Abstract
The Neisseria gonorrhoeae genome encodes a homologue of the Escherichia coli FNR protein (the fumarate and nitrate reductase regulator). Despite its similarity to E. coli FNR, the gonococcal FNR only partially complemented an E. coli fnr mutation. After error-prone PCR mutagenesis of the gonococcal fnr gene, we identified four mutant fnr derivatives carrying the same S18F substitution, and we showed that the mutant FNR could activate transcription from a range of class I and class II FNR-dependent promoters in E. coli. Prompted by the similarities between gonococcal and E. coli FNR, we made changes in gonococcal fnr that created substitutions that are equivalent to previously characterized substitutions in E. coli FNR. First, our experiments showed that cysteine, C116, in the gonococcal FNR, equivalent to C122 in E. coli FNR, is essential, presumably because, as in E. coli FNR, it binds to an iron-sulfur center. Second, the L22H and D148A substitutions in gonococcal FNR were made. These changes are equivalent to the L28H and D154A changes in E. coli FNR, which had been shown to increase FNR activity in the presence of oxygen. We show that the effects of these substitutions in gonococcal FNR are distinct from those of the S18F substitution. Similarly, substitutions in the putative activating regions of gonococcal FNR were made. We show that the activity of gonococcal FNR in E. coli can be increased by transplanting certain activating regions from E. coli FNR. The effects of these substitutions are additive to those due to S18F. From these data, we conclude that the effects of the S18F substitution in gonococcal FNR are distinct from the effects of the other substitutions. S18 is immediately adjacent to one of three N-terminal cysteine residues that coordinate the iron-sulfur center, and thus the S18F substitution is most likely to stabilize this center. Support for this came from complementary experiments in which we created the S24F substitution in E. coli FNR, which is equivalent to the S18F substitution in gonococcal FNR. Our results show that the S24F substitution changes the activity of E. coli FNR and that the changes are distinct from those due to previously characterized substitutions.
Neisseria gonorrhoeae can grow well in aerated, amino acid-rich media supplemented with glucose, lactate, or pyruvate as the major source of carbon and energy (25). It also has a limited ability to survive and grow anaerobically in the presence of nitrite as an alternative electron acceptor to oxygen (18). Under these conditions, synthesis of at least three proteins, Pan1, Pan2, and Pan3 (proteins induced during anaerobic growth), is induced and five proteins are repressed (7). Pan1 was subsequently renamed AniA (for anaerobically induced protein). The AniA protein is essential for nitrite reductase activity (24). Expression of aniA is induced strongly during anaerobic growth in the presence of nitrite relative to aerobic growth in the absence of nitrite (8, 27). The implication that gonococci respond to two environmental signals, the absence of oxygen and the presence of nitrite, was recently confirmed with the isolation and characterization of genes encoding homologues of the Escherichia coli transcription factor, FNR (fumarate and nitrate reductase regulator), and the nitrate- or nitrite-sensing two component regulatory systems, NarXL and NarQP. The corresponding gonococcal genes were insertionally inactivated and shown to be essential for the gonococcal response to oxygen limitation and the presence of nitrite (21).
The gonococcal FNR is predicted to be 37.5% identical and 50% similar in amino acid sequence to the E. coli FNR protein (Fig. 1). However, preliminary attempts to detect transcription activation by the gonococcal protein at four different E. coli FNR-dependent promoters were unsuccessful (see Table 1 in reference 21). In contrast, the gonococcal FNR repressed transcription from the E. coli ndh promoter to almost the same extent as the E. coli FNR protein. These results established that the gonococcal FNR expressed in E. coli is competent to recognize and bind to an E. coli FNR-binding site but interacts with the host RNA polymerase and activates transcription poorly.
FIG. 1.
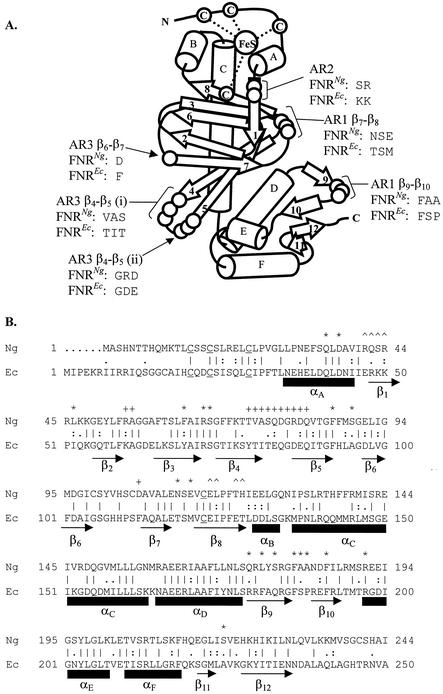
Structure of N. gonorrhoeae and E. coli FNR proteins. (A) The structure of E. coli FNR protein is modeled on the crystal structure of the related E. coli CRP protein. Alpha-helices are lettered A to F; beta-sheets are numbered 1 to 12. The four cysteine residues coordinating the iron-sulfur center and portions of the ARs altered in Fig. 7 are labeled. (B) The amino acid sequences of the gonococcal (Ng) and E. coli (Ec) FNR proteins were aligned. A vertical line denotes residue identity, a colon denotes strong residue similarity, and a dot denotes weak residue similarity. Residues comprising AR1 of E. coli FNR are marked with an asterisk, residues comprising AR2 are marked with a caret, and residues comprising AR3 are marked with a plus sign. Alpha-helices are lettered A to F and marked with rectangles; beta-sheets are numbered 1 to 12 and marked with arrows. The four cysteine residues that coordinate the iron-sulfur center of E. coli FNR are underlined.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic or relevant feature | Reference or source |
|---|---|---|
| E. coli K-12 strains | ||
| JM109 | lacZ recA | Promega |
| JCB3861 | Δ(nirB-cysG) pnirB::lacZ fnr | 23 |
| JRG1728 | ΔlacX74 galK galU rpsL Δ(ara-leu) Δ(tyrR-fnr-rac-trg)17 zdd-230::Tn9 | 29 |
| JRG1728 λ FF(−41.5) | JRG1728 pFF(−41.5)::lacZ | 20 |
| JRG1728 λ FF(−71.5) | JRG1728 pFF(−71.5)::lacZ | 20 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lac1qZΔM15 Tn10(Tetr)] | Stratagene |
| N. gonorrhoeae strains | ||
| RUG7001 | F62 proAB | 13 |
| RUG7022 | RUG7001 fnr | 13 |
| Bacterial plasmids | ||
| pBR322 | Commercially available cloning vector | Promega |
| pFNR | E. coli fnr gene, under the control of its own promoter, cloned between the HindIII and BamHI sites of pBR322 | 2 |
| pGCFNR3 | N. gonorrhoeae fnr gene, under the control of the E. coli fnr promoter, cloned between the HindIII and BamHI sites of pBR322 | This work |
| pQE60 | C-terminal His tag overexpression vector | QIAGEN |
| pRW50 | Broad-host-range lac expression vector | 22 |
Considerable progress has been made in understanding the mechanism of transcription activation by the E. coli FNR. To be effective as a transcription activator, four cysteine residues, one in the central region and three in the oxygen-sensing N-terminal domain, must bind a [4Fe-4S] iron-sulfur center (11, 16). The FNR polypeptide is synthesized during both aerobic and anaerobic growth, but the associated iron-sulfur center is degraded in aerobic cultures with a half life of about 2 min (16). The assembly of the [4Fe-4S] iron-sulfur center promotes dimerization during anaerobic growth, a prerequisite for FNR to bind to its inverted repeat target sequence at FNR-dependent promoters (19). Transcription activation then depends upon effective contacts being made between key residues of FNR in so-called activating region 1 (AR1), AR2, and AR3, and different parts of RNA polymerase (3, 5, 31, 32). The low level of transcription activation by the gonococcal FNR at E. coli FNR-dependent promoters offered an opportunity to reveal key differences between FNR proteins in the two genera, and to provide new insights into differences in gene expression and its regulation in two pathogenic bacteria. Despite the considerable similarities between FNR in E. coli and N. gonorrhoeae, there are differences in residues thought to be important in ARs that contact RNA polymerase (Fig. 1). It was anticipated that mutagenesis of these residues might generate an FNR derivative that activates transcription in E. coli. Here we describe successful experiments designed to introduce mutations into the gonococcal FNR protein that result in the production of novel derivatives able to activate transcription from a range of E. coli FNR-dependent promoters.
