Abstract
The recent completion of the human genome sequence has enabled the identification of a large fraction of our gene catalogue and their physical chromosomal position. However, current efforts lag at defining the cis-regulatory sequences that control the spatial and temporal patterns of each gene's expression. This task remains difficult due to our lack of knowledge of the vocabulary controlling gene regulation and the vast genomic search space, with greater than 95% of our genome being noncoding. Recent comparative genomic-based strategies are beginning to aid in the identification of functional sequences based on their high levels of evolutionary conservation. This has proven successful for comparisons between closely related species such as human–primate or human–mouse, but also holds true for distant evolutionary comparisons, such as human–fish or human–bird. In this review we provide support for the utility of cross-species sequence comparisons by illustrating several applications of this strategy, including the identification of new genes and functional non-coding sequences. We also discuss emerging concepts as this field matures, such as how to properly select which species for comparison, which may differ significantly between independent studies.
Biology is a discipline rooted in comparisons. Comparative physiology has assembled a detailed catalogue of the biological similarities and differences between species, revealing insights into how life has adapted to fill a wide range of environmental niches. For example, following the initial budding of limbs in every vertebrate embryo, some develop to become legs (mammals), wings (birds), fins (fish) or even atrophy into vestigial structures (some reptiles, such as snakes). Comparative studies in anatomy, biochemistry, pharmacology, immunology and cell biology have provided the fundamental paradigms from which each discipline has grown.
Genomics is the most recent branch of biology to employ comparison-based strategies. At the foundation of the evolutionary relationship of all vertebrates is conserved genetic information in the form of DNA sequence, which is assumed to underlie homologous functional and anatomical similarities between species. Technological progress in DNA cloning and sequencing has resulted in the generation of a large dataset of genomic sequence information. In the past two years, draft genome sequence has become available for six vertebrates: human, mouse, rat, zebrafish and two pufferfish (Fugu rubripes and Tetraodon nigroviridis) (Lander et al. 2001; Venter et al. 2001; Aparicio et al. 2002; Waterston et al. 2002). The sudden wealth of sequence data has allowed whole genome alignments to compare and contrast the evolution and content of vertebrate genomes. Such comparative strategies have identified pockets of DNA sequences conserved over evolutionary time, and such evolutionary conservation has been a powerful guide in sorting functional from non-functional DNA (Duret & Bucher, 1997; Hardison et al. 1997; Hardison, 2000; Loots et al. 2000; Pennacchio & Rubin, 2001; Gottgens et al. 2002). Accordingly, this review focuses on the biological insights derived from comparative sequence-based studies and their increasing utility as the amount of genome sequence data increases. Details on various computational tools used in these studies can be found in several recent reviews (Pennacchio & Rubin, 2001; Frazer et al. 2003; Pennacchio & Rubin, 2003b; Ureta-Vidal et al. 2003). A list of the most commonly used tools in comparative genomics is provided in Table 1.
Table 1.
Websites for sequence alignment tools and databases useful for comparative-based analyses
| Tool or database | Website |
|---|---|
| NCBI (BLAST) | http:/www.ncbi.nlm.nih.gov/BLAST/ |
| GenomeVISTA (AVID) | http://pipeline.lbl.gov/cg-bin/GenomeVista |
| UCSC Genome Browser (BLAT) | http://genome.uscs.edu/ |
| EMSEMBL (SSAHA) | http://www.ensembl.org |
| BLAST2 | http://www.ncbi.nlm.nih.gov/blast/bl2seq/bL2.html |
| VISTA | http://www-gsd.lbl.gov/VISTA/index.html |
| PIPMAKER | http://bio.cse.psu.edu/pipmaker/ |
| Alfresco | http://www.sanger.ac.uk/Software/Alfresco/ |
| SynPlot | http://www.sanger.ac.uk/Users/jgrg/SynPlot/ |
| Jdotter | http://athena.bioc.uvic.ca/pbr/jdotter/ |
| GLASS | http://plover.lcs.mit.edu/ |
The power of varying evolutionary distance in comparative genomics
The utility of comparative sequence analysis is based on the hypothesis that important biological sequences are conserved between species due to functional constraints. To derive insights into biology through comparative sequence analysis the first challenge is the choice of species to compare. The ideal pairwise comparison is between two organisms that share a common physiology or biology. For example, human–mouse sequence comparisons have been useful in mapping the regulation of genes involved in lipid metabolism that are shared between the two organisms, but conversely such comparisons are not useful for understanding the regulation of a given lipid gene found only in the primate lineage. These concerns must also be balanced with the amount of actual sequence conservation between two organisms; too much conservation and the functional regions are obscured, too little conservation and they are hidden. Thus a balance of biological relevance, evolutionary distance and sequence analysis provides the best opportunity for the identification of conserved sequences that appear to be evolving under evolutionary constraints in a background of sequence that has randomly diverged due to genetic drift.
