guenet JL, Hicks G, de Angelis MH, Jackson I, Jacob HJ, Jenkins N, Johnson D, Justice M, Kay S, Kingsley D, Lehrach H, Magnuson T, Meisler M, Poustka A, Rinchik EM, Rossant J, Russell LB, Schimenti J, Shiroishi T, Skarnes WC, Soriano P, Stanford W, Takahashi JS, Wurst W, Zimmer A. Sequence interpretation. Functional annotation of mouse genome sequences. Science. 2001;291:1251–1255. doi: 10.1126/science.1058244. [DOI] [PubMed] [Google Scholar]
- Nadeau JH, Frankel WN. The roads from phenotypic variation to gene discovery: mutagenesis versus QTLs. Nat Genet. 2000;25:381–384. doi: 10.1038/78051. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity. Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Paigen K, Eppig JT. A mouse phenome project. Mamm Genome. 2000;11:715–717. doi: 10.1007/s003350010152. [DOI] [PubMed] [Google Scholar]
- Patton EE, Zon LI. The art and design of genetic screens: zebrafish. Nat Rev Genet. 2001;2:956–966. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. ‘Stemness’: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Rinchik EM. Developing genetic reagents to facilitate recovery, analysis, and maintenance of mouse mutations. Mamm Genome. 2000;11:489–499. doi: 10.1007/s003350010095. [DOI] [PubMed] [Google Scholar]
- Rinchik EM, Carpenter DA. N-ethyl-N-nitrosourea mutagenesis of a 6- to 11-cM subregion of the Fah-Hbb interval of mouse chromosome 7: Completed testing of 4557 gametes and deletion mapping and complementation analysis of 31 mutations. Genetics. 1999;152:373–383. doi: 10.1093/genetics/152.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3:176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- Swank RT, Novak EK, McGarry MP, Rusiniak ME, Feng L. Mouse models of Hermansky Pudlak syndrome: a review. Pigment Cell Res. 1998;11:60–80. doi: 10.1111/j.1600-0749.1998.tb00713.x. [DOI] [PubMed] [Google Scholar]
- Tarantino LM, Gould TJ, Druhan JP, Bucan M. Behavior and mutagenesis screens: the importance of baseline analysis of inbred strains. Mamm Genome. 2000;11:555–564. doi: 10.1007/s003350010107. [DOI] [PubMed] [Google Scholar]
- Therrien M, Morrison DK, Wong AM, Rubin GM. A genetic screen for modifiers of a kinase suppressor of Ras-dependent rough eye phenotype in Drosophila. Genetics. 2000;156:1231–1242. doi: 10.1093/genetics/156.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JW, LaMantia C, Magnuson T. X-ray-induced mutations in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1998;95:1114–1119. doi: 10.1073/pnas.95.3.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC, Barnard JA, Yuspa SH, Coffey RJ, Magnuson T. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Hunter KW, Williams RW. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm Genome. 2002;13:175–178. doi: 10.1007/s00335-001-4001-Y. [DOI] [PubMed] [Google Scholar]
- Vivian JL, Chen Y, Yee D, Schneider E, Magnuson T. An allelic series of mutations in Smad2 and Smad4 identified in a genotype-based screen of N-ethyl-N- nitrosourea-mutagenized mouse embryonic stem cells. Proc Natl Acad Sci USA. 2002;99:15542–15547. doi: 10.1073/pnas.242474199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisey J, van Daal A. Agouti: from mouse to man, from skin to fat. Pigment Cell Res. 2002;15:10–18. doi: 10.1034/j.1600-0749.2002.00039.x. [DOI] [PubMed] [Google Scholar]
- Wade CM, Kulbokas EJ, 3rd, Kirby AW, Zody MC, Mullikin JC, Lander ES, Lindblad-Toh K, Daly MJ. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- Welscher P, Zuniga A, Kuijper S, Drenth T, Goedemans HJ, Meijlink F, Zeller R. Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science. 2002;298:827–830. doi: 10.1126/science.1075620. [DOI] [PubMed] [Google Scholar]
- Williams RW, Flaherty L, Threadgill DW. The math of making mutant mice. Genes Brain Behav. 2003;2:191–200. doi: 10.1034/j.1601-183x.2003.00017.x. [DOI] [PubMed] [Google Scholar]
- Yang Y, Beyer BJ, Otto JF, O'Brien TP, Letts VA, White HS, Frankel WN. Spontaneous deletion of epilepsy gene orthologs in a mutant mouse with a low electroconvulsive threshold. Hum Mol Genet. 2003;12:975–984. doi: 10.1093/hmg/ddg118. [DOI] [PubMed] [Google Scholar]
- You Y, Bergstrom R, Klemm M, Lederman B, Nelson H, Ticknor C, Jaenisch R, Schimenti J. Chromosomal deletion complexes in mice by radiation of embryonic stem cells. Nat Genet. 1997;15:285–288. doi: 10.1038/ng0397-285. [DOI] [PubMed] [Google Scholar]
- Zheng B, Sage M, Cai WW, Thompson DM, Tavsanli BC, Cheah YC, Bradley A. Engineering a mouse balancer chromosome. Nat Genet. 1999;22:375–378. doi: 10.1038/11949. [DOI] [PubMed] [Google Scholar]