Abstract
The single amino acid replacement of Tyr52 with Leu drastically increased the activity of Lactobacillus pentosus NAD-dependent d-lactate dehydrogenase toward larger aliphatic or aromatic 2-ketoacid substrates by 3 or 4 orders of magnitude and decreased the activity toward pyruvate by about 30-fold, converting the enzyme into a highly active d-2-hydroxyisocaproate dehydrogenase.
NAD-dependent l- and d-lactate dehydrogenases (l- and d-LDH, EC 1.1.1.27 and EC 1.1.1.28, respectively) convert pyruvate into l- and d-lactates, respectively, at the final step of anaerobic glycolysis, concomitantly oxidizing NADH into NAD+ (16). Lactic acid bacteria possess at least one of the two types of LDHs, fermenting the corresponding stereoisomer of lactic acid (12). In spite of the similarity in their catalytic reactions, the two types of enzymes are evolutionally separate from each other, belonging to distinct protein superfamilies (4, 21, 33). d-LDHs share a common protein structure not only with various d-2-hydroxyacid dehydrogenases (2, 4, 10, 13, 14, 21, 25, 26, 29, 32, 33) but also other dehydrogenases such as formate (26) and l-alanine (3) dehydrogenases.
l- and d-LDHs are highly divergent enzymes in lactic acid bacteria, showing great variety in both their amino acid sequences and catalytic properties or substrate specificities (1, 2, 4, 7, 12, 21, 33). There is only 40 to 50% amino acid identity among known d-LDHs of different Lactobacillus species, which show significantly different kinetic parameters, such as kcat and Km for substrates (4, 7, 21, 33). Instead of or together with d-LDH, some lactobacilli such as Lactobacillus casei (17, 19, 25) and L. delbrueckii (5) have d-hydroxyisocaproate dehydrogenases (d-HicDHs), which exhibit high activity not toward pyruvate but 2-ketoacids with larger aliphatic or aromatic side chains at the C-3 position, while L. confusus has l-HicDH, an l-LDH-related enzyme (30). d-HicDHs show 40 to 50% amino acid identity with known Lactobacillus d-LDHs (5, 25, 33), which is comparable to the identity among the d-LDHs, suggesting that these two types of enzymes are particularly related evolutionally. Since optically active 2-hydroxyacids are valuable for the synthesis of useful compounds (17-19), d-HicDHs are promising enzymes for industrial application, although their actual physiological role remains uncertain.
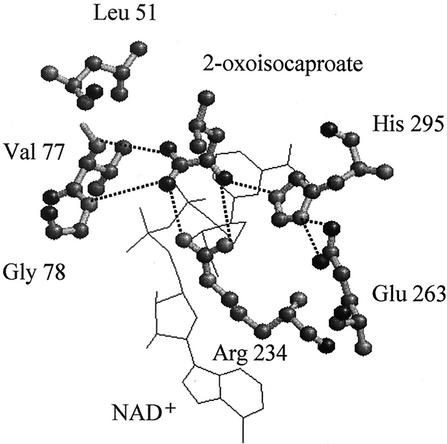
The substrate recognition by an enzyme has been extensively studied in the case of l-LDH, mostly through alteration of the substrate specificity by means of protein engineering (1, 8, 11, 15, 37, 38), but much less has been learnt about d-LDH or related enzymes. Recently, however, the three-dimensional structures of L. pentosus apo (32) and L. bulgaricus holo (27) d-LDHs, and the ternary complex of L. casei d-HicDH (10) were determined, implying their substrate recognition sites. Figure 1 shows the position of Leu51 in the substrate binding site of the L. casei d-HicDH ternary complex structure (10), together with those of Arg234, Glu263, and His294, which were indicated to be residues involved in the catalytic function of d-LDH by amino acid replacement studies (20, 22, 33-35). Since Leu51 is located very near the substrate C-3 position and is consistently replaced by conserved Tyr (Tyr52) in Lactobacillus d-LDHs (4, 21, 33), it is easily imaginable that the amino acid at this position defines the size or shape of the hydrophobic pocket for the C-3 groups of 2-ketoacid substrates (10). In this paper, we show how L. pentosus d-LDH is sufficiently converted into a d-HicDH through only 1 amino acid replacement of Tyr52 to Leu.
FIG. 1.