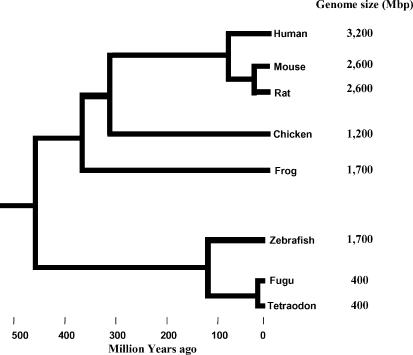
In recent years, the availability of sequence from numerous species has allowed multiple species comparisons to aid in calibrating the ideal evolutionary distance required for the optimal identification of functionally conserved sequences (Koop & Hood, 1994; Hood et al. 1995; Dubchak et al. 2000; Pennacchio & Rubin, 2001; Gottgens et al. 2002). In this review we adopt a human-centric focus of comparative genomics, describing strategies where sequence-based analyses alone have been used to enable better understanding of functional sequences in the human genome. Examples are provided of the most commonly used vertebrate genomes in cross-species sequence comparisons (Fig. 1), highlighting the uniqueness, usefulness and limitations of each.
Figure 1. Phylogenetic tree of a subset of vertebrate species.
The approximate divergence time of each of the eight vertebrate species whose genome sequences are currently available is represented (not drawn to scale). Haploid genome sizes are indicated in million base pairs.
Human–mouse sequence comparisons
The evolutionary distance between humans and mice places these species at strategic positions for the identification of shared functionally conserved sequences. It has been estimated that the rate of divergence in independently evolving vertebrate genomes is on average 0.1–0.5% per million years, supporting the premise that the ∼80 million years separating humans and mice from their last common ancestor is sufficient for functionally important sequences to be identified (Tautz, 2000). A number of recent studies have reported the identification of functional sequences solely through the use of human–mouse genomic comparisons, thereby further validating this assumption (Loots et al. 2000; Pennacchio et al. 2001; Gottgens et al. 2002; Kappen & Yaworsky, 2003). The most standard applications of human–mouse comparative sequence analyses involve: (1) the annotation of previously undefined genes; (2) the identification of large (80–1000 bp) functional gene-regulatory elements; and (3) the detailed characterization of transcription factor binding sites (‘phylogenetic footprints’) present in larger conserved non-coding regions.
Identification of new genes
The first application of human–mouse comparative genomics relates to the discovery of new genes within the human and mouse genomes, which have previously been invisible to extensive computational and experimental investigation. Since coding sequences of active genes are commonly under strong negative selection, human–mouse sequence comparisons are expected to unveil sequences corresponding to previously unidentified genes, thus expanding the complete gene catalogue of each organism. The discovery of the apolipoprotein A5 gene (APOA5) exemplifies this principle. Solely through the use of human–mouse genomic sequence comparisons, this evolutionary paralog (a related gene or sequence arisen from the duplication of an ancestral gene or sequence) of the neighbouring APOA4 gene was identified based on its high degree of sequence conservation within a previously well-studied cluster of apolipoproteins (Pennacchio et al. 2001). Transcripts from this corresponding interval were identified in human and mouse liver tissue, serving as evidence that these conserved sequences correspond to a previously missed gene. Further studies in transgenic and knockout mice revealed that the newly described APOA5 gene is a pivotal determinant of plasma triglyceride levels (Pennacchio et al. 2001). In addition, these findings were extended to human physiology when strong genetic associations between common APOA5 polymorphisms and plasma triglyceride levels were uncovered in a wide range of studies (reviewed by Pennacchio & Rubin, 2003a). Similar strategies will be useful in identifying un-annotated genes that are still predicted to exist in the human and mouse genomes.
