Abstract
All Escherichia coli strains so far examined possess a chromosomally encoded nanATEK-yhcH operon for the catabolism of sialic acids. These unique nine-carbon sugars are synthesized primarily by higher eukaryotes and can be used as carbon, nitrogen, and energy sources by a variety of microbial pathogens or commensals. The gene nanR, located immediately upstream of the operon, encodes a protein of the FadR/GntR family that represses nan expression in trans. S1 analysis identified the nan transcriptional start, and DNA footprint analysis showed that NanR binds to a region of ∼30 bp covering the promoter region. Native (nondenaturing) polyacrylamide gel electrophoresis, mass spectrometry, and chemical cross-linking indicated that NanR forms homodimers in solution. The region protected by NanR contains three tandem repeats of the hexameric sequence GGTATA. Gel shift analysis with purified hexahistidine-tagged or native NanR detected three retarded complexes, suggesting that NanR binds sequentially to the three repeats. Artificial operators carrying different numbers of repeats formed the corresponding number of complexes. Among the sugars tested that were predicted to be products of the nan-encoded system, only the exogenous addition of sialic acid resulted in the dramatic induction of a chromosomal nanA-lacZ fusion or displaced NanR from its operator in vitro. Titration of NanR by the nan promoter region or artificial operators carrying different numbers of the GGTATA repeat on plasmids in this fusion strain supported the binding of the regulator to target DNA in vivo. Together, the results indicate that GGTATA is important for NanR binding, but the precise mechanism remains to be determined.
The sialic acids are a unique family of >40 natural derivatives of the nine-carbon monosaccharide N-acetylneuraminic acid (Neu5Ac) that function in mammalian and avian immune system regulation, other cell-cell and cell-molecule signaling events, and central nervous system development (31). Despite the recent identification of sialate biosynthesis in certain insect larvae and some fungal pathogens (2), plants, most protists, Archaea, and most eubacteria do not synthesize sialic acids. Although a few pathogenic bacteria have acquired the ability to synthesize sialic acid, this capability is generally confined to higher eukaryotes. In contrast to the limited phylogenetic distribution of sialate biosynthesis, many bacteria use sialic acids as sources of carbon, nitrogen, and energy or as sources of amino sugars for cell wall and membrane components, and some decorate their surfaces with sialic acid as a mechanism for avoiding host innate immunity. Sialic acid metabolism thus is central to a variety of host-microbe interactions (36). Utilization of sialic acid in Escherichia coli K-12 requires the nanATEK operon (26). Here, we show that expression of the operon is controlled by a repressor protein encoded by the upstream gene nanR. These results provide the first characterization of a sialate transcriptional regulatory system in any organism and establish an experimental scaffold for understanding the functions of sialate metabolism in a variety of host-pathogen interactions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Unless otherwise indicated, all bacterial strains were grown in Luria-Bertani medium (Fisher, Pittsburgh, Pa.) with vigorous aeration at 37°C or on plates solidified with 1.5% agar. Terrific broth was purchased from MoBio (Solana Beach, Calif.). E. coli strain BL21(DE3) Star (Invitrogen, Carlsbad, Calif.) was used for T7 polymerase-dependent expression of nanR cloned as a C-terminal histidine-tagged fusion in pET21c (Novagen, Madison, Wis.). Strains JM101 (wild type), IBPC1016 (nanR6; previously ama-6), and IBPC1017 (nanR7; previously ama-7) have been described (26). The nanR7 mutation was introduced into JM101 carrying the nanA-lacZ fusion using Tn10cam11 (26). Artificial operators containing various numbers of GGTATA repeats were cloned into the pGEM-T Easy vector (Promega Corp., Madison, Wis.) and designated pGEM-1 through pGEM-6 to indicate the number of GGTATA repeats carried within each vector. All plasmids were maintained by the inclusion of ampicillin in the media to a final concentration of 100 μg/ml. Unless otherwise indicated, all sugars were in their d conformations and were purchased from Sigma Chemical Co. (St. Louis, Mo.). Purified catabolite activator protein (CAP) was the kind gift of Annie Kolb.
Construction of the nanA-lacZ translational fusion.
A 438-bp PCR fragment was amplified with oligonucleotides Nan1 and Nan2 (26), generating a DNA fragment including 232 bp with the TAA stop codon from the 3′ end of nanR, the 121-bp nanR-nanA intergenic region, and 85 bp of nanA that was inserted into the SmaI site of the fusion vector pRS/NM482 (pRS414 with its cloning region replaced by the in-phase codons of pNM482) to produce a translational nanA-lacZ fusion. The fusion gene was transferred to bacteriophage lambda by in vivo recombination as described previously (33) and used to lysogenize JM101 and the nanR6 derivative IBPC1016. A DNA fragment covering the nanA insert and lacZ fusion junction was PCR amplified from chromosomal DNA of a lysogen by using oligonucleotide Nan2 and a primer specific for the lacZ gene. The sequence of the insert and nanA-lacZ junction was verified using the Amersham Thermosequenase kit.
Purification of NanR.
The nanR (yhcK) open reading frame (ORF) was engineered with 5′ NdeI and 3′ BamHI cleavage sites using primers 5′-GGGAATTCCATATGGGCCTTATGAACGCATTTG-3′ and 5′-CGCGGATCCCGTTTCTTTTTGTTGGTGGTCTG-3′, respectively (restriction sites are underlined). Amplicons generated by PCR from the pTZ(Nan1-3) NanR-overproducing plasmid (26) were digested and cloned into the pET21c vector, fusing the ORF to a C-terminal hexahistidine tag. This plasmid was transformed into strain BL21(DE3) Star, and the culture was induced at an A600 of 0.5 with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The cells were harvested after 2 h of further incubation, and the pellets were frozen overnight at −20°C. Cell lysis was accomplished by thawing the pellets and sonicating them in 50 mM Tris (pH 8.0)-500 mM NaCl- 15 mM imidazole. Hexahistidine-tagged NanR (NanR-His6) was purified to electrophoretic homogeneity by nickel-nitrilotriacetic acid chromatography (Qiagen, Valencia, Calif.) as described by the manufacturer. Cloning was carried out by C&P Biotech, Thornhill, Ontario, Canada.