The substrate binding site of L. casei d-HicDH. The figure was drawn by using RasMol, according to the substrate binding site of the L. casei d-HicDH ternary complex structure (10). The two bound ligands, NAD+ and 2-ketoisocaproate, are shown as wire frame models. Leu51, Arg234, Glu263, and His294, which correspond to Tyr52, Arg235, Glu264, and His295 in d-LDHs, respectively, are indicated by ball-and-stick models.
An oligodeoxynucleotide, 5′-C GGT GCC GAT GTA CTG CAG CAA AAG GAC TAT ACT GC-3, was purchased from Takara Shuzo in order to construct a mutant L. pentosus d-LDH (Y52L) in which Tyr52 was replaced with Leu. Site-directed mutagenesis was performed with a MUTA-GENE in vitro mutagenesis kit (Bio-Rad), according to the method described by Kunkel (23), and the DNA fragments were sequenced by the dideoxy chain terminator procedure (28) with a DNA sequencer model 4000L (LI-COR) to prove that only the mutation expected had occurred. The wild-type and Y52L mutant d-LDHs of L. pentosus JCM1558 (ATCC 8041), previously called L. plantarum, were produced in Escherichia coli MV1184, and the cultivation of E. coli cells harboring expression plasmids for the d-LDH genes was performed essentially according to the previously described procedure (34). Enzyme purification was performed with both the previous procedure and a slight modification of it, which involves hydrophobic column chromatography on Butyl Toyopearl 650 M gel (Tosoh, Tokyo, Japan) instead of affinity chromatography on a 5′-AMP Sepharose gel (Amersham-Pharmacia Biotech). In the modified procedure, the Butyl Toyopearl column is equilibrated with 25 mM Tris-HCl (pH 7.4) buffer containing ammonium sulfate (45% saturation) and is eluted with a linear gradient of ammonium sulfate, from 45 to 0% saturation, at room temperature with a BioLogic system (Bio-Rad). The enzyme sample obtained with the modified procedure contained no significant NADH or NAD+ contamination as judged by spectroscopic analysis (data not shown) and gave a single protein band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis according to the method described by Laemmli (24) and exhibited essentially the same specific activity as that of the sample obtained by the previously described procedure (data not shown). During purification of the enzyme, protein concentrations were determined according to the method described by Bradford (6) with Bio-Rad Protein Assay protein reagent (Bio-Rad) by using bovine serum albumin as a standard protein. For kinetic and fluorescence analyses, the concentration of the purified enzyme was determined by using extinction coefficients at 280 nm of 27,045 and 25,855 M−1∋3χ · cm−1 for the coenzyme-free wild-type and Y52L enzymes, respectively, as determined from the amino acid composition and molecular weight of L. pentosus d-LDH (33). The enzyme activity toward various 2-ketoacids was assayed at 30°C in 100 mM sodium MES (2-[N-morpholino]ethanesulfonic acid) buffer (pH 5.5) containing 0.1 mM NADH and various concentrations of 2-ketoacids (sodium salts). One unit was defined as the conversion of 1 μmol of substrate per min. Kinetic parameters were calculated from plots of v/[S] versus [S], where [S] is substrate concentration.
The binding of NADH and oxamate to the enzymes was followed as the change in NADH fluorescence intensity (ΔF) essentially according to the method used for l-LDHs (31) with excitation and emission wavelengths of 340 and 460 nm, respectively, by using a JASCO FP-750 spectrofluorophotometer. ΔF was determined by comparing the fluorescence intensities of NADH in the presence and absence of the enzymes (15 μM) at 30°C in 50 mM sodium MES buffer (pH 6.0). Oxamate binding was determined in the presence of the enzymes (15 μM) and 0.1 mM NADH as the quenching of NADH fluorescence caused by various concentrations of sodium oxamate and was corrected by comparison with the nonspecific quenching in the absence of the enzymes. The dissociation constants for the enzymes with NADH and oxamate were determined according to the procedure for l-LDHs (9, 31) by curve fitting with KaleidaGraph.