MATERIALS AND METHODS
Strains, plasmids, and oligonucleotide primers.
Bacterial strains and plasmids used are listed in Table 1; oligonucleotide primers are listed in Table 2.
TABLE 2.
PCR and mutagenic primers
| Primer name | Substitution introduced | Nucleotide sequencea (5′ to 3′) |
|---|---|---|
| NgFNRNcoI | TACGCCATGGCTTCGCATAATACTACAC | |
| NgFNRBamHI | ATCACGGATCCCAAAATGGCATACAACCCC | |
| NgFNRBglII | ATTAAGATCTGGCGTGCGAACAACCGG | |
| EcpFNRHindIII | CGCATAAGCTTCGTGAATATTTTGCC | |
| EcpFNRNcoI | TATTACCATGGGTCTGCTCAAGCCG | |
| NgFNR1 | S18X | CGCTGTGTTCTTCCTGTNNNTTGCGGGAACTCTGCCTGCC |
| NgFNR2 | S18X | GGCAGGCAGAGTTCCCGCAANNNACAGGAAGAACACAGCG |
| NgFNR3 | S18P | CGCTGTGTTCTTCCTGTCCNTTGCGGGAACTCTGCCTGCC |
| NgFNR4 | S18C | CGCTGTGTTCTTCCTGTTGTTTGCGGGAACTCTGCCTGCC |
| NgFNR5 | S18M | CGCTGTGTTCTTCCTGTATGTTGCGGGAACTCTGCCTGCC |
| NgFNR6 | L22H | CCTGTTCTTTGCGGGACATCTGCCTGCCTGTCGGG |
| NgFNR7 | L22H | CCCGACAGGCAGGCAGATGTCCCGCAAAGAACAGG |
| NgFNR8 | S18F L22H | CCTGTTTTTTGCGGGACATCTGCCTGCCTGTCGGG |
| NgFNR9 | S18F L22H | CCCGACAGGCAGGCAGATGTCCCGCAAAAAACAGG |
| NgFNR10 | C99A | GGCATGGACGGCATCGCTTCCTATGTGC |
| NgFNR11 | C99A | GCACATAGGAAGCGATGCCGTCCATGCC |
| NgFNR12 | C105A | CCTATGTGCACAGTGCCGACGCGGTCGCC |
| NgFNR13 | C105A | GGCGACCGCGTCGGCACTGTGCACATCGG |
| NgFNR14 | C116A | CAGCGAGGTAGCCGAGTTGCCGTTTACC |
| NgFNR15 | C116A | GGTAAACGGCAACTCGCATACCTCGCTG |
| NgFNR16 | D148A | CGCGAAATTGTGCGCGCTCAAGGTGTTATGC |
| NgFNR17 | D148A | GCATAACACCTTGAGCGCGCACAATTTCGCG |
| NgFNR18 | V74T A75I S76T | GGCTTCTTCAAAACAACCACAATCACACAGGACGGCCG |
| NgFNR19 | V74T A75I S76T | CGGCCGTCCTGTGTGATTGTGGTTGTTTTGAAGAAGCC |
| NgFNR20 | R80D D81E | CCAGTCAGGACGGCGATGAACAGGTAACGG |
| NgFNR21 | R80D D81E | CCGTTACCTGTTCATCGCCGTCCTGACTGG |
| NgFNR22 | D106F | GCACAGTTGCTTCGCGGTCGCCTTGG |
| NgFNR23 | D106F | CCAAGGCGACCGCGAAGCAACTGTGC |
| NgFNR24 | S43K R44K | CCGTCATCCGCCAAAAGAAGCGCCTGAAAAAAGGC |
| NgFNR25 | S43K R44K | GCCTTTTTTCAGGCGCTTCTTTTGGCGGATGACGG |
| NgFNR26 | N112T E114M | CGCCTTGGAAACCAGCATGGTATGCGAGTTGC |
| NgFNR27 | N112T E114M | GCAACTCGCATACCATGCTGGTTTCCAAGGCG |
| NgFNR28 | A181S A182P | CCGAGGTTTCTCCCCCAACGACTTCATC |
| NgFNR29 | A181S A182P | GATGAAGTCGTTGGGGGAGAAACCTCGG |
| EcFNR1 | S24F | CCATTGCCAGGATTGCTTCATCAGCCAGC |
| EcFNR2 | S24F | GCTGGCTGATGAAGCAATCCTGGCAATGG |
| EcFNR3 | L28H | CATCAGCCAGCATTGCATCCCGTTCAC |
| EcFNR4 | L28H | GTGAACGGGATGCATCGCTGGCTGATG |
| EcFNR5 | D154A | GGTGAAATCAAAGGCGCTCAGGACATGATCC |
| EcFNR6 | D154A | GGTAAACGGCAACTCGCATACCTCGCTG |
Engineered restriction sites are underlined. Mutations are shown in boldface type.
Growth conditions.
E. coli cultures were grown in Luria-Bertani broth (LB) supplemented with 0.4% glucose and appropriate antibiotics. Aerobic cultures (8 ml) were grown in 50-ml conical flasks with vigorous aeration; anaerobic 10-ml cultures were grown in narrow test tubes without aeration.
Construction of pGCFNR3.
Plasmid pGCFNR3 was constructed by three-way ligation. The pBR322-based vector fragment was prepared by restricting pFNR (30) with HindIII and BamHI. This fragment was dephosphorylated with calf intestinal phosphatase and purified from an agarose gel. The E. coli fnr promoter was cloned as a 500 bp PCR product amplified from pFNR using primers EcpFNRHindIII and EcpFNRNcoI into pGEM T-Easy (Promega) with E. coli JM109 as the host. Plasmid DNA was isolated, restricted with HindIII and NcoI and the 500 bp band was purified from a 1% agarose gel. The gonococcal FNR coding sequence was amplified using primers NgFNRNcoIDS and NgFNRBamHIUP2 and cloned into pGEM T-Easy. Plasmid DNA was isolated, restricted with HindIII and NcoI and the 1.5-kb band was purified from a 1% agarose gel. The three fragments were ligated together and transformed into E. coli strain JM109 with selection for ampicillin resistance.
Error-prone PCR mutagenesis.
Pools of gonococcal fnr genes containing random nucleotide changes were generated by error-prone PCR using BioTaq (Bioline). The fidelity of the DNA polymerase was decreased by increasing the concentration of magnesium ions in the reaction mixture to 3 mM. The PCR product was cloned into the vector pGEM-T Easy, transferred as an NcoI-BamHI fragment into the vector fragment of pGCFNR3, and the resulting plasmids were transformed into ultra-competent E. coli strain JM109. Plasmid DNA was extracted from the pooled transformants and transformed into the fnr strain JCB3861. Transformants able to activate transcription from the nirB promoter were then identified as red colonies after overnight growth on MacConkey-lactose-ampicillin agar.
Random and site-specific mutagenesis of codon S18 of the gonococcal fnr gene.
The QuikChange site-directed mutagenesis system (Stratagene) and degenerate primers NgFNR1 and NgFNR2 were used to replace the S18 codon of the fnrNg gene carried on pGCFNR3 with random sequences. Codons for proline, cysteine and methionine not generated by this random procedure were then introduced using the specific PCR primers NgFNR3, NgFNR4, and NgFNR5 (Table 2). Plasmids expressing all 20 possible derivatives at codon 18 were transformed into the fnr mutant, E. coli strain JRG1728 with an FNR-dependent promoter, FF(−41.5), fused to lacZ and inserted at the λ attachment site.
Site-directed mutagenesis of the gonococcal and E. coli fnr genes.
Site-specific mutations were introduced into the gonococcal fnr gene (on plasmid pGCFNR3) or the E. coli fnr gene (on plasmid pFNR) using the QuikChange site-directed mutagenesis system (Stratagene). Table 2 lists the primers used to create specific mutations.
β-Galactosidase assay.
β-Galactosidase activities of bacteria that had been grown either aerobically or anaerobically, in duplicate, were determined as previously described (15). Units are nanomoles of o-nitrophenol-β-d-galactoside (ONPG) hydrolyzed minute−1 (milligram of dry mass)−1. Each experiment was repeated at least three times with freshly transformed E. coli.
Western blotting.