To construct a plasmid that would permit overproduction of native NanR, nanR was amplified using a forward (5′-ATGAACGCATTTGATTCGCAAACCG-3′) and reverse (5′-GCCGGCCCCGGGTTATTTCTTTTTGTTGGTGGTCTGACC-3′) primer pair (the engineered XmaI site is underlined). The pBAD24 (14) expression vector was digested with NcoI, blunt ended, and then digested a second time with XmaI. The purified amplicon was ligated to the doubly digested pBAD24 vector, and the transformed plasmids expressed in strain JM109 were screened for overproduction of NanR by polyacrylamide gel electrophoresis after induction with 0.2% l-arabinose. One construct was sequenced by the W. M. Keck Center for Comparative and Functional Genomics, University of Illinois, to verify insertion of the wild-type nanR gene and saved as pSX675. To purify native NanR, JM109 harboring pSX675 was grown in 1 liter of Terrific broth to an A600 of 0.4 and induced with 0.2% l-arabinose for 3 h with vigorous aeration. The cell pellet was resuspended in binding buffer that contained 20 mM Tris-HCl, pH 8.0, 1 mM EDTA, 5% glycerol, 1 mM dithiothreitol, 0.01% Triton X-100, and 100 mM NaCl. The cells were disrupted by sonication, and particulate material was removed by centrifugation at 50,000 × g in a TLA 100.5 rotor of a Beckman TL-100 ultracentrifuge.
Target DNA (246 bp), including 39 bp to the end of nanR with its terminating codon, 86 bp to the beginning of the translational start site of nanA, and the entire 121-bp intergenic region, was amplified with a forward (5′-CGACGCAGACTCGCTTTATC-3′) and reverse (biotin-labeled 5′-GGCACGCTTTCGGTCAGACC-3′) primer pair. The soluble protein extract containing overproduced native NanR was incubated for 10 min at room temperature with the biotinylated DNA, and the protein-DNA complexes were subsequently bound to μMACS Streptavidin MicroBeads (Miltenyi Biotec, Auburn, Calif.). The iron filings contained within a μ column trapped the paramagnetic complexes, and the pure native NanR was eluted with high salt after the column was stringently washed to remove unbound polypeptides according to the manufacturer's instructions. N-terminal amino acid sequencing of the purified native NanR was carried out by the Protein Services Facility, University of Illinois, using automated Edman chemistry.
Determination of NanR molecular weight.
Determination of the NanR-His6 molecular weight was carried out under nondenaturing polyacrylamide gel electrophoresis conditions essentially as described by Ferguson (11). Nondenatured protein molecular weight markers were purchased from Sigma, and the analysis was conducted according to Sigma Technical Bulletin MKR-137.
To determine the molecular mass of NanR-His6 by gel permeation chromatography, a column containing Sephacryl S-100 was equilibrated in 10 mM Tris buffer, pH 8.0, containing 100 mM NaCl and used to obtain a standard curve from the elution volumes of RNase A (13.7 kDa), chymotrypsin A (25 kDa), ovalbumin (43 kDa), and bovine serum albumin (66 kDa). The void volume of the column was determined using Blue Dextran. Matrix-assisted laser desorption-ionization- time of flight (MALDI-TOF) spectrometry was performed with 100 μg of NanR-His6 on an Applied Biosciences Voyager System 4066 mass spectrometer equipped with a nitrogen laser operating at 337 nm for desorption-ionization at an accelerating voltage of 25 kV and a grid voltage at 90 to 93% of the acceleration. The analysis was performed by the Mass Spectrometry Facility, University of Illinois. Cross-linking was initiated by the addition of ethylene glyco-bis succinimidylsuccinate (Pierce Chemical Co., Rockford, Ill.) in dimethyl sulfoxide to a final concentration of 10 mM, with 2.5 μg of purified native or hexahistidine-tagged NanR in a total volume of 20 μl. The reaction mixtures were incubated at room temperature for 1 h and quenched by adding 0.1 M glycine (final concentration) prior to analysis by denaturing polyacrylamide gel electrophoresis.
β-Galactosidase assay.
Cells were usually grown in E minimal salts medium (9) with 0.4% glycerol and 0.5% Casamino Acids (NZ amine; Sigma). β-Galactosidase activity was measured as described previously (21), using 0.1% sodium dodecyl sulfate and 2 drops of chloroform for cell permeabilization. Data from at least two independent experiments are presented in Miller units.
S1 nuclease protection and DNase I footprint analysis.
Total RNA was purified by the hot-phenol method, and S1 nuclease transcript mapping was carried out as previously described (25). The probe used was the 438-bp Nan1-2 PCR fragment in which the Nan1 oligonucleotide had been labeled with [32P]ATP and polynucleotide kinase. DNase I footprint analysis was carried out as described previously (25) using the same probe as for the S1 experiment.
Mobility gel shift analysis.
The 246-bp fragment described above was labeled at both ends with [32P]ATP and polynucleotide kinase and purified by centrifugation through Micro Bio-Spin 30 chromatography columns (Bio-Rad, Hercules, Calif.). Typical 10-μl (total volume) reaction mixtures contained binding buffer (25 mM HEPES and 50 mM sodium glutamate, pH 8.0), 0.1 ng of labeled DNA, 20 ng of poly(dA-dT), and either purified NanR or extract at the concentrations indicated. After the mixture was incubated at room temperature for 10 min, 1.5 μl of a 50% (wt/vol) sucrose solution was added, and the sample was loaded onto a 5% polyacrylamide gel in 0.5× TBE (45 mM Tris, pH 8.3, 45 mM sodium borate, 1 mM EDTA). Samples were run at a constant 100 V for 1.5 h, and the dried gels were exposed overnight at −80°C against Kodak X-OMAT AR film and Fisher Biotech Intensifying Screen for detection of radiolabeled bands by autoradiography. The dissociation constant (Kd) of the interaction between NanR (monomer concentration) and the labeled 246-bp DNA fragment was determined by liquid scintillation spectrometry of excised free DNA according to the method of Carey (8).