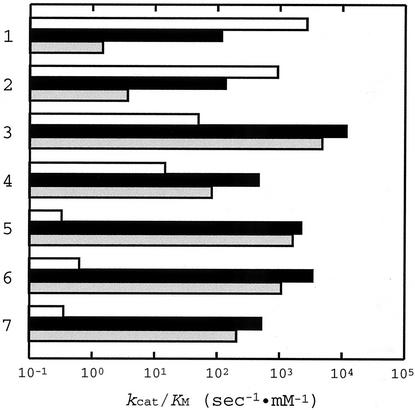
Table 1 summarizes the Km and kcat values of the wild-type and Y52L mutant L. pentosus d-LDHs for various 2-ketoacid substrates, and Fig. 2 compares the catalytic efficiencies (kcat/Km) of these two types of enzyme and L. casei d-HicDH (19) for these substrates. Compared with the wild-type L. pentosus d-LDH, the Y52L enzyme exhibited slightly reduced catalytic activity toward pyruvate and hydroxypyruvate, which are favorable substrates for the wild-type enzyme, by about 23- and 7-fold in terms of kcat/Km, respectively. These reductions in kcat/Km were mostly due to the increased Km values (15- and 10-fold higher, respectively), while the kcat values for these two substrates were not markedly affected by this amino acid replacement (Table 1). In contrast to the cases of these two substrates, the Y52L enzyme exhibited drastically increased catalytic activity toward 2-ketovalerate, 2-ketocaproate, and 2-ketoisocaproate by 3 or 4 orders of magnitude in kcat/Km. In these cases, the increases in activity resulted from both the reduction in Km (6- to 50-fold) and the increases in kcat (more than 100-fold) (Table 1). The Tyr52-to-Leu replacement also increased the catalytic activity of the enzyme toward 2-ketobutyrate and phenylpyruvate by 30- and 240-fold in kcat/Km, respectively, by both reducing Km and increasing kcat. Consequently, the Y52L mutant enzyme exhibited comparable levels of activity toward these five substrates to the activity that the wild-type enzyme showed toward pyruvate or hydroxypyruvate (Fig. 2A and B).
TABLE 1.
Kinetic parameters for various substrates for L. pentosus d-LDH
| Substrate | Wild-type Km (mM) | kcat (s−1) | Y52L Km (mM) | kcat (s−1) |
|---|---|---|---|---|
| Pyruvate | 0.12 ± 0.03 | 321 ± 12 | 1.8 ± 0.1 | 213 ± 4 |
| Hydroxypyruvate | 0.28 ± 0.03 | 257 ± 10 | 3.0 ± 0.1 | 407 ± 4 |
| 2-Ketobutyrate | 8.1 ± 0.6 | 118 ± 4 | 0.38 ± 0.01 | 175 ± 2 |
| 2-Ketovalerate | 17.0 ± 1.0 | 5.7 ± 0.2 | 0.15 ± 0.01 | 334 ± 6 |
| 2-Ketoisovalerate | 27.0 ± 0.8 | 5.5 ± 0.1 | 11.7 ± 0.4 | 18.6 ± 0.4 |
| 2-Ketocaproate | 20.9 ± 0.6 | 13.3 ± 0.2 | 0.056 ± 0.003 | 189 ± 3 |
| 2-Ketoisocaproate | 31.0 ± 2.0 | 11.1 ± 0.1 | 0.12 ± 0.01 | 65.7 ± 0.5 |
| Phenylpyruvate | 0.8 ± 0.2 | 40 ± 2 | 0.067 ± 0.001 | 778 ± 34 |
FIG. 2.
Comparison of the substrate specificities of the wild-type and Y52L L. pentosus d-LDHs, and L. casei d-HicDH. The kcat/Km values for rows follow: 1, pyruvate; 2, hydroxypyruvate; 3, phenylpyruvate; 4, 2-ketobutyrate; 5, 2-ketovalerate; 6, 2-ketocaproate; and 7, 2-ketoisocaproate. They are logarithmically plotted for the wild-type (white boxes) and Y52L (black boxes) L. pentosus d-LDHs and L. casei d-HicDH (19) (gray boxes).
The Y52L enzyme exhibited less enhanced activity toward 2-ketoisovalerate than the other substrates with a large hydrophobic side chain, with only an approximately 2.5-fold-smaller Km and a 3.5-fold-increased kcat compared to the wild-type enzyme (Table 1). In addition, benzoylformate or d-mandelate was also inert for this mutant enzyme, as in the case of the wild-type enzyme (data not shown), suggesting that the Tyr-to-Leu replacement is less effective for substrates with a C-3-branched side chain. These results may not be so strange, since there is no known Lactobacillus d-HicDH that exhibits high activity toward 2-ketoisovalerate or benzoylformate. It is known that benzoylformate is a favorable substrate for NAD-dependent d-mandelate dehydrogenases (d-ManDH) that have been purified from L. curvatus (18) and Enterococcus faecalis (36). However, much less information has been obtained about the structures of these d-ManDHs than has been obtained about d-LDHs or d-HicDHs.