The E. coli fnr mutant strain JRG1728 was transformed with pFNR or derivatives of pGCFNR3 and the transformants were grown with aeration to late stationary phase in LB supplemented with ampicillin. N. gonorrhoeae strains RUG7001 (fnr+) and RUG7022 (fnr) were grown microaerobically to exponential phase in gonococcal broth. Bacteria were harvested and lysed in sample buffer. Proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis were blotted onto a polyvinylidene fluoride membrane, and the presence of FNR protein was detected using an antibody raised against E. coli FNR kindly supplied by Jeff Green.
Purification of FNR proteins.
Gonococcal and E. coli fnr genes were cloned into the C-terminal His tag overexpression vector pQE60 (QIAGEN). QuikChange mutagenesis was used to introduce an S24F substitution into an fnrEc (D154A) gene carried on pQE60, generating a doubly substituted fnrEc gene. Two gonococcal fnr derivatives, D148A and S18F D148A, were amplified by PCR using primers NgFNRNcoI and NgFNRBglII. The resultant fragments were ligated into NcoI-BglII digested pQE60. FNR proteins were purified aerobically as described in (35), except that XL1-Blue was used as an expression strain, and FNR proteins were purified aerobically by affinity chromatography using HiTrap chelating columns charged with nickel ions and an ÄKTA prime high-performance liquid chromatography machine (Amersham Biosciences). The iron-sulfur centers of FNR proteins were not reconstituted.
Gel retardation assays.
Gel retardation assays were used to examine the binding of FNR derivatives to DNA as previously described (4), except that FF(−41.5) promoter DNA was used.
RESULTS
Expression of the gonococcal FNR in E. coli.
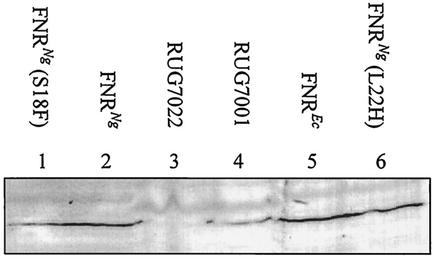
Plasmid pFNR contains the E. coli FNR-coding sequence cloned into pBR322 and expressed under the control of the E. coli fnr promoter (2). The coding sequence of the fnr gene from N. gonorrhoeae strain F62 was amplified by PCR and cloned into the vector fragment of pFNR to generate plasmid pGCFNR3. This plasmid was transformed into E. coli fnr mutant strain JRG1728. Western blotting using an antiserum to E. coli FNR detected a protein of similar size in E. coli strain JRG1728 transformed with pFNR or pGCFNR3, and in N. gonorrhoeae strain RUG7001, but not in the fnr mutant, N. gonorrhoeae RUG7022 (Fig. 2).
FIG. 2.
Western blotting was used to detect the expression of E. coli and N. gonorrhoeae FNR proteins. E. coli cultures were grown to late exponential phase; N. gonorrhoeae cultures were grown microaerobically. Lanes were loaded with equal biomass as follows: lane 1, E. coli JRG1728 transformed with pGCFNR3 carrying an S18F substitution; lane 2, E. coli JRG1728 transformed with pGCFNR3; lane 3, N. gonorrhoeae RUG7022; lane 4, N. gonorrhoeae RUG7001; lane 5, E. coli JRG1728 transformed with pFNR; lane 6, E. coli JRG1728 transformed with pGCFNR3 carrying an L22H substitution. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto polyvinylidene difluoride membrane. FNR protein was detected using antisera raised against E. coli FNR as a primary antibody and an alkaline phosphatase-conjugated secondary antibody. A band corresponding to FNR protein is present in lanes containing protein from E. coli transformed with plasmids expressing E. coli or gonococcal FNR proteins, and in N. gonorrhoeae strain RUG7001. No band is present in lane 3, which contains proteins from N. gonorrhoeae fnr mutant strain RUG7022.
Plasmid pGCFNR3 was transformed into two derivatives of the E. coli fnr strain JRG1728 in which class II and class I FNR-dependent promoters fused to a lacZ reporter gene had been integrated into the chromosome at the λ attachment site. The class II promoter, FF(−41.5), included a consensus FNR-binding site located at position −41.5 (i.e., the axis of symmetry of the FNR-binding site is located between bases 41 and 42 upstream from the transcription start) (34). The class I promoter, FF(−71.5), has a consensus FNR binding site at position −71.5. A small activity above the background was detected following anaerobic growth of either transformant. Plasmid pGCFNR3 was also transformed into E. coli JCB3861, an fnr strain carrying a nirB::lacZ fusion. Little β-galactosidase activity above background was detected following anaerobic growth. It was concluded that gonococcal FNR is expressed from pGCFNR3 at levels comparable to the E. coli FNR from pFNR but activates transcription poorly in E. coli.
Mutations in the gonococcal fnr gene resulting in enhanced transcription activation in E. coli.
In attempts to improve the ability of the gonococcal FNR to activate transcription at FNR-dependent promoters in E coli, the fnr coding sequence was amplified by error-prone PCR using N. gonorrhoeae strain F62 chromosomal DNA as template. The resulting library was cloned into the vector fragment of pGCFNR3 and transformed into E. coli strain JCB3861. Transformants able to activate transcription from the nirB promoter were then identified as red colonies after overnight growth on MacConkey-lactose-ampicillin agar. Four such colonies were isolated from approximately 80,000 colonies screened. Three of the colonies contained plasmids encoding FNR proteins with a single amino acid substitution, serine 18 replaced with phenylalanine (S18F). However, these three plasmids also carried silent mutations elsewhere within the fnr sequence, establishing that they were the result of independent mutations. The fourth plasmid also encoded an FNR protein with the S18F substitution, but there were two additional amino acid substitutions.
Comparison of transcription activation by gonococcal FNR proteins at different consensus FNR-dependent E. coli promoters.
The ability of the S18F variant to activate transcription at the FF(−41.5) and FF(−71.5) promoters was compared with transcription activation, or the lack of it, by both the unsubstituted gonococcal FNR and FNR from E. coli.
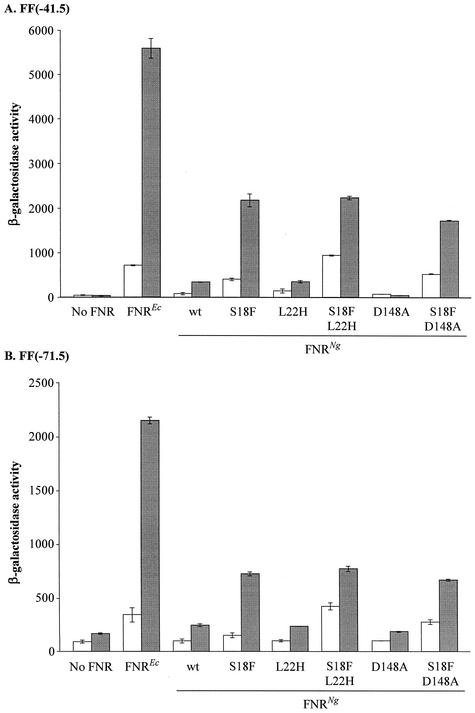
At the class II FF(−41.5) promoter, the E. coli FNR induced transcription 9.3-fold, giving a β-galactosidase activity during anaerobic growth of about 7,000 units (Fig. 3A). Only very low β-galactosidase activity (250 units) was detected with the gonococcal FNR expressed from pGCFNR3 at this promoter. The S18F variant of the gonococcal FNR activated transcription to a level above 2,000 units, with an anaerobic to aerobic induction ratio of 6.8-fold. Western blotting demonstrated that the S18F derivative of gonococcal FNR was expressed in E. coli at a similar or slightly lower level to the wild-type FNRNg protein (Fig. 2).
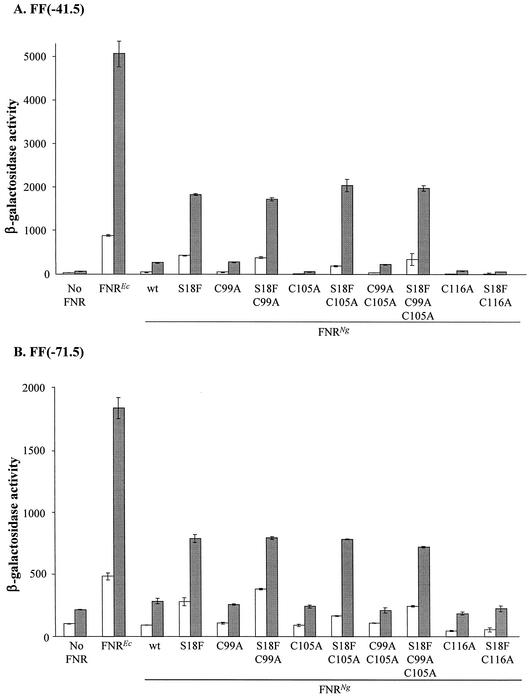
FIG. 3.