Computer-assisted (in silico) analyses.
The National Center for Biotechnological Information public database was screened for homologues of nan gene products using the BLAST algorithm (1). Molecular modeling of FadR (protein database number 1HW2) against the N terminus of NanR and estimation of dimer width were kindly performed by James Murray (Oxford University, Oxford, United Kingdom) using the RasMol program (available at www.umass.edu).
RESULTS
Induction of the nan operon.
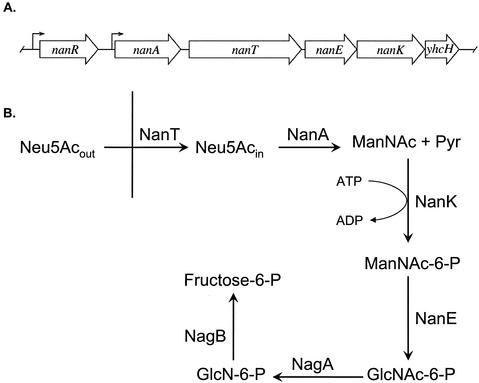
Figure 1A shows the genetic organization of the nanATEK-yhcH operon and the upstream locus encoding the transcriptional repressor of the operon, NanR, in E. coli K-12 (26). The known or proposed functions of NanA (sialate aldolase), NanT (sialate permease), NanE (N-acetylmannosamine 6-phosphate [ManNAc-6-P] to N-acetylglucosamine 6-phosphate [GlcNAc-6-P] 2-epimerase), and NanK (ATP-dependent ManNAc kinase) are shown in Fig. 1B. The function of yhcH is unknown, but deleting it had no apparent effect on sialate metabolism, whereas loss of nanA, -T, -E, or -K prevents growth on Neu5Ac as a sole carbon or nitrogen source (26, 38). A composite operon composed of orthologues of nanA, -E, and -K and the N-acetylglucosamine utilization genes nagA and nagB has been identified and partially characterized in Haemophilus influenzae (26, 37). In addition to H. influenzae and E. coli, unpublished BLAST comparisons of E. coli nan gene products identified putative nan loci in a variety of gram-negative and gram-positive organisms, suggesting that the ability to catabolize sialic acid is widespread in bacteria.
FIG. 1.
Genetic organization of E. coli nan pathway of sialate catabolism. (A) ORFs and their transcription directions are indicated by large arrows. The bent arrows indicate known or putative promoters. (B) Abbreviations for products, reactants, and enzymes are given in the text. The vertical line represents the E. coli inner membrane.
We previously showed that aldolase (NanA)-specific activity was increased up to 1,000-fold in cells grown on Neu5Ac (38). To further quantify the induction of the nan operon, we used a nanA-lacZ fusion. Growth on Neu5Ac produced a >200-fold increase in activity (Table 1). We previously isolated several strains carrying mutations within the nanR gene that increased expression of the downstream nanATEK operon (26). The introduction of two of these mutations, nanR6 or nanR7, into JM101 produced an equivalent large increase in nanA-lacZ activity (Table 1). These observations support the idea that growth on Neu5Ac is responsible for the displacement of NanR and induction of the nan operon. The absence of any strong inductive effect by sugars metabolically related to sialic acid (Fig. 1B), N-acetylglucosamine (GlcNAc), N-acetylmannosamine (ManNAc), and glucosamine (GlcN) (Table 1), is consistent with earlier conclusions that Neu5Ac, instead of one of its metabolic breakdown products, is the physiological inducer (38, 39). There is a four- to fivefold difference in β-galactosidase activity between glycerol and glucose, indicating that the nan operon is subject to catabolite repression, which agrees with previous results that showed decreased nanA expression when wild-type cells were grown in the presence of glucose compared to expression in the presence of glycerol (38). Similarly, in the nanR mutants, expression is higher in glycerol- than in glucose-grown cultures.
TABLE 1.
Effect of carbon source on nanA induction
| Carbon sourcea | Relevant genotype | β-Galactosidase activity (Miller units) | Fold induction |
|---|---|---|---|
| None | nanR+ | 25 ± 9 | 1 |
| Glucose | nanR+ | 5 ± 3 | 0.2 |
| GlcNAc | nanR+ | 20 ± 2 | 0.7 |
| GlcN | nanR+ | 50 ± 20 | 2 |
| ManNAc | nanR+ | 50 ± 15 | 2 |
| Man | nanR+ | 100 ± 30 | 4 |
| Neu5Ac | nanR+ | 5,300 ± 950 | 212 |
| Glucoseb | nanR6 | 3,300 ± 350 | 132 |
| Glycerolb | nanR6 | 7,850 ± 450 | 314 |
| Glucoseb | nanR7 | 2,500 ± 160 | 100 |
| Glycerolb | nanR7 | 8,300 ± 990 | 332 |
Unless otherwise indicated, strain JM101 bearing a nanA-lacZ fusion was grown in minimal E medium and 0.5% Casamino Acids with glycerol as the carbon source at 0.4% final concentration or with the indicated sugar added to a final concentration of 2 mg/ml. The results are the averages of three independent measurements±standard deviations.
JM101 carrying the nanA-lacZ fusion and either nanR6 or nanR7 was grown in MOPS (morpholine propanesulfonic acid) medium at 30°C with 0.2% glucose or 0.4% glycerol. The results are the means of two independent cultures. Abbreviations are defined in the text.