Comparison of the kcat/Km values showed that the L. pentosus Y52L d-LDH and L. casei d-HicDH (19) exhibit quite similar trends in their substrate specificities, although the former enzyme exhibits still markedly higher activity toward pyruvate and hydroxypyruvate than the latter one (Fig. 1B and C). This comparison clearly indicates that L. pentosus d-LDH was sufficiently converted into a d-HicDH through only 1 amino acid replacement of Tyr52 for Leu, demonstrating that only the difference in the amino acid residue at position 52 distinguishes the enzyme functions of L. pentosus d-LDH and L. casei d-HicDH, although these two enzymes have only 46% identical amino acid residues (33).
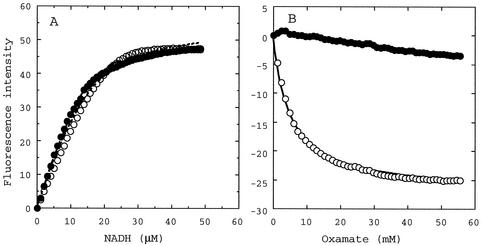
For both the wild-type and Y52L mutant enzymes, the fluorescence intensity of NADH markedly increased when NADH was bound into the enzymes, giving the same apparent dissociation constant (Kd) level of 3.4 ± 0.3 and 1.4 ± 0.1 μM, respectively (Fig. 3A). On the other hand, the fluorescence of the enzyme-bound NADH was markedly quenched on the addition of oxamate, a pyruvate analogue, in the case of the wild-type enzyme (Fig. 3B), giving an apparent oxamate Kd of about 5 mM, which is in good agreement with the reported competitive inhibitor constant (Ki) of oxamate (5 mM) at the same pH (35). In the case of the Y52L enzyme, in contrast, the NADH fluorescence was not markedly quenched by oxamate up to 60 mM. These results indicate that the Tyr52-to-Leu replacement markedly affects substrate binding but not coenzyme binding.
FIG. 3.
Change in NADH fluorescence intensity showing NADH binding (A) and oxamate binding (B) to the wild-type (open circles) and Y52L (closed circles) L. pentosus d-LDHs. The lines in panel A indicate the calculated curves obtained with the equation of Stinson and Holbrook (31) by using maximal ΔF of 53.8 and 49.0 and Kd values of 3.4 and 1.4 μM for the wild-type and Y52L enzymes, respectively. The line for oxamate binding to the wild-type enzyme (B) indicates the curve using a maximal fluorescence change of −27.5 and a Kd of 5 mM.
It is known that some lactobacilli possess both d-LDH and d-HicDH (5), although it is uncertain whether L. pentosus JCM1558 cells has d-HicDH besides d-LDH or not, since no, if any, marked d-HicDH activity was detected in the cell extract (data not shown). Our results demonstrate that Lactobacillus d-LDH and d-HicDH were readily converted into each other, through a small structural change such as a single amino acid replacement. From the aspect of protein engineering, the results strongly suggest that various enzyme functions can be introduced into the framework of d-LDHs, or related enzymes, by means of only a few amino acid replacements, as already demonstrated for l-LDH (8, 37).