Effects of the substitutions L22H and D148A in the presence or absence of the S18F substitution on transcription activation by the gonococcal FNR at class I and class II FNR-dependent promoters. Mutations were introduced into the gonococcal fnr gene on plasmid pGCFNR3 using the QuikChange method. The mutated plasmids were transformed into strain JRG1728 carrying the consensus FNR-dependent promoter-lacZ fusions FF(−41.5) (A) or FF(−71.5) (B) integrated into the phage λ attachment site. Each transformant was assayed in duplicate from two independent cultures; units of activity are nanomoles of ONPG hydrolyzed · minute−1 · (milligram of bacteria [dry mass])−1. Error bars, standard deviations.
At the class I FF(−71.5) promoter, the E. coli FNR activated transcription to a level of 2,200 units with a 7-fold anaerobic to aerobic induction ratio (Fig. 3B). It should be noted that this promoter was also partially active in the absence of an FNR protein. Only 215 units of β-galactosidase activity were detected in the presence of gonococcal FNR. This increased to 680 units in the presence of the S18F form of the gonococcal FNR, with an induction ratio of 6.8. Clearly, therefore, the S18F form of gonococcal FNR can act as a class I transcription activator, though it is only 30% as effective as the E. coli FNR. Similar results were obtained with a second class I consensus, synthetic promoter, FF(−61.5) carried on the chromosome, and also with multicopy plasmids expressing lacZ under the control of either the FF(−41.5) or FF(−71.5) promoters (data not shown). In each situation, the gonococcal FNR either failed to activate transcription, or gave very low levels of β-galactosidase activity, but the S18F form gave significant transcription activation and induction ratios intermediate between the gonococcal and E. coli FNR. This established not only that the S18F form of the gonococcal FNR is able to activate transcription at both class I and class II FNR-dependent promoters, but also that it is still able to detect and respond to the presence or absence of oxygen.
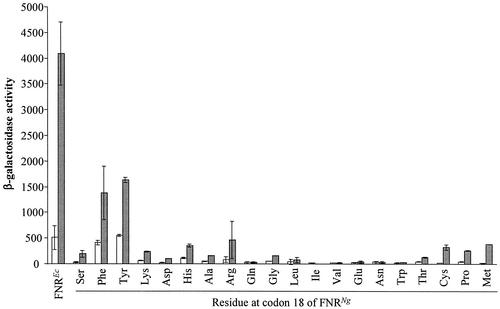
To investigate whether other substitutions at codon 18 would also result in transcription activation in E. coli, 20 derivatives of gonococcal fnr were the generated, each bearing a different amino acid at position 18. The ability of the 20 gonococcal FNR derivatives to activate transcription at the FF(−41.5) promoter was assessed (Fig. 4). Only two substitutions, S18F and S18Y, significantly increased transcription activation by gonococcal FNR. Furthermore, both of these mutated FNR proteins also activated transcription significantly during aerobic growth. These data indicate that the gonococcal FNR with either the S18F or the S18Y substitution is competent to interact with the E. coli RNA polymerase.
FIG. 4.
Effect on transcription activation at a class II consensus FNR-dependent promoter of replacing serine 18 of the gonococcal FNR in plasmid pGCFNR3 with different amino acids. The gonococcal fnr genes carried on pGCFNR3 were mutated using the QuikChange method and the resulting plasmids transformed into E. coli strain JRG1728λFF(−41.5), which carries a chromosomal consensus FNR-binding site centered between 41 and 42 bases upstream from lacZ integrated into the phage λ attachment site (20). Purified transformants, together with pFNR expressing the E. coli FNR as a positive control, were grown aerobically (open bars) and anaerobically (filled bars) in LB-glucose and assayed for β-galactosidase activity. Other details are as explained in the legend to Fig. 3.
Comparison of the gonococcal and E. coli FNR proteins.
Although the structure of E. coli FNR has not been determined, models based upon the crystal structure of the homologous E. coli CRP protein have proved useful in designing and interpreting mutagenesis data, and in studies of the mechanism of transcription activation by both proteins (6). Different classes of substitution have been identified that affect different stages in transcription activation by the E. coli FNR. Substitutions in the cysteine residues that coordinate the iron-sulfur center of FNR, C20, C23, C29, and C122, reduce transcription activation (9). Substitutions such as L28H and D154A result by independent mechanisms in increased transcription during aerobic growth. The L28H substitution (located adjacent to the third cysteine residue involved in the formation of an iron-sulfur center) stabilizes the [4Fe-4S] center, and D154A strengthens subunit interactions at the dimerization interface of FNR (1, 17, 19). Substitutions in the three ARs have led to the identification of residues that interact with the α and σ subunits of RNA polymerase (5). The primary sequence was used to model the gonococcal FNR, and to compare its proposed structure with that of the E. coli FNR (Fig. 1). This revealed not only that all four cysteine residues required to bind the iron-sulfur center are conserved, but also that L28 and D154 are conserved as L22 and D148. However, there are significant differences in the three ARs (Fig. 1). To further investigate the basis for increased FNR-dependent transcription due to the S18F form of gonococcal RNA polymerase, further substitutions were introduced into the gonococcal FNR.
Identification of the central cysteine residue essential for transcription activation by the gonococcal FNR protein.
In the gonococcal FNR, there are only three N-terminal cysteines corresponding to C20, C23, and C29 in the E. coli FNR that provide ligands to bind the iron-sulfur center (11). However, there are three cysteines in the central region that might provide the fourth ligand, corresponding to C122 of E. coli FNR (Fig. 1). The central cysteine essential for oxygen sensing by gonococcal FNR was therefore determined.
Alteration of C116, which corresponds to C122 of E. coli FNR, to alanine effectively abolished transcription activation by the wild-type or S18F gonococcal FNR at the FF(−41.5) promoter (Fig. 5A). In contrast, substitution of C99, C105, or both residues to alanine had no effect on transcription activation, though the C105A substitution decreased the very low activity in the absence of S18F. Essentially identical results were obtained with the class I FF(−71.5) promoter (Fig. 5B). It is concluded that C116 provides the fourth, essential ligand for binding the oxygen-sensing iron-sulfur center.
FIG. 5.
Identification of the central cysteine residue of the gonococcal FNR that is essential for transcription activation by FNRNg (S18F). The QuikChange method was used to change three cysteine residues on the central part of the gonococcal FNR to alanine, and the effects of the substitutions on transcription activation by FNRNg and FNRNg (S18F) in E. coli was determined. Each derivative gonococcal FNR was assayed in strain JRG1728 carrying the consensus FNR-dependent promoter-lacZ fusions FF(−41.5) (A) or FF(−71.5) (B) integrated into the phage λ attachment site. Open bars: aerobic cultures; hatched bars, anaerobic cultures. Other details are as explained in the legend to Fig. 3.
Effects of an L22H substitution in the gonococcal FNR on transcription activation in E. coli.
The S18F substitution is located adjacent to the second cysteine that is involved in binding the oxygen-sensing iron-sulfur center of FNR (Fig. 1). Superficially this is reminiscent of substitutions of residues adjacent to N-terminal cysteines of the E. coli FNR such as L28H that results in increased transcription activation during aerobic growth due to the stabilization of the [4Fe-4S] iron-sulfur center (1, 17). As a first step to probe the molecular basis for increased transcription activation, the effects of the S18F substitution were compared with that of an L22H derivative, which corresponds to the L28H substitution in E. coli FNR.