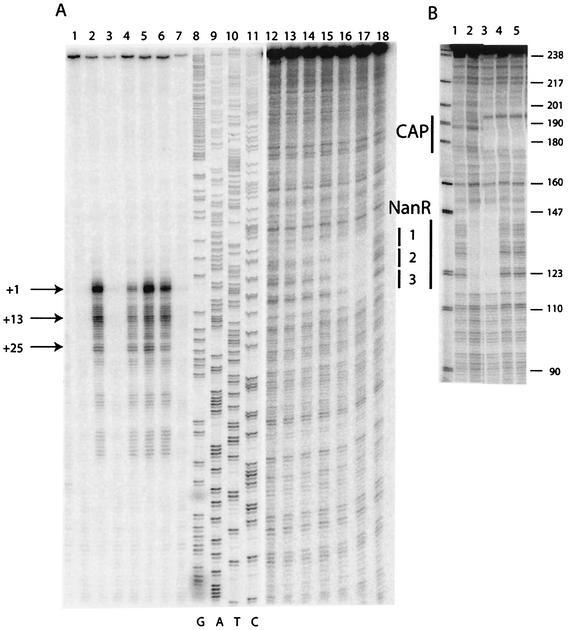
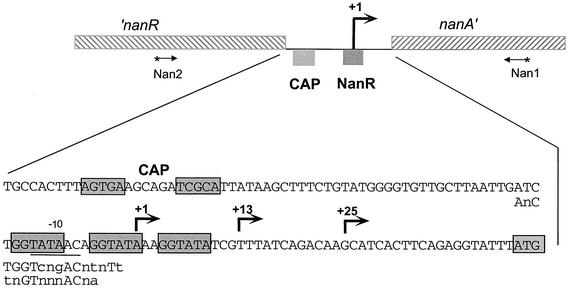
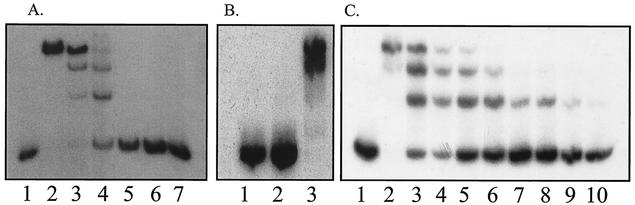
The transcriptional start site for nanA was mapped on mRNA prepared from IBPC1016 carrying the nanR6 mutation, as well as the parental wild-type strain (JM101), during growth on different carbon sources. A series of strong signals were detected with mRNA from mutant cells grown with each of the different carbon sources (Fig. 2A, lanes 2, 4, and 6) and also in the wild-type strain grown on Neu5Ac (Fig. 2A, lane 5). No nanA mRNAs were detected in the wild type during growth on glucose or glycerol. These results confirm those with the nanA-lacZ fusion showing that expression of nanA is strongly induced by growth on Neu5Ac and is constitutive in the nanR6 mutant strain. The major start site is located just 44 bp upstream of the nanA translational start codon (Fig. 3). It corresponds to a −10 sequence of TATAAC but with no recognizable −35 consensus sequence. The series of shorter transcripts with strong signals at +13 and +25 could be de novo transcriptional start sites. There is a putative −10 sequence (TATAAA) upstream of +13 which was originally proposed by Ohta et al. (23) as +1. Alternatively, the series of transcripts could be due to slippage of the RNA polymerase along the DNA or to posttranscriptional processing. A detailed mutagenesis study is needed to understand this rather unusual transcriptional pattern. Note that the amount of mRNA is distinctly lower for the nanR6 mutant grown on glucose than for that grown on glycerol, consistent with lower β-galactosidase levels in glucose than in glycerol medium. A consensus binding site for the CAP transcriptional regulator is recognizable on the DNA sequence ∼60 bp upstream of the longest nanA transcript (Fig. 3). This distance is typical for a classical class I CAP-controlled operon (7).
FIG. 2.
S1 nuclease transcript mapping and DNA footprint analysis of the nanR-nanA intergenic region. (A) The DNA fragment used for the analyses was the 439-bp PCR Nan1-2 (Fig. 3) labeled at the Nan1 end within the nanA ORF. Lanes 1 to 7, S1 mapping of nanA transcripts (25 μg of total RNA) isolated from JM101 (lanes 1, 3, and 5) and IBPC1016 (nanR6) (lanes 2, 4, and 6) grown on glycerol (lanes 1 and 2), glucose (lanes 3 and 4), and Neu5Ac (lanes 5 and 6). Lane 7, control (25 μg of tRNA). Lanes 8 to 11, DNA sequencing reactions on the Nan1-2 fragment using the Nan1 oligonucleotide as a primer. Lanes 12 to 18, DNase I footprinting on the Nan1-2 fragment (lanes 12 to 17 show the binding of purified NanR-His6 at 0.9, 1.7, 3.5, 7.0, 14, and 41 nM; lane 18 shows the free DNA). (B) The DNA fragment used for DNase I footprinting was the 245-bp Nan1-BstN1 fragment labeled at the Nan1 end. Lane 1, free DNA; lane 2, DNA with crude extract of NanR-overproducing strain (∼50 μg of total protein/ml); lane 3, same as lane 2 with 20 nM CAP and 200 μM cyclic AMP (cAMP); lane 4, 5 nM CAP and 200 μM cAMP; lane 5, 20 nM CAP and 200 μM cAMP. The numbers on the right are sizes (in base pairs) of pBR322 after digestion (shown at left) with MspI.
FIG. 3.
Intergenic region of the E. coli chromosome between nanR and nanA. The nucleotide sequence between the end of nanR and the translational start codon (boxed) of nanA is indicated. The bent arrows indicate transcriptional start site(s). The CAP and NanR binding sites are indicated, and the nucleotides known or proposed to constitute these sites are boxed. The −10 region is underlined. The first line of nucleotides below the intergenic region gives the FadR consensus pseudopalindrome, and the second line gives the FadR subfamily consensus binding site (29). Lowercase letters indicate bases that are variable within the consensus sequence. The oligonucleotides (Nan1 and Nan2) were previously described (26) and were used to amplify the 439-bp DNA fragment that includes the 121-bp intergenic region.