Identification of gene regulatory sequences
While it is intuitive that comparative sequence analysis is suitable to identify exons based on conservation, its ability to uncover conserved gene regulatory sequences is less obvious owing to the small size of transcription factor binding sites (∼6–12 bp in size). Nevertheless, the architecture of the majority of characterized enhancers in metazoan genomes is thought to be determined by a combination of multiple transcription factor binding sites, arranged in a modular fashion within large clusters. Thus, the size of these enhancer elements is expected to be similar to many exons. An early study of human–mouse comparative sequence analysis as a starting point to identify gene regulatory elements was performed on a human interleukin gene cluster, which has long been known to harbour genes involved in several human inflammatory conditions (Noguchi et al. 1997; Rioux et al. 2001). In this work, human–mouse comparisons revealed a highly conserved 401 bp non-coding sequence within a genomic interval containing the interleukin-4, -5 and -13 genes (Loots et al. 2000). Subsequent deletion of this conserved non-coding sequence from mice revealed inappropriate expression of all three interleukins upon TH2 cytokine stimulation (Loots et al. 2000; Mohrs et al. 2001), thus demonstrating that the 401 bp conserved element corresponds to a regulatory element able to coordinately modulate the expression of three interleukin genes spread over 120 kb of sequence. This coordinated expression of interleukins had been previously proposed, but several studies using traditional approaches failed to uncover the sequence so clearly revealed by a comparative approach (Noguchi et al. 1997; Lacy et al. 2000).
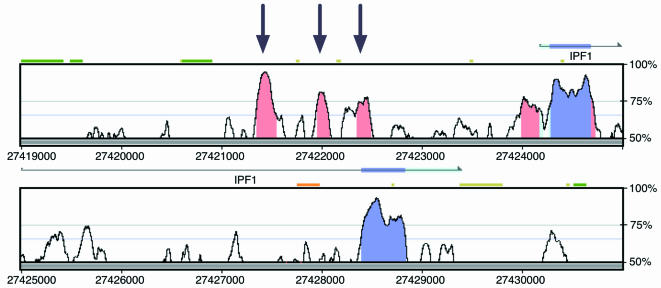
Human–mouse sequence comparisons are thus expected to represent a powerful tool in the puzzle of the decoding of gene-regulatory sequence. A paucity of published studies reporting the identification of functional gene regulatory sequences through traditional approaches highlights the difficulty in defining functional non-coding sequences. With the availability of large amounts of human–mouse genomic sequence, cross-species comparisons are poised to dramatically increase our ability to decipher non-coding DNA. Nevertheless, a handful of characterized enhancers, originally identified through labourious experimental strategies, have been retrospectively shown to be highly conserved between human and mouse. Several studies using standard enhancer-trapping strategies identified and characterized three regulatory sequences within a segment 1.5–3.0 kb upstream of the human pancreatic duodenal homeobox 1 (PDX-1) gene promoter (Sharma et al. 1996; Ben-Shushan et al. 2001). As shown in Fig. 2, both exons of the PDX-1 gene as well as several non-coding sequences are well conserved between humans and mice in this interval. Inspection of the sequence upstream of PDX-1 shows three distinct segments of sequence conservation located approximately 1.6–2.8 kb upstream of the promoter, corresponding to the three enhancers previously shown to regulate PDX-1 expression. These sequences, easily highlighted by direct genomic comparisons, would probably have been prioritized for characterization of biological function based solely on a comparative strategy. Similar strategies will probably identify many human gene regulatory elements in the genome.
Figure 2. Human versus mouse PDX-1 (IPF1) genomic sequence comparison.
VISTA Genome Browser output in which human is the reference sequence with per cent similarity to mouse plotted on the vertical axis (http:/pipeline.lbl.gov). Vertical arrows correspond to highly conserved non-coding sequences that coincide with previously defined gene regulatory elements. Gene orientation (horizontal arrows) and exon location (coloured bars) are provided above each panel.