The L22H substitution slightly increased activity from the class II FF(−41.5) promoter from 100 to 200 units during aerobic growth, but had no effect during anaerobic growth (Fig. 3A). However, the same substitution did not increase the rate of transcription at the class I FF(−71.5) promoter during aerobic or anaerobic growth (Fig. 3B). The effects of combining the S18F and L22H substitutions were also determined. At the class II promoter, FF(−41.5), the double mutant carrying both the S18F and L22H substitutions activated transcription to a level of 900 units during aerobic growth, approximately double the activity of the S18F single mutant. The anaerobic activity remained at 2,000 units, typical of the S18F single substitution (Fig. 3A). At the class I promoter, FF(−71.5), aerobic transcription in the presence of the S18F L22H double substitution was increased from 100 units due to L22H alone or 200 units due to S18F alone to more than 400 units (Fig. 3B). These data suggest that the effects of the S18F and L22H substitutions are distinct and additive and therefore essentially independent of each other. Western blot analysis showed that the level of expression of the L22H derivative of the gonococcal FNR protein was similar to that of the wild-type protein (Fig. 2).
Effects of a D148A substitution in the gonococcal FNR on transcription activation in E. coli.
Substitutions such as D154A in the E. coli FNR strengthen subunit interactions at the dimerization interface, providing a second mechanism for increasing transcription activation. As D148 is conserved in the gonococcal FNR, it was of interest to construct a derivative carrying the substitution D148A, and to compare its properties with E. coli FNR. In marked contrast to the S18F substitution, the D148A substitution decreased the basal rate, especially at the FF(−41.5) promoter, during anaerobic growth (Fig. 3).
At both the FF(−41.5) and FF(−71.5) promoters, the S18F D148A substituted gonococcal FNR protein had slightly higher aerobic and slightly lower anaerobic activity than the S18F derivative (Fig. 3). In summary, these studies revealed differences between the effects of the D148A substitution on the gonococcal FNR and the corresponding substitution in the E. coli FNR. Increased transcription activation due to S18F was independent of the mutation at D148.
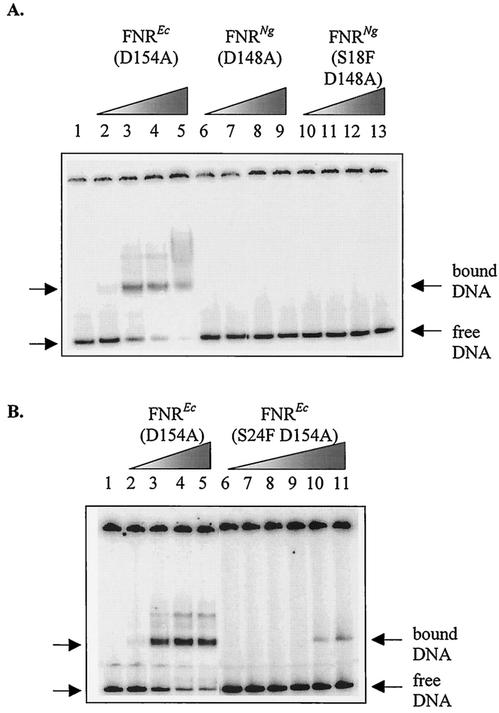
Inability of the S18F or D148A substitutions to promote binding of gonococcal FNR to model FNR-dependent promoters in vitro.
The ability of the purified gonococcal FNR proteins to bind to the FF(−41.5) promoter was assessed by gel retardation assays. E. coli FNR (D154A) was included as a positive control. Known concentrations, between 0.05 μM and 3 μM, of purified FNREc (D154A), FNRNg (D148A) and FNRNg (S18F D148A) protein were incubated with terminally 32P-labeled FF(−41.5) promoter DNA and separated by polyacrylamide gel electrophoresis (Fig. 6A). In the absence of FNR, the promoter DNA migrated to the bottom of the gel, but was retarded by the E. coli FNR (D154A) at all concentrations tested. In the presence of up to 3 μM of the gonococcal FNR proteins, the FF(−41.5) promoter DNA was not retarded during electrophoresis; the D148A and S18F D148A substituted gonococcal FNR proteins did not bind to the promoter DNA. Higher concentrations of gonococcal FNR (7.8 μM for the D148A derivative, 9.6 μM in the case of the S18F D148A derivative) also did not bind to the promoter DNA (data not shown). The inability of either substituted gonococcal FNR proteins to bind promoter DNA in vitro signifies that the D148A substitution does not, either alone or in combination with S18F, allow FNRNg to dimerize aerobically in the absence of an iron-sulfur center. This is in direct contrast to the corresponding D154A substitution in E. coli FNR, which allows aerobic DNA binding and transcription activation in vitro (19). Consequently, further biochemical characterization of the gonococcal FNR in vitro will require the anaerobic reconstitution of the iron-sulfur center for each of the substituted forms of the protein.
FIG. 6.
Gel retardation assays to study the effects of the S18F and S24F substitutions alone or in combination with D148A or D154A substitutions on binding of gonococcal or E. coli FNR to DNA in vitro. End labeled FF(−41.5) promoter DNA was incubated with various concentrations of FNR proteins. (A) The concentration of FNR protein in each reaction was as follows: lane 1, no protein; lanes 2, 6 and 10, 0.05 μM; lanes 3, 7 and 11, 0.1 μM; lanes 4, 8 and 12, 1 μM; and lanes 5, 9 and 13, 3 μM. (B) The concentration of FNR protein in each reaction mixture was as follows: lane 1, no protein; lane 2, 0.05 μM; lane 3, 0.1 μM; lane 4, 0.5 μM; lane 5, 1 μM; lane 6, 0.025 μM; lane 7, 0.05 μM; lane 8, 0.25 μM; lane 9, 0.5 μM; lane 10, 1.5 μM; lane 11, 2.1 μM.
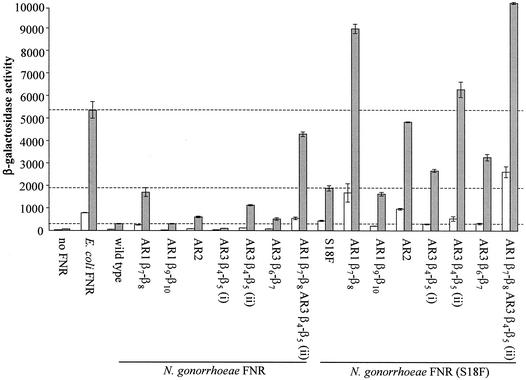
Substitution of parts of the ARs of gonococcal FNR with the corresponding residues of the E. coli FNR protein.
Substitutions in the three ARs have led to the identification of residues that interact with the α and σ subunits of RNA polymerase (5). At two patches of AR1, one patch of AR2, and three patches of AR3, residues of the N. gonorrhoeae FNR protein were substituted for the corresponding residues of the E. coli FNR protein (Fig. 1). QuikChange site directed mutagenesis (Stratagene) was used, with the oligonucleotide primers listed in Table 2, to introduce the AR substitutions into the N. gonorrhoeae fnr gene on pGCFNR3.
Five AR-substituted FNRNg proteins activated transcription to a greater extent than the wild-type FNRNg protein at the FF(−41.5) promoter (Fig. 7). Substitutions in AR1, AR2, and AR3 increased the ability of FNRNg to activate transcription at the FF(−41.5) promoter. These substitutions did not significantly alter the ratio of aerobic to anaerobic activation, implying that they did not alter the oxygen-sensing iron-sulfur center. Two AR-substituted FNRNg proteins did not activate transcription to a greater extent than the wild-type protein: the β9-β10 substitution in AR1 did not increase the activity of FNRNg, whereas the β4-β5 (i) substitution in AR3 decreased activity. When combined, the two substitutions that most increased the activity of FNRNg, AR1 β7-β8 and AR3 β4-β5 (ii), further enhanced transcription activation by FNRNg.
FIG. 7.
Effects of AR substitutions on transcription activation by the gonococcal FNR at the FF(−41.5) promoter. Mutations detailed in Fig. 1 were introduced into the gonococcal fnr gene on plasmid pGCFNR3 using the QuikChange method. The mutated plasmids were transformed into strain JRG1728 carrying the consensus FNR-dependent promoter-lacZ fusion FF(−41.5) integrated into the phage λ attachment site. Open bars: aerobic cultures; hatched bars, anaerobic cultures. The upper horizontal line marks transcription activity due to E. coli FNR during anaerobic growth; the lower and middle horizontal lines mark transcription activity due to wild-type and S18F substituted gonococcal FNR during anaerobic growth, respectively. Other details are as explained in the legend to Fig. 3.