Purification of NanR.
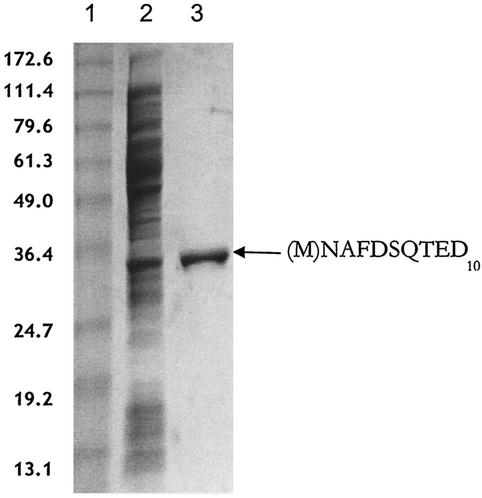
To facilitate the characterization of NanR and its binding to the putative nan operator (Fig. 3), the repressor was purified in two molecular forms. Initially, for ease of purification, we overproduced NanR-His6 and separated it from contaminating proteins in a single step by metal ion affinity chromatography as described in Materials and Methods. However, because peptide tags have been shown to interfere with the normal functions of some DNA binding proteins (6), we also sought to purify NanR without a tag. Figure 4 shows the purification of native NanR using paramagnetic microbead technology from an extract of E. coli transformed with the l-arabinose-inducible nanR plasmid pSX675. The molecular identity of NanR was confirmed by its expected monomer molecular mass (30 kDa) after denaturing polyacrylamide gel electrophoresis and by its observed N-terminal amino acid sequence (Fig. 4, lane 3). This result indicates that the second of two closely spaced in-frame ATG start sites (accession number P45427) is the one actually used for translation.
FIG. 4.
Purification of native NanR. Lane 1, molecular mass markers with masses (in kilodaltons) on the left. Lane 2, 31 μg of induced soluble extract. Lane 3, 3.5 μg of purified native NanR with the amino acid sequence of the first 10 residues on the right. The N-terminal methionine shown in parentheses is apparently removed in vivo.
Solution quaternary structure of NanR.
Most helix-turn-helix (HTH) regulators function as homodimers binding to palindromic or pseudopalindromic operators. We used nondenaturing (native) polyacrylamide gel electrophoresis to determine the molecular weight of purified NanR-His6. The Ferguson plot (11) graphs the relative migrations of marker proteins and NanR in gels with different polyacrylamide concentrations. Plotting the negative slopes of these curves against molecular weight yielded a straight line with a regression coefficient of 0.9972 (not shown). From this standard curve, the molecular mass of tagged NanR was calculated to be 53,315 Da, which is most consistent with NanR existing as a homodimer in solution. The lower-than-expected (theoretical dimer molecular mass, 63,081 Da) mass of the NanR-His6 homodimer may indicate aberrant migration if the shape of NanR differs from that of a sphere. A similar decrement from the predicted dimer molecular weight was obtained by gel permeation chromatography, whereas MALDI-TOF and chemical cross-linking indicated that NanR is a homodimer in solution with a molecular weight close to the theoretical value (data not shown). Chemical cross-linking showed that purified native NanR also behaved like a homodimer in solution, with a molecular mass near 60 kDa. The range of molecular weights determined for native and His-tagged NanR (53,315 to 66,991) using a variety of physical methods indicates that the homodimer is the most likely form to bind target DNA both in vivo and in vitro.
Identification of the NanR binding site.
When an extract containing overproduced NanR was incubated with a 246-bp radiolabeled DNA fragment covering 39 bp 5′ from the termination codon of nanR to 86 bp 3′ from the translational codon of nanA and including the 121-bp intergenic region (Fig. 3), a concentration-dependent ladder of three retarded bands was detected by gel mobility shift analysis (Fig. 5A, lanes 2 to 4). An extract prepared from the nanR null mutant IBPC1017 carrying the nanR7 mutation did not confer a mobility shift (Fig. 5B, lane 2). Adding purified NanR-His6 to the IBPC1017 nanR extract reestablished gel shift activity (Fig. 5A, lane 3). The same result was observed when native NanR was used for the reconstitution (not shown). Incubating purified NanR-His6 in the presence of radiolabeled promoter without added extract produced the same gel shift pattern (Fig. 5C) as the crude extract containing native NanR (Fig. 5A). The apparent Kd (5.6 ± 3.4 nM) of the interaction between NanR-His6 and its operator (measured by quantifying the disappearance of free DNA) is within the range found for other transcriptional regulators. Purified native NanR produced the same pattern of gel-shifted bands as its tagged derivative and was found to have a Kd of 2.5 nM, which is within the range calculated for NanR-His6.
FIG. 5.
Gel mobility shift analysis of NanR. (A) Extract of JM101 harboring pTZ(Nan1-3) was added to the 246-bp labeled DNA fragment and subjected to gel shift analysis. Lane 1, DNA fragment alone; lanes 2 to 7, serial 10-fold dilutions of the extract (0.5 mg of total protein/ml) starting at a 1/10 dilution. (B) Gel shift analysis with extract of the nanR7 mutant IBPC1017 (lane 2) and with the exogenous addition of 26 nM NanR-His6 (lane 3). Lane 1, DNA fragment alone. (C) Gel shift activity using purified NanR-His6 was determined at different dilutions. Lane 1, DNA fragment alone. Lanes 2 to 10 contained 44.2, 17.7, 8.8, 6.6, 4.5, 2.2 (duplicate lanes), 1.7, and 0.8 nM NanR-His6, respectively.