Acknowledgments
This work was supported by a Grant-in-Aid for Science Research to H.T. from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Arai, K., T. Kamata, H. Uchikoba, S. Fushinobu, H. Matsuzawa, and H. Taguchi. 2001. Some Lactobacillus l-lactate dehydrogenases exhibit comparable catalytic activities for pyruvate and oxaloacetate. J. Bacteriol. 183:397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, M., C. Molinas, S. Dutka-Malen, and P. Courvalin. 1991. Structural relationship between the vancomycin resistance protein VanH and 2-hydroxycarboxylic acid dehydrogenases. Gene 103:133-134. [DOI] [PubMed] [Google Scholar]
- 3.Baker, P. J., Y. Sawa, H. Shibata, S. E. Sedelnikova, and D. W. Rice. 1998. Analysis of the structure and substrate binding of Phormidium lapideum alanine dehydrogenase. Nat. Struct. Biol. 5:561-567. [DOI] [PubMed] [Google Scholar]
- 4.Bernard, N., T. Ferain, D. Garmyn, P. Hols, and J. Delcour. 1991. Cloning of the d-lactate dehydrogenase gene from Lactobacillus delbrueckii subsp. bulgaricus by complementation in Escherichia coli. FEBS Lett. 290:61-64. [DOI] [PubMed] [Google Scholar]
- 5.Bernard, N., K. Johnsen, T. Ferain, D. Garmyn, P. Hols, J. J. Holbrook, and J. Delcour. 1994. NAD+-dependent d-2-hydroxyisocaproate dehydrogenase of Lactobacillus delbrueckii subsp. bulgaricus. Gene cloning and enzyme characterization. Eur. J. Biochem. 224:439-446. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Bugg, T. D., G. D. Wright, S. Dutka-Malen, M. Arthur, P. Courvalin, and C. T. Walsh. 1991. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30:10408-10415. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, A. R., T. Atkinson, and J. J. Holbrook. 1989. From analysis to synthesis: new ligand binding sites on the lactate dehydrogenase framework. Trends Biochem. Sci. 14:101-105, 145-148. [DOI] [PubMed]
- 9.Clarke, A. R., D. B. Wigley, D. A. Barstow, W. N. Chia, T. Atkinson, and J. J. Holbrook. 1987. A single amino acid substitution deregulates a bacterial lactate dehydrogenase and stabilizes its tetrameric structure. Biochim. Biophys. Acta 913:72-80. [DOI] [PubMed] [Google Scholar]
- 10.Dengler, U., K. Niefind, M. Kieβ, and D. Schomburg. 1997. Crystal structure of a ternary complex of d-2-hydroxyisocaproate dehydrogenase from Lactobacillus casei, NAD+ and 2-oxoisocaproate at 1.9 Å resolution. J. Mol. Biol. 267:640-660. [DOI] [PubMed] [Google Scholar]
- 11.El Hawrani, A. S., R. B. Sessions, K. M. Moreton, and J. J. Holbrook. 1996. Guided evolution of enzyme with new substrate specificities. J. Mol. Biol. 264:97-110. [DOI] [PubMed] [Google Scholar]
- 12.Garvie, E. I. 1980. Bacterial lactate dehydrogenases. Microbiol. Rev. 44:106-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldberg, J. D., T. Yoshida, and P. Brick. 1994. Crystal structure of an NAD-dependent d-glycerate dehydrogenase at 2.4 Å resolution. J. Mol. Biol. 236:1123-1140. [DOI] [PubMed] [Google Scholar]
- 14.Grant, G. A. 1989. A new family of 2-hydroxyacid dehydrogenase. Biochem. Biophys. Res. Commun. 165:1371-1374. [DOI] [PubMed] [Google Scholar]
- 15.Hogan, J. K., C. A. Pittle, J. B. Jones, and M. Gold. 1995. Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the l-lactate dehydrogenase of Bacillus stearothermophilus. Biochemistry 34:4225-4230. [DOI] [PubMed] [Google Scholar]
- 16.Holbrook, J. J., A. Liljas, S. J. Steindel, and M. G. Rossmann. 1975. Lactate dehydrogenase, p. 191-292. In P. D. Boyer (ed.), The enzymes, 3rd ed., vol. 11. Academic Press, New York, N.Y.