The AR substitutions were also combined with the S18F substitution. At the FF(−41.5) promoter, six AR-substituted FNRNg (S18F) proteins activated transcription to a greater extent than the FNRNg (S18F) protein (Fig. 7). As in the wild-type background, the β9-β10 substitution of AR1 did not increase the ability of the FNRNg (S18F) protein to activate transcription. The β4-β5 (i) substitution increased transcription activation by FNRNg (S18F), but not the wild-type protein, suggesting that the S18F substitution causes this patch of AR3 to be better aligned so as to make better contacts with E. coli σ70.
The ability of the AR-substituted FNRNg proteins to activate transcription from the class I FNR-dependent promoter, FF(−71.5), was also assessed (data not shown). Only two AR-substituted FNRNg proteins, both substituted at the β7-β8 loop of AR1, activated transcription to a greater extent than the wild-type protein. Substitutions in AR2 or AR3 did not increase transcription activation by FNRNg at pFF(−71.5) because AR2 and AR3 do not make contacts with RNAP during class I FNR-dependent transcription activation.
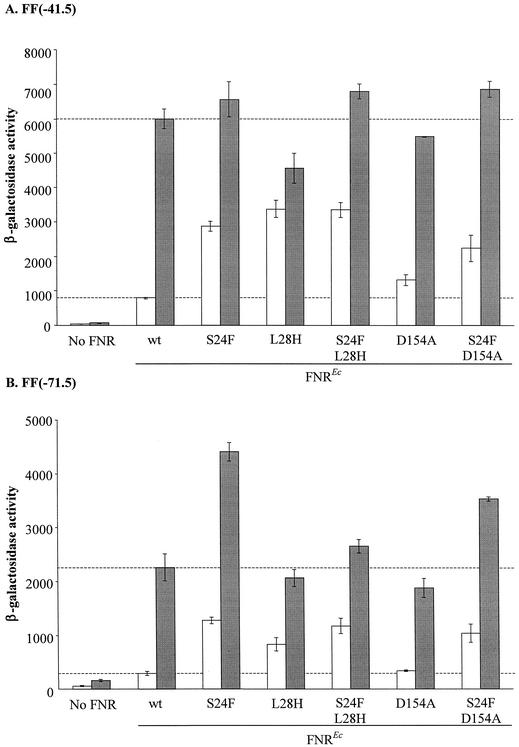
Effects of an S24F mutation, corresponding to the S18F change in the gonococcal FNR, on transcription activation by the E. coli FNR protein.
Mutations that increase the ability of E. coli FNR to activate transcription during aerobic growth have been isolated in several laboratories, and many of these mutations map to codons for residues adjacent to the cysteines essential for binding the iron-sulfur center (17). However, in none of these previous studies has an S24F substitution been identified.
As expected from many previous studies, introduction of the substitution D154A into the E. coli FNR protein resulted in a twofold increase in transcription activation at the class II FF(−41.5) promoter during aerobic growth, but slightly decreased activity during anaerobic growth (Fig. 8A). Similarly, introduction of the L28H substitution increased transcription activation during aerobic growth, but the level of anaerobic transcription was slightly lower than that of the wild-type FNR. By contrast, an S24F substitution increased aerobic transcription activation 3.5-fold to a level comparable with FNR (L28H), but had no effect on anaerobic transcription activation (Fig. 8A). Enhanced transcription activation relative to the wild type was also found with the S24F L28H and S24F D154A forms of FNR, especially during aerobic growth. Thus, at the class II FNR-dependent promoter, effects of previously described substitutions were enhanced by the additional S24F substitution.
FIG. 8.
Effects of an S24F substitution, corresponding to S18F in the gonococcal FNR, on transcription activation by the E. coli FNR protein. Mutations were introduced into the E. coli fnr gene on plasmid pFNR using the QuikChange method. The mutated plasmids were transformed into strain JRG1728 carrying the consensus FNR-dependent promoter-lacZ fusions FF(−41.5) (A) or FF(−71.5) (B) integrated into the phage λ attachment site. The upper horizontal line marks transcription activity due to the unsubstituted E. coli FNR during anaerobic growth (hatched bars); the lower line shows the corresponding activity for the aerobic culture (open bars). Other details are as explained in the legend to Fig. 3.
At the class I promoter, the S24F substitution alone caused a fivefold increase in transcription activation during aerobic growth and a twofold increase during anaerobic growth (Fig. 8B). Under both growth conditions, these rates were appreciably above those observed with the L28H or D154A substituted E. coli FNR. In combination with the S24F substitution, rates of transcription with the S24F L28H and S24F D154A forms of FNR were similar during aerobic growth, but decreased during anaerobic growth, compared with the S24F substitution alone. This again suggests that the S24F substitution activates transcription by a mechanism distinct from that induced by either L28H or D154A.
The effect of the S24F substitution on the DNA binding properties of E. coli FNR (D154A) in vitro was also assessed. Radiolabeled FF(−41.5) promoter DNA was incubated with FNREc (D154A) or FNREc (S24F D154A) and separated by polyacrylamide gel electrophoresis (Fig. 6B). As in the previous experiment, when FNREc (D154A) was present at concentrations between 0.05 and 1 μM, it bound to and retarded the migration of the promoter fragment. The doubly substituted FNREc (S24F D154A) protein also bound to and retarded the DNA fragment, but only when present at concentrations of 1.5 and 2.1 μM.
Effects of the S18F and S24F substitutions on transcription repression by the gonococcal and E. coli FNR proteins.
E. coli FNR represses transcription from different promoters by two distinct mechanisms. The synthetic model promoter, FFgalΔ4, is a derivative of the promoter of the E. coli galactose operon in which there is an engineered consensus FNR-binding site located between the −10 and −35 elements (33). At this promoter, binding of FNR prevents access of RNA polymerase to its binding sites and repression is dependent upon DNA binding by FNR. In contrast, at the natural E. coli ndh promoter, FNR binds as a dimer during anaerobic growth (but not during aerobic growth) to two sites, centered at −50.5 and −94.5, forming a repression loop that prevents transcription initiation by RNA polymerase (10). The two FNR dimers interact with each other via the repression patch, which is a surface-exposed region on the same face of FNR as AR1 (12). Thus, unlike at the FFgalΔ4 promoter, repression is dependent upon both DNA binding and FNR dimer-dimer interactions via the repression patch. The effects of the S18F and S24F substitutions on transcription from each of these promoters were determined using strains in which these two promoters were coupled to a lacZ reporter gene.
At both the ndh and FFgalΔ4 promoters, β-galactosidase expression during anaerobic growth was lower in the presence of either the gonococcal or E. coli FNR than in their absence, confirming that both proteins bind to repress transcription (Table 3). However, higher expression was observed in the presence of either the unsubstituted or the S18F substituted gonococcal FNR than with E. coli FNR, indicating that both forms of the gonococcal FNR bind less tightly than their E. coli counterparts. The S18F and S24F derivatives of gonococcal and E. coli FNR repressed transcription at pndh during anaerobic growth to a greater extent than the wild-type proteins (Table 3). Although during aerobic growth neither the gonococcal nor the wild-type E. coli FNR affected expression from the ndh promoter, the S24F substitution resulted in decreased expression, indicating that some FNR-binding occurred even during aerobic growth.
TABLE 3.
Effects of the S18F and S24F substitutions in gonococcal and E. coli FNR on repression at the FFgalΔ4 and ndh promoters
| Promoter | FNR proteina | Mean β-galactosidase activity ± SD under growth conditionb
|
Ratioc | |
|---|---|---|---|---|
| Aerobic | Anaerobic | |||
| pndh | None | 1,950 ± 250 | 6,250 ± 350 | |
| FNREc | 2,020 ± 300 | 310 ± 15 | 20 | |
| FNREc (S24F) | 940 ± 90 | 160 ± 25 | 39 | |
| FNRNg | 2,220 ± 160 | 3,150 ± 210 | 2 | |
| FNRNg (S18F) | 2,220 ± 80 | 2,150 ± 115 | 3 | |
| pFFgalΔ4 | None | 1,035 ± 21 | ||
| FNREc | 14 ± 1 | 75 | ||
| FNREc (S24F) | 18 ± 1 | 58 | ||
| FNRNg | 45 ± 2 | 23 | ||
| FNRNg (S18F) | 35 ± 4 | 29 | ||
Superscripts Ec and Ng refer to FNR from E. coli and N. gonorrhoeae.