DNase I footprint analysis using a crude extract from a strain overproducing NanR from the vector pTZ(Nan1-3) indicated protection of an ∼30-bp region (Fig. 2B, lane 2). Purified CAP protected a region corresponding to the consensus CAP binding motif shown in Fig. 3 (Fig. 2B, lanes 4 and 5), and both CAP and NanR can bind simultaneously (Fig. 2B, lane 3). To further investigate the interaction of NanR with its operator, we used purified NanR-His6 for footprint analysis. As shown in Fig. 2A (lanes 12 to 18), the purified protein protected a region of DNA that includes the three direct repeats of the sequence GGTATA separated by 3 or 2 bp (Fig. 3). Testing dilutions of the repressor shows that NanR binds sequentially to three sites, which is consistent with the presence of the three retarded bands in the gel mobility shift assay. The region protected corresponds to the three direct GGTATA repeats bound by native NanR and shows that the distal site 1 has the highest affinity. The 5′ end of the major nanA transcript lies within the middle repeat, so that bound NanR would be expected to prevent RNA polymerase binding.
The simplest interpretation of the gel shift and protection assays is that NanR binds its operator by covering approximately three turns of the helix with a stoichiometry of three NanR molecules (presumably homodimers) per target DNA molecule. Assuming that the three bands in the gel shift shown in Fig. 5 result from the successive binding of NanR to target DNA, a logarithmic plot of the molecular weights of these complexes (assuming either a monomeric or dimeric form of NanR and no effect on mobility from DNA bending) versus the relative mobilities of the complexes yields a straight line. This logarithmic relationship between mobility and predicted stoichiometry has been observed for other DNA binding proteins, although it does not rigorously exclude conformational isomerization of the complexes as an alternative explanation (12). Direct determination of the NanR stoichiometry in each band or a cocrystal structure of NanR with its operator will be necessary to confirm the mechanism of NanR binding.
To further demonstrate that NanR specifically recognizes the nanA promoter region in vivo, we cloned the 246-bp DNA fragment used for gel shift analysis into pGEM to generate plasmid pSX676. This construct was transformed into the nanA-lacZ derivative of wild-type strain JM101, resulting in a 50-fold induction compared to an untransformed control (Table 2). Transformation with vector alone resulted in a twofold induction, suggesting minimal nonspecific interaction between NanR and the relatively high intracellular concentration of plasmid DNA. We conclude that vector containing the nanA promoter region titrates intracellular NanR, leading to induction of the nanA fusion.
TABLE 2.
In vivo titration of NanR
| Plasmida | No. of GGTATA repeats | β-Galactosidase activity (Miller units) | Fold induction |
|---|---|---|---|
| None | 0 | 60 ± 20 | 1 |
| pGEM-Teasy | 0 | 120 ± 40 | 2 |
| pSX676 | 3 | 3,200 ± 330 | 53 |
| pGEM-1 | 1 | 90 ± 20 | 1.5 |
| pGEM-2 | 2 | 1,820 ± 240 | 30 |
| pGEM-3 | 3 | 3,050 ± 750 | 51 |
| pGEM-4 | 4 | 3,260 ± 420 | 54 |
| pGEM-6 | 6 | 5,130 ± 1,750 | 86 |
The indicated plasmids were transformed into JM101 carrying the nanA-lacZ reporter fusion. Data are the averages of seven independent experiments ± standard deviations. Cells were grown in E minimal salts with 0.4% glycerol and 0.5% Casamino Acids.
Binding of NanR to artificial operators.
Additional evidence for the importance of the tandem GGTATA repeat to NanR binding came from an analysis of its interactions with artificial operators. When we tested a DNA fragment carrying a single GGTATA motif flanked by 10 or 12 arbitrarily chosen residues, one or two gel-shifted bands were detected using 30 ng (∼100 nM) of purified native NanR (Table 3). Binding was not detected at the relatively low NanR concentration used for the previous gel shift analysis (Fig. 5C); thus, it was unclear whether the interaction of NanR and a single motif reflects specific binding. However, operators with two or three tandem repeats produced the same number of gel-shifted bands as the number of repeats of the motif (Table 3). When two different DNA fragments with three repeats were tested, the number of residues flanking the GGTATA motifs did not influence NanR binding. Thus, operators ranging in length from 49 to 246 bp with three copies of the GGTATA motif invariably produced three gel-shifted bands (Table 3 and Fig. 5). When operators with four or six repeats were tested, the number of bands detected was equal to or one greater than the number of repeats (Table 3). Binding of purified native NanR to operators with four or six repeats displayed the same concentration dependence as binding to the natural operator shown in Fig. 5C, with Kds near 1 and 0.5 nM, respectively. The combined results of these artificial-operator-binding experiments indicate that the GGTATA motif is an important component of NanR target recognition under in vitro conditions.
TABLE 3.
Binding of NanR to artificial operators with various numbers of tandem GGTATA repeats
| Operatora | Length (bp) | No. of tandem repeats | No. of bands in gel shift |
|---|---|---|---|
| 5′-TAATTGATCTGGTATATCGTTTATCAGA-3′ | 28 | 1 | 1 or 2 |
| 5′-TAATTGATCTGGTATAACAGGTATATCGTTTATCAGA-3′ | 37 | 2 | 2 |
| 5′-CTTAATTGATCTGGTATAACAGGTATAAAGGTATATCGTTTATACGACA-3′ | 49 | 3 | 3 |
| 5′-GGTGTTGCTTAATTGATCTGGTATAACAGGTATAAAGGTATATCGTTTATCAGACAAGCA-3′ | 60 | 3 | 3 |
| 5′-TTGATCTGGTATAACAGGTATAAAGGTATAAAGGTATATCGTTTA-3′ | 45 | 4 | 4 or 5 |
| 5′-TTGATCTGGTATAACAGGTATAAAGGTATAAAGGTATAAAGGTATAAAGGTATATCGTTT-3′ | 60 | 6 | 6 or 7 |
Only the 5′ strand of each operator is shown, with the GGTATA motifs underlined.