Identification of transcription factor binding sites through ‘phylogenetic footprinting’
Human–mouse sequence conservation has also proved a useful guide in the detailed characterization of regulatory elements identified through cross-species sequence comparisons. For instance, an enhancer responsible for the expression of nestin in the ventral midbrain neuroepithelium of mice was recently identified through the use of human–mouse–rat genomic comparisons (Kappen & Yaworsky, 2003). To deduce the critical transcription factor binding sites responsible for the activity of this enhancer, further analysis by ‘phylogenetic footprinting’ of conserved sequences coupled with reporter gene assays were employed. ‘Phylogenetic footprinting’ uses multispecies sequence alignments to identify highly conserved motifs at a fine scale (6–12 bp), comparable to the size of transcription factor binding sites (Gumucio et al. 1996). Following the identification of such ‘footprints’ in the nestin enhancer, nucleotide substitutions were introduced into two sites of a putative transcription factor binding site (RXR-β) that was a candidate for mediating the enhancer activity. Transgenic mice harbouring these mutations lost the tissue-specific gene expression compared to the normal version of these binding sites, indicating that ‘phylogenetic footprinting’ of the enhancer had identified transcription factor binding sites of biological importance (Kappen & Yaworsky, 2003). Thus, in addition to the identification of enhancer elements, comparative genomics is useful for the detailed characterization of their composition.
Though a wealth of important examples may reinforce the notion that humans and mice occupy a privileged position for cross-species sequence comparisons, they alone cannot capture all biologically active sequences. First, it has been well established that the degree of sequence conservation is heterogeneous among different genomic segments in human and mouse. For instance, the T-cell receptor locus has been shown to be extremely conserved in human–mouse sequence comparisons (Koop & Hood, 1994), while the α-globin locus has been found to be highly divergent (Hardison et al. 1991). Such interspecies variation is due to wide-ranging differences in the human–mouse nucleotide substitution rates across the genome. The result is a set of genomic regions with vast amounts of conservation (though probably not functional), and a set lacking significant conservation (though still containing functional elements). Such an observation carries significant implications for cross-species sequence comparisons since this strategy assumes that natural selection has constrained functional sequences to evolve at slower rates than non-functional sequences. In practice, human–mouse comparisons are not always feasible for deriving biological insights for a given genomic region.
To study regions of the human genome where human–mouse sequence comparisons are not ideal, examination of species occupying different evolutionary distances may be useful. In regions that are too well conserved between human and mouse, the comparison of humans to more distantly related species is warranted (i.e. birds, reptiles, amphibians), while in regions that are poorly conserved between humans and mice, the comparison of humans to closer species can be beneficial (i.e. primates, dogs, rabbits). In the remainder of this review, we will describe the utility of human genomic comparisons to species other than mouse.
Human–chicken sequence comparisons
The shared ancestor that gave rise to birds and mammals existed approximately 300 million years ago during the vertebrate radiation (Kumar & Hedges, 1998), placing the distance between humans and chickens at approximately 3–4 times that of humans and mice (Fig. 1). A deeper phylogenetic relationship suggests that most neutrally evolving sequences in humans and birds will have diverged significantly more than those between humans and mice. In general, conserved DNA between humans and birds is more likely to be functional than that found between humans and mice. While no entire avian genome sequence is currently available, small genomic fragments of chicken DNA have been sequenced for comparative studies.
As an example, the identification and characterization of a human cardiac-specific enhancer regulating the homeobox gene Nkx2-5 was aided by the addition of orthologous chicken sequence (i.e. related sequences in chicken that started to diverge after a speciation event). Initial examination of the region 10 kb upstream of Nkx2-5 between humans and mice revealed five conserved non-coding sequences, but the addition of the orthologous chicken sequence revealed that only one of these five was also conserved in chickens. Functional studies in transgenic mice confirmed that this segment corresponds to a cardiac-specific enhancer regulating Nkx2-5 expression (Lien et al. 2002). Further dissection of this enhancer through ‘phylogenetic footprinting’ revealed the precise transcription factor binding sites responsible for the enhancer activity, aided by having the chicken genomic sequence. While human–mouse enhancer sequence comparisons revealed between 90 and 100% identity throughout the segment, making the identification of conserved ‘footprints’ difficult, human–mouse–chicken enhancer sequence comparison decreased the overall conservation in the region to 70% and revealed that four Smad binding sites were conserved in all three species. A combination of mouse transgenics and mutagenesis later confirmed that one of the conserved Smad sites mediates the enhancer activation of Nkx2-5 in the developing heart (Lien et al. 2002).