Each transformant was assayed in duplicate from two independent cultures; units of activity are nanomoles of ONPG hydrolyzed · min−1·(milligram of bacteria [dry mass])−1.
Ratio of the anaerobic activity in the absence of FNR to that in the presence of the FNR protein specified.
Similar data were obtained with the FFgalΔ4 promoter. During anaerobic growth, expression was lower in the presence of either the gonococcal or E. coli FNR. However, at this promoter, the S18F substitution in the gonococcal FNR resulted in a slight increase in repression, but the S24F substitution in the E. coli FNR resulted in less severe repression than observed with the unsubstituted protein.
DISCUSSION
Expression of the gonococcal FNR in E. coli.
Previous studies in this laboratory with the gonococcal fnr gene were based upon the DNA sequence in the Oklahoma Gonococcal DNA Database (URL http://www.genome.ou.edu/gono.html), which predicted the gonococcal FNR protein to be 12 amino acids longer than its E. coli counterpart (21). Subsequently, however, fnr homologues have been identified in the published sequences of the genomes of two meningococcal strains. Despite overall similarities of 97% between FNR from meningococci and gonococci, the meningococcal counterparts lack the extra 12 N-terminal amino acids. Immediately downstream of the previously assumed start codon for translation of the gonococcal FNR sequence is a second in-frame ATG that coincides with the translation start codon of the meningococcal FNR, and is preceded by a plausible ribosome binding site. This alternative start codon was used to design plasmid pGCFNR3, from which gonococcal FNR protein, and the derivatives generated in this study, were shown by Western blotting to be expressed at levels similar to the E. coli FNR (Fig. 2 and data not shown). Furthermore, antisera to E. coli FNR detected proteins of identical size expressed in N. gonorrhoeae strain RUG7001, in E. coli JRG1728 transformed with pGCFNR3, and in wild-type E. coli (Fig. 2). This confirmed that the downstream AUG codon is the translation start for gonococcal FNR, and that pFNR is a suitable vector for studying gonococcal FNR in E. coli.
Although many key features of E. coli FNR, including all four cysteine residues that bind the [4Fe-4S] iron-sulfur center, are conserved in the gonococcal FNR, the ARs differ considerably between the two proteins (Fig. 1). For example, only 8 of the 19 residues that form AR1 are identical in the two proteins, although six of the 11 differences are conservative substitutions. Even greater differences are seen in AR3, where only three residues are identical and four of the differences are conservative substitutions. In view of these differences, it was not surprising that the gonococcal FNR activates transcription poorly in E. coli. Despite this, comparison of the sequences of the gonococcal and E. coli rpoA and rpoD genes, encoding the α and σ70 subunits of RNA polymerase, respectively, reveals that there are only minor differences in the residues thought to contact FNR. This might indicate that interactions between the gonococcal FNR and RNAP are somewhat different to their E. coli counterparts.
It was anticipated that the inability of the gonococcal FNR to activate transcription in E. coli is due to its inability to interact productively with the host RNA polymerase, and that mutagenesis of the ARs would be required for the gonococcal FNR to function in E. coli. The isolation of the S18F substitution was therefore unexpected and surprising for several reasons. First, although about 80,000 colonies were screened, only four contained active FNR derivatives, and in each case, the altered FNR carried the S18F mutation. Secondly, the S18F variant of the gonococcal FNR activated transcription similarly at both class I and class II FNR-dependent promoters, suggesting that the S18F mutation improves FNR activity in a nonspecific way. Thirdly, significant rates of transcription were detected during aerobic as well as during anaerobic growth. Note that the above statements assume that the steady state levels of β-galactosidase activity reflect rates of transcription initiation at the promoter assayed. Most of the β-galactosidase activities detected were less than those dependent upon the E. coli FNR protein, so this assumption is likely to be valid.
Mechanism of transcription activation by the S18F derivative of gonococcal FNR.
Superficially, the S18F variant of the gonococcal FNR is similar to the D154A substituted form of the E. coli FNR, notably in the increased rate of transcription during aerobic growth. The primary effect of the D154A substitution is to stabilize dimerization of the FNR monomers, thereby increasing the affinity of FNR for its binding sites. Several lines of evidence indicate that this is not the primary effect of the S18F substitution in the gonococcal FNR, or of the corresponding S24F substitution in the E. coli FNR. First, the D154A substitution in the E. coli FNR increased transcription activation independently of the S24F substitution: the effects of the D154A and S24F substitutions are essentially additive. Secondly, the D154A substitution in E. coli FNR (and the D148A equivalent in gonococcal FNR) decreased the rate of transcription during anaerobic growth and hence decreased the anaerobic to aerobic induction ratio. In contrast, the S24F substitution in E. coli FNR (and the S18F equivalent in gonococcal FNR) increased transcription activation both aerobically and anaerobically. Finally, in vitro gel retardation assays failed to reveal increased affinity of purified gonococcal FNR S18F for its binding sites. We cannot explain why the corresponding S24F substitution in E. coli FNR decreased promoter binding.
Similar arguments indicate that the S18F substitution in gonococcal FNR is unlikely to realign contacts between the ARs and E. coli RNA polymerase. Thus, modifications to all three of the ARs of gonococcal FNR to make them more like E. coli FNR resulted in increased transcription activation, but when combined with the S18F substitution, the effects were again essentially additive, indicating that they modify the gonococcal FNR in different ways.
The mechanism by which the S18F substitution increases transcription activation by gonococcal FNR remains unknown. However, we envisage two possibilities. One is that the S18F substitution in gonococcal FNR improves or creates a direct contact with RNA polymerase that activates transcription. Although this region of FNR has not been previously implicated in direct contacts with RNA polymerase, this would be consistent with the phenotype conferred by the S18F substitution, which is similar to that of the AR1 β7-β8 substituted protein (Fig. 7). An alternative possibility is that the S18F substitution stabilizes the FNR iron-sulfur center and that this results in enhanced dimerization, DNA binding and transcription activation. Thus, the S18F substitution would be a general gain of function mutation, increasing transcription activation both aerobically and anaerobically. The location of the S18F substitution argues strongly for this second possibility. Such a mechanism would be consistent with chemical studies, which have established that Fe-S clusters synthesized in the absence of protein are extremely oxygen-labile, but can be stabilized by association with protein (14). In particular, adjacent hydrophobic residues can stabilize iron-sulfur centers in model peptides (26). Hence, the FNR iron-sulfur center may be stabilized by aromatic amino acids located near to the key cysteine residues (cited by Bates et al. [1] in the context of their studies of the L28H substitution of the E. coli FNR). Interestingly, replacement of an aromatic residue close to the Fe-S center of the high potential iron-sulfur protein, HiPIP, increased its accessibility to oxygen and subsequent degradation (28). Thus, our observation that the S18F and S18Y substitutions were the most effective in increasing transcription activation by gonococcal FNR, argues for a model in which changes at residue 18 in gonococcal FNR (and the corresponding residue 24 in E. coli FNR) exert their effects by modulating the stability of the FNR iron-sulfur cluster.
Acknowledgments
Financial support from the UK Biotechnology and Biological Sciences Research Council in the form of project grant 6/PRS12198 and a research studentship to T.O. is gratefully acknowledged. DNA was sequenced at the Birmingham Functional Genomics Laboratory, supported by BBSRC grant 6/JIF13209.
We thank A. Barnard for a gift of purified FNREc (D154A) protein and J. Green for a gift of anti-FNR antisera. We are also grateful to two referees for their perceptive reviews of the manuscript and their constructive comments concerning the possible mechanism by which the S18F substitution increases transcription activation.