To determine the effects of the artificial operators on titration of NanR in vivo, we cloned the DNA fragments with 1, 2, 3 (60 bp), 4, or 6 repeats of the GGTATA motif into pGEM-T and transformed JM101 containing the reporter nanA-lacZ fusion. Table 2 shows that despite detection of a mobility shift with a DNA fragment carrying one repeat, this artificial operator had no effect on β-galactosidase production beyond that caused by the pGEM-T vector alone. In contrast, two repeats produced about half the amount of enzyme activity as the three repeats did (either as the 60- or 246-bp operator), whereas four or six repeats produced activities about the same as or greater than the operators with three repeats (Table 2). The amount of β-galactosidase stimulated by transformation by pGEM-6 approaches the level of the nanR mutants (Table 1), suggesting that increasing the number of tandem repeats beyond the usual three GGTATA motifs results in an increased affinity of NanR for its operator in vivo. The results shown in Table 2 establish the importance of at least two repeated GGTATA motifs to NanR binding in vivo.
Inefficient displacement of NanR by Neu5Ac in vitro.
On the basis of induction of aldolase (NanA) by exogenous addition of Neu5Ac to the medium or induction of the permease encoded by nanT in a nonpolar nanA mutant strain that produces Neu5Ac biosynthetically (endogenous induction), it was previously concluded that sialic acid was the physiological nan inducer (38, 39). This conclusion is supported by the present results shown in Table 1. On the basis of these results, we expected Neu5Ac to efficiently displace NanR from its operator in vitro. However, Neu5Ac, ManNAc, GlcNAc, GlcN, and Man at 8 mM concentration in the binding reaction did not appreciably affect the gel shift pattern (not shown). Similarly, adding 1 mM Neu5Ac to the upper buffer chamber had little effect on NanR binding, indicating that the lack of displacement of NanR from its sites is not due to the negatively charged Neu5Ac being entirely stripped from NanR during electrophoresis.
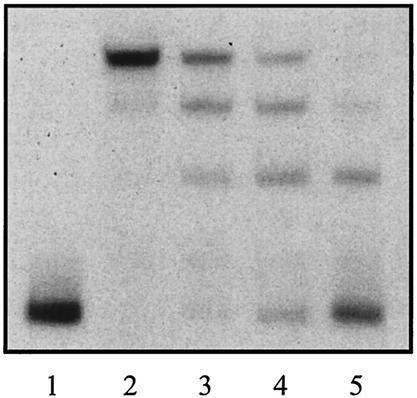
More than 90% of Neu5Ac in solution exists as the β isomer with axial C-2 hydroxyl extending up from the ring. Mutarotation (ring opening and closing) slowly interconverts this form to the α isomer, which is the form that exists in all known glycoconjugates except the sialyl donor, CMP-sialic acid, where the sugar is in the β configuration. A variety of physical techniques have shown that an enzyme like sialate aldolase (NanA) “chooses” the α isomer from solution as its substrate, effectively driving the mutarotation of the predominant β form (10). We therefore preincubated Neu5Ac with native NanR, and as shown in Fig. 6, this treatment resulted in at least partial displacement of NanR from the DNA. If the NanT permease is also specific for the α isomer, then under physiological conditions, this form would be preferentially delivered into the cell, which could explain the rapid induction kinetics of the operon (38, 39) and our inability to duplicate the process in vitro. A physical analysis of the actual form of Neu5Ac bound to NanR or present intracellularly will be necessary to confirm our hypothesis that the α isomer of Neu5Ac is the physiological inducer.
FIG. 6.
Gel mobility shift analysis after preincubation with Neu5Ac. Purified native NanR at 2.3 (lanes 2 and 3) or 0.5 (lanes 4 and 5) nM was incubated (lanes 3 and 5) or not (lanes 2 and 4) with 10 mM Neu5Ac for 1 h at room temperature prior to gel shift analysis using the 246-bp DNA fragment. Lane 1 shows the DNA fragment alone.
DISCUSSION
NanR belongs to the FadR/GntR family of transcriptional regulators that includes activators, repressors, and molecules which both activate and repress a wide range of bacterial operons (17, 29). Although homologues of the nan catabolic operon genes are found in many bacterial species, none except enteric organisms closely related to E. coli are associated with a nanR-like gene. The absence of NanR orthologues in the other bacteria indicates the evolution of alternative regulatory mechanisms for controlling nan expression in these organisms.
Members of the FadR/GntR family of “winged” HTH proteins consists of two domains, a highly conserved N-terminal DNA binding domain containing the HTH motif (17) and a more divergent C-terminal domain involved in dimerization and effector binding (29). FadR is the only member of the GntR superfamily whose structure has been solved in three dimensions in complex with its DNA operator (34, 41). This structure demonstrates an unusual interaction of FadR with its 17-bp pseudopalindromic DNA binding site, in which the recognition helices of both HTH motifs of the FadR homodimer interact with the same major groove. This is in contrast to other prokaryotic HTH proteins, like CAP (32) or LacI (20), which contact two adjacent major grooves. In addition, FadR uses a winged-helix (WH) motif to contact the adjacent minor grooves. Although most HTH transcriptional regulators recognize palindromic or pseudopalindromic targets, we have provided evidence that NanR recognizes three tandem GGTATA repeats. Though relatively uncommon in bacteria, this type of interaction is common in eukaryotic regulators of the WH class, where the wing confers specificity, usually by stabilizing the interaction between the DNA residues present in the major groove of the target and the so-called recognition helix through interactions between the wing and an adjacent minor groove (5, 13). However, the WH family is extremely versatile (18), including monomers, homodimers, and heterodimeric protein-DNA complexes. One member of the family, RFX1, has been shown to use its wing to make contact with the major groove, and the recognition helix was found to be overlaid on the minor groove (13). The WH motif also may participate in protein-protein interactions (13).