An interesting observation from these data is that genome sequences obtained from organisms with little or no use as model organisms for experimental biology represent extremely important resources for annotating the human genome. This underscores the importance of prioritizing the choices for sequencing further vertebrate genomes based not simply on a hierarchical list of experimentally suitable models, but also on a composite of factors that take into account the potential uses of the data generated for applications such as comparative genomics. Indeed, while the chicken, honeybee and chimpanzee are not standard experimental models, their genomes have been prioritized for the next round of DNA sequencing (Boguski, 2002).
Human–fish sequence comparisons
Human–fish comparisons also provide a useful evolutionary position for comparative sequence-based discovery. Several species of fish have been fully sequenced, which include working drafts for zebrafish, and the two pufferfish, Fugu rubripes and Tetraodon nigroviridis (Aparicio et al. 2002). The phylogenetic relationship between fish and humans dates back 400–450 million years, making fish the most distant vertebrates with available genomic sequence for comparison with humans (Fig. 1). Although this large evolutionary distance implies that only a fraction of the functional sequences in the human genome are still shared, comparison has revealed that most known human genes are also conserved in fish. Importantly, the annotation of conserved sequences between the human and Fugu rubripes genomes led to the rapid identification of over 1000 previously unidentified human genes (Abrahams et al. 2002; Aparicio et al. 2002). While the majority of conserved orthologous sequences between human and pufferfish represent coding sequences, thousands of conserved sequences that do not appear to correspond to genes are also present. This suggests that human–pufferfish genomic comparisons may result in the discovery of functionally important non-coding sequences in the human genome. However, these comparisons are likely to miss gene regulatory functions that are no longer preserved across such large evolutionary distances.
One of the most attractive features of pufferfish for its use in cross-species sequence comparisons is the compact size of its genome, totaling a mere 365 million bp (one-eighth the size of the human genome; Brenner et al. 1993). This compactness predicts that regulatory sequences shared between humans and pufferfish will be found much closer to a given pufferfish gene than its human ortholog, so human–pufferfish comparisons may identify distant regulatory elements in the human genome (Gilligan et al. 2002). A recent comparison of a 3.7 million bp sequence from humans with pufferfish identified 195 kb of sequence with orthology, revealing several genes in the region shared between the two species (Bagheri-Fam et al. 2001). Moreover, eight conserved sequences which were not predicted to be exons were identified within 750 kb of the human SOX9 transcription factor gene. In the pufferfish genome these conserved sequences are located within less than 80 kb of SOX9, suggesting that these may represent distant sequences that regulate SOX9 expression (Bagheri-Fam et al. 2001). Thus, the use of Fugu rubripes sequences in genomic comparisons may be a useful tool for the identification of both local and distant regulatory elements. However, given the extreme evolutionary distance between fish and mammals, and the extensive biological differences that separate these species, it is likely that these conserved sequences represent only a subset of the functional elements in the human genome. Since many aspects of embryologic development are extremely well conserved in all vertebrates, it is possible that the catalogue of conserved sequences between mammals and fish will be enriched for elements regulating the expression of genes involved in these developmental processes, and represent sequences whose biological impact in the organism is the most dramatic.
Interprimate sequence comparisons
Finally, comparison of the human sequence to that of other primate species is a strategy likely to identify functional regions of the human genome. The overall strategy previously described for cross-species sequence comparisons is based on using species of relatively distant phylogenetic positions to maximize the identification of functionally conserved sequences in the human genome. However, this strategy is limited in that it does not allow studies aimed at identifying primate-specific genes or regulatory sequences. For instance, the comparison of the human and mouse genomes identified ∼1% of mouse genes without a human ortholog (Waterston et al. 2002). In addition, this estimate does not take into account the numerous examples where tandem duplications lead to the formation and expansion of gene families in one species but not the other. To this end, only 80% of human–mouse genes have a 1:1 orthologous relationship (Waterston et al. 2002). Therefore, there is a need to develop strategies to characterize the catalogue of the 20% of genes and regulatory elements that do not have a true ortholog in both humans and mice. For these studies, comparing human sequences to those of closer evolutionary species, such as primates, may prove essential. However, the use of primate sequences for cross-species sequence comparisons poses a paradox, in that while primates are likely to share most genes present in the human genome, their close phylogenetic relationship results in high levels of sequence identity between orthologous sequences. For example, humans, chimpanzees and gorillas shared a common ancestor approximately 6.0–8.0 million years ago and their average rate of sequence conservation is 98–99% even in non-coding intervals (Hacia, 2001).