REFERENCES
- 1.Bates, D. M., C. V. Popescu, N. Khoroshilova, K. Vogt, H. Beinert, E. Münck, and P. J. Kiley. 2000. Substitution of leucine 28 with a histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S]2+ cluster to oxygen. J. Biol. Chem. 275:6234-6240. [DOI] [PubMed] [Google Scholar]
- 2.Bell, A., K. L. Gaston, J. A. Cole, and S. J. W. Busby. 1989. Cloning of binding sequences for the Escherichia coli transcription activators, FNR and CRP: location of bases involved in discrimination between FNR and CRP. Nucleic Acids Res. 17:3865-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake, T., A. Barnard, S. J. Busby, and J. Green. 2002. Transcription activation by FNR: evidence for a functional activating region 2. J. Bacteriol. 184:5855-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Browning, D. F., C. M. Beatty, A. J. Wolfe, J. A. Cole, and S. J. Busby. 2002. Independent regulation of the divergent Escherichia coli nrfA and acsP1 promoters by a nucleoprotein assembly at a shared regulatory region. Mol. Microbiol. 43:687-701. [DOI] [PubMed] [Google Scholar]
- 5.Browning, D. F., D. Lee, J. Green, and S. J. Busby. 2002. Secrets of bacterial transcription initiation taught by the Escherichia coli FNR protein, p. 127-142. In D. A. Hodgson and C. M. Thomas (ed.), Signals, switches, regulons, and cascades: control of bacterial gene expression. Proceedings of the 150th ordinary meeting. Society for General Microbiology, Reading, United Kingdom.
- 6.Busby, S. J. W., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 7.Clark, V. L., L. A. Campbell, D. A. Palermo, T. M. Evans, and K. W. Klimpel. 1987. Induction and repression of outer membrane proteins by anaerobic growth of Neisseria gonorrhoeae. Infect. Immun. 55:1359-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark, V. L., J. S. Knapp, S. Thompson, and K. W. Klimpel. 1988. Presence of antibodies to the major anaerobically induced gonococcal outer membrane protein in sera from patients with gonococcal infections. Microb. Pathog. 5:381-390. [DOI] [PubMed] [Google Scholar]
- 9.Green, J., A. D. Sharrocks, B. Green, M. Geisow, and J. R. Guest. 1993. Properties of FNR proteins substituted at each of the five cysteine residues. Mol. Microbiol. 8:61-68. [DOI] [PubMed] [Google Scholar]
- 10.Green, J., and J. R. Guest. 1994. Regulation of transcription at the ndh promoter of Escherichia coli by FNR and novel factors. Mol. Microbiol. 12:433-444. [DOI] [PubMed] [Google Scholar]
- 11.Green, J., B. Bennett, P. A. Jordan, E. T. Ralph, A. J. Thomson, and J. R. Guest. 1996. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem. J. 316:887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green, J., and F. A. Marshall. 1999. Identification of a surface of FNR overlapping activating region I that is required for repression of gene expression. J. Biol. Chem. 274:10244-10248. [DOI] [PubMed] [Google Scholar]
- 13.Householder, T. C., W. A. Belli, S. Lissenden, J. A. Cole, and V. Clark. 1999. cis- and trans-acting elements involved in regulation of aniA, the gene encoding the major anaerobically induced outer membrane protein in Neisseria gonorrhoeae. J. Bacteriol. 181:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibers, J. A., and R. H. Holm. 1980. Modeling coordination sites in metallobiomolecules. Science 209:223-235. [DOI] [PubMed] [Google Scholar]
- 15.Jayaraman, P.-S., T. Peakman, S. J. W. Busby, R. Quincey, and J. A. Cole. 1987. Location and sequence of the promoter of the gene for the NADH-dependent nitrite reductase of Escherichia coli and its regulation by oxygen, the FNR protein and nitrite. J. Mol. Biol. 196:781-788. [DOI] [PubMed] [Google Scholar]
- 16.Khoroshilova, N., C. V. Popescu, E. Münck, H. Beinert, and P. J. Kiley. 1997. Iron-sulphur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc. Nat. Acad. Sci. USA 94:6087-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiley, P. J., and W. S. Reznikoff. 1991. Fnr mutants that activate gene expression in the presence of oxygen. J. Bacteriol. 173:16-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knapp, J. S., and V. Clark. 1984. Anaerobic growth of Neisseria gonorrhoeae coupled to nitrite reduction. Infect. Immun. 46:176-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazazzera, B. A., D. M. Bates, and P. J. Kiley. 1993. The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev. 7:1993-2005. [DOI] [PubMed] [Google Scholar]
- 20.Lee, D. J., H. J. Wing, N. J. Savery, and S. J. W. Busby. 2000. Analysis of interactions between activating region 1 of Escherichia coli FNR protein and the C-terminal domain of the RNA polymerase α subunit: use of alanine scanning and suppression genetics. Mol. Microbiol. 37:1032-1040. [DOI] [PubMed] [Google Scholar]
- 21.Lissenden, S., S. Mohan, T. Overton, T. Regan, H. Crooke, J. A. Cardinale, T. C. Householder, P. Adams, C. D. O'Connor, V. Clark, H. Smith, and J. A. Cole. 2000. Identification of transcription activators which regulate gonococcal adaptation from aerobic to anaerobic or oxygen-limited growth. Mol. Microbiol. 37:839-855. [DOI] [PubMed] [Google Scholar]
- 22.Lodge, J., J. Fear, S. J. W. Busby, P. Gunasekaren, and N. R. Kamini. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 75:271-276. [DOI] [PubMed] [Google Scholar]
- 23.Macdonald, H., and J. Cole. 1985. Molecular cloning and functional analysis of the cysG and nirB genes of Escherichia coli K12, two closely-linked genes required for NADH-dependent nitrite reductase activity. Mol. Gen. Genet. 200:328-334. [DOI] [PubMed] [Google Scholar]
- 24.Mellies, J., J. Jose, and T. F. Meyer. 1997. The Neisseria gonorrhoeae gene aniA encodes an inducible nitrite reductase. Mol. Gen. Genet. 256:525-532. [DOI] [PubMed] [Google Scholar]
- 25.Morse, S. A., and L. Bartenstein. 1974. Factors affecting autolysis of Neisseria gonorrhoeae. Proc. Soc. Exp. Biol. Med. 145:1418-1421. [DOI] [PubMed] [Google Scholar]
- 26.Mulholland, S. E., B. R. Gibney, F. Rabanal, and P. L. Dutton. 1999. Determination of nonligand amino acids critical to [4Fe-4S]2+/+ assembly in ferredoxin maquettes. Biochemistry 38:10442-10448. [DOI] [PubMed] [Google Scholar]
- 27.Silver, L. E., and V. L. Clark. 1995. Construction of a translational lacZ fusion system to study gene regulation in Neisseria gonorrhoeae. Gene 166:101-104. [DOI] [PubMed] [Google Scholar]
- 28.Soriano, A., D. Li, S. Bian, A. Agarwal, and J. A. Cowan. 1996. Factors influencing redox thermodynamics and electron self-exchange for the [Fe4S4] cluster in Chromatium vinosum high potential iron protein: the role of core aromatic residues in defining cluster redox chemistry. Biochemistry 35:12479-12486. [DOI] [PubMed] [Google Scholar]
- 29.Spiro, S., and J. R. Guest. 1987. Regulation and overexpression of the fnr gene of Escherichia coli. J. Gen. Microbiol. 133:3279-3288. [DOI] [PubMed] [Google Scholar]
- 30.Spiro, S., R. E. Roberts, and J. R. Guest. 1989. FNR-dependent repression of the ndh gene of Escherichia coli and metal ion requirement for FNR-regulated gene expression. Mol. Microbiol. 3:601-608. [DOI] [PubMed] [Google Scholar]
- 31.Williams, R., A. Bell, G. Sims, and S. J. W. Busby. 1991. The role of two surface exposed loops in transcription activation by the Escherichia coli CRP and FNR proteins. Nucleic Acids Res. 19:6705-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams, S. M., N. J. Savery, S. J. W. Busby, and H. J. Wing. 1997. Transcription activation at the class I FNR-dependent promoter: identification of the activating surface of FNR and the corresponding contact site in the C-terminal domain of the RNA polymerase α subunit. Nucleic Acids Res. 25:4028-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams, S. M., H. J. Wing, and S. J. W. Busby. 1998. Repression of transcription initiation by Escherichia coli FNR protein: repression by FNR can be simple. FEMS Microbiol. Lett. 163:203-208. [DOI] [PubMed] [Google Scholar]
- 34.Wing, H. J., S. M. Williams, and S. J. W. Busby. 1995. Spacing requirements for transcription activation by Escherichia coli FNR protein. J. Bacteriol. 177:6704-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wing, H. J., J. Green, J. R. Guest, and S. J. W. Busby. 2000. Role of activating region 1 of Escherichia coli FNR protein in transcription activation at class II promoters. J. Biol. Chem. 275:29061-29065. [DOI] [PubMed] [Google Scholar]