- 17.Hummel, W., H. Schütte, and M.-R. Kula. 1985. d-2-Hydroxyisocaproate dehydrogenase from Lactobacillus casei. A new enzyme suitable for stereospecific reduction of 2-ketocarboxylic acids. Appl. Microbiol. Biotechnol. 21:7-15. [Google Scholar]
- 18.Hummel, W., H. Schütte, and M.-R. Kula. 1988. d-(-)-Mandelic acid dehydrogenase from Lactobacillus curvatus. Appl. Microbiol. Biotechnol. 28:433-439. [Google Scholar]
- 19.Kallwass, H. K. W. 1992. Potential of R-2-hydroxyisocaproate dehydrogenase from Lactobacillus casei for stereospecific reductions. Enzyme Microb. Technol. 14:28-35. [Google Scholar]
- 20.Kochhar, S., N. Chuard, and H. Hottinger. 1992. Glutamate-264 modulates the pH dependence of the NAD+-dependent d-lactate dehydrogenase, J. Biol. Chem. 267:20298-20301. [PubMed] [Google Scholar]
- 21.Kochhar, S., P. E. Hunziker, P. Leong-Morgenthaler, and H. Hottinger. 1992. Primary structure, physicochemical properties, and chemical modification of NAD+-dependent d-lactate dehydrogenase. Evidence for the presence of Arg-235, His-303, Tyr-101, and Trp-19 at or near the active site. J. Biol. Chem. 267:8499-8513. [PubMed] [Google Scholar]
- 22.Kochhar, S., V. S. Lamzin, A. Razeto, M. Delley, H. Hottinger, and J. E. Germond. 2000. Roles of his205, his296, his303 and Asp259 in catalysis by NAD+-specific d-lactate dehydrogenase. Eur. J. Biochem. 267:1633-1639. [DOI] [PubMed] [Google Scholar]
- 23.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-49229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lerch, H. P., H. Blocker, H. K. W. Kallwass, J. Hoppe, H. Tsai, and J. Collins. 1989. Cloning, sequencing and expression in Escherichia coli of the d-2-hydroxyisocaproate dehydrogenase gene of Lactobacillus casei. Gene 78:47-57. [DOI] [PubMed] [Google Scholar]
- 26.Popov, V. O., and V. S. Lamzin. 1994. NAD+-dependent formate dehydrogenase. Biochem. J. 301:625-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Razeto, A., S. Kochhar, H. Hottinger, M. Dauter, K. S. Wilson, and V. S. Lamzin. 2002. Domain closure, substrate specificity and catalysis of d-lactate dehydrogenase from Lactobacillus bulgaricus. J. Mol. Biol. 318:109-119. [DOI] [PubMed] [Google Scholar]
- 28.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 742:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuller, D. J., G. A. Grant, and L. J. Banaszak. 1995. Allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase. Nat. Struct. Biol. 2:68-76. [DOI] [PubMed] [Google Scholar]
- 30.Schütte, H., W. Hummel, and M.-R. Kula. 1984. l-2-Hydroxyisocaproate dehydrogenase. A new enzyme from Lactobacillus confusus for the stereospecific reduction of 2-ketocarboxylic acid. Appl. Microbiol. Biotechnol. 19:167-176. [Google Scholar]
- 31.Stinson, R. A., and J. J. Holbrook. 1973. Equilibrium binding of nicotinamide nucleotides to lactate dehydrogenases. Biochem. J. 131:719-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoll, V. S., M. S. Kimber, and E. F. Pai. 1996. Insights into substrate binding by d-2-ketoacid dehydrogenases from the structure of Lactobacillus pentosus d-lactate dehydrogenase. Structure 4:437-447. [DOI] [PubMed] [Google Scholar]
- 33.Taguchi, H., and T. Ohta. 1991. d-Lactate dehydrogenase is a member of the d-isomer-specific 2-hydroxyacid dehydrogenase family. Cloning, sequencing, and expression in Escherichia coli of the d-lactate dehydrogenase gene of Lactobacillus plantarum. J. Biol. Chem. 266:12588-12594. [PubMed] [Google Scholar]
- 34.Taguchi, H., and T. Ohta. 1993. Histidine 296 is essential for the catalysis in Lactobacillus plantarum d-lactate dehydrogenase. J. Biol. Chem. 268:18030-18034. [PubMed] [Google Scholar]
- 35.Taguchi, H., T. Ohta, and H. Matsuzawa. 1997. Involvement of Glu-264 and Arg-235 in the essential interaction between the catalytic imidazole and substrate for d-lactate dehydrogenase. J. Biochem. 122:802-809. [DOI] [PubMed] [Google Scholar]
- 36.Tamura, Y., A. Ohkubo, S. Iwai, Y. Wada, T. Shinoda, K. Arai, S. Mineki, M. Iida, and H. Taguchi. 2002. Two forms of NAD-dependent d-mandelate dehydrogenase in Enterococcus faecalis IAM 10071. Appl. Environ. Microbiol. 68:947-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilks, H. M., K. W. Hart, R. Feeney, C. R. Dunn, H. Muirhead, W. N. Chia, D. A. Barstow, T. Atkinson, A. R. Clarke, and J. J. Holbrook. 1988. A specific, highly active malate dehydrogenase by redesign of a lactate dehydrogenase framework. Science 242:1541-1544. [DOI] [PubMed] [Google Scholar]
- 38.Wilks, H. M., D. J. Halsall, T. Atkinson, W. N. Chia, A. R. Clarke, and J. J. Holbrook. 1990. Designs for a broad substrate specificity ketoacid dehydrogenase. Biochemistry 29:8587-8591. [DOI] [PubMed] [Google Scholar]