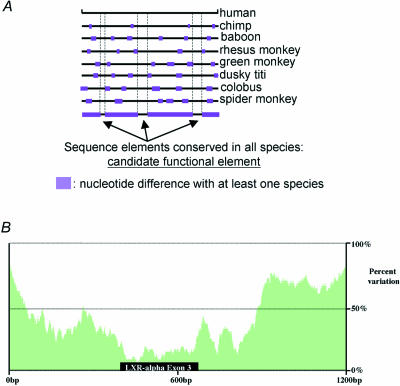
Recently, a strategy named ‘phylogenetic shadowing’ was introduced to overcome the excessive sequence identity shared by primates, which makes their use in cross-species sequence comparisons possible (Boffelli et al. 2003). The foundation of this approach is to analyse orthologous sequence from numerous primate species to increase the evolutionary distance of the sequence comparisons. Rather than performing only pairwise comparisons between human and mouse, human and chicken, or human and pufferfish, ‘phylogenetic shadowing’ compares a dozen or more different primate species. The summation of these primate comparisons robustly identifies regions of increased variation and ‘shadows’ representing conserved segments (Fig. 3A).
Figure 3. Phylogenetic shadowing of primate species.
A, the alignment and comparison of sequences from multiple species sequences reveal which sequences have been conserved in most species, making them likely candidates for being functionally relevant. B, a sequence variation plot of numerous aligned primate sequences flanking an exon of the LXR-á gene. On the x axis 1200 bp of sequence is depicted, while on the y axis the per cent variation is plotted. Note that the lack of sequence diversity (‘phylogenetic shadow’) corresponds closely with the functional exon interval.
As a proof of principle, ‘phylogenetic shadowing’ proved successful for the identification of both exons as well as putative gene regulatory elements (Boffelli et al. 2003). In this study, 13–17 primate sequences of several orthologous genomic segments were generated and compared. For a single exon from four independent genes, highly conserved ‘shadows’ coincided strongly with these functionally important protein-encoding regions (see Fig. 3B for one example). In addition, analysis of the human apolipoprotein (a) gene (apo(a)) revealed highly conserved intervals embedded within the upstream promoter region, and functional studies of these ‘phylogenetic shadows’ compared to more variable flanking DNA supported their role in regulating apo(a) expression (Boffelli et al. 2003). The success of this approach suggests that a genome-wide comparison of a handful of primate species will aid in the identification of both human exons and gene regulatory elements.
Conclusions and future perspective
Comparative genomics is a relatively new field that complements a long history of comparison-based disciplines in biology. The recent development of a large dataset of vertebrate genomic sequences has aided in global gene predictions as well as in the identification of sequences important in gene regulation. In addition, vertebrate comparative sequence analysis is poised to contribute to the exploration of the genetic bases for differences and similarities among species. In combination with areas of study such as comparative physiology or comparative biochemistry, we are likely at last to understand the genetic explanation for how species have adapted to perform their shared or unique biological functions.
The number of cross-species sequence comparisons will undoubtedly increase in use as additional genomes are sequenced. While we currently have access to a handful of vertebrate genome sequences and our tools for dealing with these datasets are rapidly improving, the computational challenges ahead are formidable. Current efforts have focused primarily on pairwise comparisons to annotate and explore a single species of interest (such as humans), but future methods will require the simultaneous analysis of sequence data from numerous species in the form of multiple alignments in order to catalogue the evolutionary extent of sequence conservation and divergence. In addition, the area of high-throughput experimental biology is a quickly evolving field with vast opportunities to exploit comparative sequence data.
For the biologist, the application of cross-species sequence analyses requires flexibility. It should be emphasized that no single pairwise comparison is sufficient to capture all biologically functional sequences based on conservation. Thus, the primary decision in the process of designing a comparative genomic-based study is the biological question under investigation and which two (or more) species are most appropriate for comparison. While there is no clear way to predict which repertoire of species is ideally suited for each cross-species comparison, the analysis of aligned human–mouse orthologous sequences provides an initial starting point for most biological studies. However, for example, if the study aims to identify regulatory elements of a primate-specific gene, it will not be useful to compare human-mouse or other lower vertebrates. In contrast, the study of basic vertebrate biological processes may be aided by distant species sequence comparisons (human-bird, human-amphibian, human-fish, etc.). Therefore, the biologist must make logical predictions about which species to compare and should readily adopt additional species as warranted based on their initial comparative analysis. The currently available vertebrate genome sequences are immediate resources for the community and additional vertebrate genomes are in the pipeline.
Acknowledgments
We thank E. Rubin, J. Priest, D. Boffelli and J. Wang for thoughtful discussions. This work was supported in part by the NIH-NHLBI Programs for Genomic Application Grant HL66681 and NIH Grant HL071954A through the US Department of Energy under contract no. DE-AC03-76SF00098.
References
- Abrahams BS, Mak GM, Berry ML, Palmquist DL, Saionz JR, Tay A, Tan YH, Brenner S, Simpson EM, Venkatesh B. Novel vertebrate genes and putative regulatory elements identified at kidney disease and NR2E1/fierce loci. Genomics. 2002;80:45–53. doi: 10.1006/geno.2002.6795. [DOI] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Bagheri-Fam S, Ferraz C, Demaille J, Scherer G, Pfeifer D. Comparative genomics of the SOX9 region in human and Fugu rubripes: conservation of short regulatory sequence elements within large intergenic regions. Genomics. 2001;78:73–82. doi: 10.1006/geno.2001.6648. [DOI] [PubMed] [Google Scholar]
- Ben-Shushan E, Marshak S, Shoshkes M, Cerasi E, Melloul D. A pancreatic beta-cell-specific enhancer in the human PDX-1 gene is regulated by hepatocyte nuclear factor 3beta (HNF-3beta), HNF-1alpha, and SPs transcription factors. J Biol Chem. 2001;276:17533–17540. doi: 10.1074/jbc.M009088200. [DOI] [PubMed] [Google Scholar]
- Boffelli D, Mcauliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, Pachter L, Rubin EM. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- Boguski MS. Comparative genomics: the mouse that roared. Nature. 2002;420:515–516. doi: 10.1038/420515a. [DOI] [PubMed] [Google Scholar]
- Brenner S, Elgar G, Sandford R, Macrae A, Venkatesh B, Aparicio S. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature. 1993;366:265–268. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- Dubchak I, Brudno M, Loots GG, Pachter L, Mayor C, Rubin EM, Frazer KA. Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 2000;10:1304–1306. doi: 10.1101/gr.142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Bucher P. Searching for regulatory elements in human noncoding sequences. Curr Opin Struct Biol. 1997;7:399–406. doi: 10.1016/s0959-440x(97)80058-9. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Elnitski L, Church DM, Dubchak I, Hardison RC. Cross-species sequence comparisons: a review of methods and available resources. Genome Res. 2003;13:1–12. doi: 10.1101/gr.222003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilligan P, Brenner S, Venkatesh B. Fugu and human sequence comparison identifies novel human genes and conserved non-coding sequences. Gene. 2002;294:35. doi: 10.1016/s0378-1119(02)00793-x. [DOI] [PubMed] [Google Scholar]
- Gottgens B, Barton LM, Chapman MA, Sinclair AM, Knudsen B, Grafham D, Gilbert JG, Rogers J, Bentley DR, Green AR. Transcriptional regulation of the stem cell leukemia gene (SCL) – comparative analysis of five vertebrate SCL loci. Genome Res. 2002;12:749–759. doi: 10.1101/gr.45502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumucio DL, Shelton DA, Zhu W, Millinoff D, Gray T, Bock JH, Slightom JL, Goodman M. Evolutionary strategies for the elucidation of cis and trans factors that regulate the developmental switching programs of the beta-like globin genes. Mol Phylogenet Evol. 1996;5:18–32. doi: 10.1006/mpev.1996.0004. [DOI] [PubMed] [Google Scholar]
- Hacia JG. Genome of the apes. Trends Genet. 2001;17:637–645. doi: 10.1016/s0168-9525(01)02494-5. [DOI] [PubMed] [Google Scholar]
- Hardison RC. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- Hardison R, Krane D, Vandenbergh D, Cheng JF, Mansberger J, Taddie J, Schwartz S, Huang XQ, Miller W. Sequence and comparative analysis of the rabbit alpha-like globin gene cluster reveals a rapid mode of evolution in a G + C-rich region of mammalian genomes. J Mol Biol. 1991;222:233–249. doi: 10.1016/0022-2836(91)90209-o. [DOI] [PubMed] [Google Scholar]
- Hardison RC, Oeltjen J, Miller W. Long human-mouse sequence alignments reveal novel regulatory elements: a reason to sequence the mouse genome. Genome Res. 1997;7:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- Hood L, Rowen L, Koop BF. Human and mouse T-cell receptor loci: genomics, evolution, diversity, and serendipity. Ann N Y Acad Sci. 1995;758:390–412. doi: 10.1111/j.1749-6632.1995.tb24844.x. [DOI] [PubMed] [Google Scholar]
- Kappen C, Yaworsky PJ. Mutation of a putative nuclear receptor binding site abolishes activity of the nestin midbrain enhancer. Biochim Biophys Acta. 2003;1625:109–115. doi: 10.1016/s0167-4781(02)00588-2. [DOI] [PubMed] [Google Scholar]
- Koop BF, Hood L. Striking sequence similarity over almost 100 kilobases of human and mouse T-cell receptor DNA. Nat Genet. 1994;7:48–53. doi: 10.1038/ng0594-48. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Lacy DA, Wang ZE, Symula DJ, Mcarthur CJ, Rubin EM, Frazer KA, Locksley RM. Faithful expression of the human 5q31 cytokine cluster in transgenic mice. J Immunol. 2000;164:4569–4574. doi: 10.4049/jimmunol.164.9.4569. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lien CL, McAnally J, Richardson JA, Olson EN. Cardiac-specific activity of an Nkx2-5 enhancer requires an evolutionarily conserved Smad binding site. Dev Biol. 2002;244:257–266. doi: 10.1006/dbio.2002.0603. [DOI] [PubMed] [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Mohrs M, Blankespoor CM, Wang ZE, Loots GG, Afzal V, Hadeiba H, Shinkai K, Rubin EM, Locksley RM. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat Immunol. 2001;2:842–847. doi: 10.1038/ni0901-842. [DOI] [PubMed] [Google Scholar]
- Noguchi E, Shibasaki M, Arinami T, Takeda K, Maki T, Miyamoto T, Kawashima T, Kobayashi K, Hamaguchi H. Evidence for linkage between asthma/atopy in childhood and chromosome 5q31-q33 in a Japanese population. Am J Respir Crit Care Med. 1997;156:1390–1393. doi: 10.1164/ajrccm.156.5.9702084. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Olivier M, Hubacek JA, Cohen JC, Cox DR, Fruchart JC, Krauss RM, Rubin EM. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Rubin EM. Genomic strategies to identify mammalian regulatory sequences. Nat Rev Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Rubin EM. Apolipoprotein A5: a newly identified gene impacting plasma triglyceride levels in humans and mice. Arteriosclerosis, Thrombosis, Vascular Biol. 2003a;23:529–534. doi: 10.1161/01.ATV.0000054194.78240.45. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Rubin EM. Comparative genomic tools and databases: providing insights into the human genome. J Clin Invest. 2003b;111:1099–1106. doi: 10.1172/JCI17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, Kocher K, Miller K, Guschwan S, Kulbokas EJ, O'Leary S, Winchester E, Dewar K, Green T, Stone V, Chow C, Cohen A, Langelier D, Lapointe G, Gaudet D, Faith J, Branco N, Bull SB, McLeod RS, Griffiths AM, Bitton A, Greenberg GR, Lander ES, Siminovitch KA, Hudson TJ. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet. 2001;29:223–228. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- Sharma S, Leonard J, Lee S, Chapman HD, Leiter EH, Montminy MR. Pancreatic islet expression of the homeobox factor STF-1 relies on an E-box motif that binds USF. J Biol Chem. 1996;271:2294–2299. doi: 10.1074/jbc.271.4.2294. [DOI] [PubMed] [Google Scholar]
- Tautz D. Evolution of transcriptional regulation. Curr Opin Genet Dev. 2000;10:575–579. doi: 10.1016/s0959-437x(00)00130-1. [DOI] [PubMed] [Google Scholar]
- Ureta-Vidal A, Ettwiller L, Birney E. Comparative genomics: genome-wide analysis in metazoan eukaryotes. Nat Rev Genet. 2003;4:251–262. doi: 10.1038/nrg1043. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]