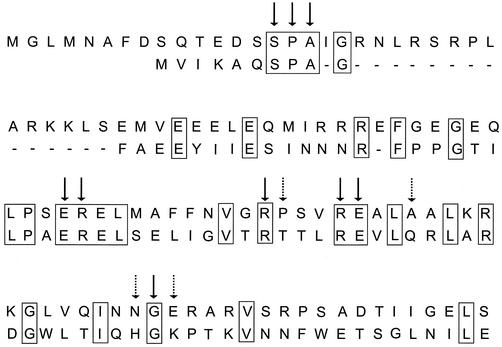
Three-dimensional modeling of the NanR N terminus with the equivalent FadR domain indicates topological similarity between the predicted structure of NanR and the corresponding structural elements of FadR. This alignment had an overall relatively poor root mean square deviation of 2.4 Å, which is consistent with ∼26% sequence identity. However, we note that 9 of the 13 FadR amino acid residues lying within hydrogen-bonding distance of the DNA target are potentially shared with the NanR N terminus (Fig. 7). This could imply that NanR makes contacts with DNA similar to those made by FadR, with the nonidentical amino acids conferring specificity on the NanR operator. However, our in vitro data suggest that the basic unit of NanR recognition is the GGTATA repeat. The three models that can most plausibly account for our results are that (i) NanR specifically recognizes each of three tandem direct repeats, (ii) each GGTATA site is itself a pseudo-inverted repeat (GGTA:TAnc; the colon indicates the location of the pseudo-inverted repeat; “nc” indicates nonconserved residues), and (iii) each adjacent pair of GGTATA repeats contributes an inverted repeat, forming a FadR-like pseudopalindrome (Fig. 3) so that NanR binds to two overlapping inverted repeats.
FIG. 7.
Alignment of FadR and NanR N termini. The N termini of NanR (top sequence) and FadR (bottom sequence) were aligned by eye, and identities are boxed. The dashes represent an arbitrary gap to maximize the alignment. The arrows indicate FadR residues within hydrogen-bonding distance of DNA targets in the cocrystal (35). The solid arrows indicate amino acids that are identical in FadR and NanR, while the dotted arrows indicate those amino acids without homology to NanR that could be responsible for specific recognition, as described in the text.
Models 1 and 2 predict that the three gel-shifted bands suggest the binding of 1, 2, or 3 mol of NanR per mol of DNA. Since the GGTATA motif is repeated after 8 or 9 bp (i.e., less than a single turn of the B form of DNA), this would imply that three dimers of NanR coat the operator by binding around the DNA and are not lying in the same plane. The maximum distance between the two edges of the FadR homodimer is 64 Å, but this is reduced to 30 Å at the level of DNA. This would provide just about sufficient room for three NanR homodimers binding to three turns of the DNA helix of ∼100 Å. Note that the footprint of the NanR binding to the operator shows protection of >30 bp, implying that the sequence protected by each NanR molecule extends beyond the GGTATA repeat. Model 1 also includes the possibility that NanR binds to DNA as a monomer, as in the case of WH proteins of the OmpR family, which bind to tandem repeats with a monomer binding to each successive major groove (4, 15, 16). Since we know nothing about the kinetics of NanR dimerization, we cannot be certain that the functional (binding) form of NanR is the homodimer. Possibly three monomers bind and oligomerize on the DNA with a trimer as the functional unit. Ferguson analysis (11) or the use of radiolabeled NanR should allow us to distinguish the possible binding stoichiometries of each complex. In model 2, the GGTATA motif is considered a pseudopalindrome, GGTA:TAnc, so that NanR should bind as a dimer but possibly interacts asymmetrically with one side of the major groove.
In model 3, the specific NanR recognition sequence covers two GGTATA motifs. The observation that a FadR consensus pseudopalindromic operator can be identified overlapping the first two GGTATA repeats (Fig. 3) could argue that this palindrome is the biologically relevant motif. This idea is supported by the observation that a minimum of two repeats are necessary for efficient band shift in vitro or for operator titration of the nanA-lacZ fusion in vivo. However, it is difficult to reconcile this model with the rest of our in vitro results, which clearly implicate all three repeats in binding. Recently, revised models for Fur binding to the previously defined Fur Box (19 bp) have been proposed (3, 19). These models are consistent with the cocrystal structure of the related protein DtxR (28, 40) and the structure of the recently determined FurR homodimer (27), indicating that both bind as homodimers to overlapping 15-bp palindromes on opposite sides of the DNA helix. If this model is relevant to NanR binding (model 3), then the FadR pseudopalindrome could constitute one of the overlapping palindromes (Fig. 3). However, no second overlapping palindrome is obvious in the nanA operator. Understanding the diversity of the FadR WH family clearly requires additional cocrystal structures.
Using site-directed mutagenesis of the repeats and the promoter titration system described here, altering any of the three repeats should decrease NanR binding, while mutagenesis of any two repeats should abrogate activity. It will also be interesting to determine the effects of increasing the distance or altering the orientation between repeats to further test our hypotheses about the NanR binding mechanism. In this regard, note that it has been shown that the same protein (CytR) can bind to both direct and inverted repeats when the spacing between the repeats is altered (24).
The inability of one GGTATA motif to titrate NanR indicates that the repressor does not recognize a single repeat unit under in vivo conditions. As this hexameric sequence is expected to occur many times throughout a >4-Mb genome, failure to recognize a single motif makes sense physiologically. However, if each retarded band in the mobility shift analysis represents the corresponding addition of one extra NanR molecule, the complex with the greatest mobility (the smallest number of NanR molecules bound) should correspond to binding of one equivalent of NanR. Possibly the “cage effect” in the band shift experiment produces sufficiently high concentrations to allow detection of the first NanR complex (Fig. 5C). Alternatively, binding of the first NanR molecule could require either recognition of at least two direct repeats or, perhaps, participation of the FadR subfamily-type consensus palindrome (Fig. 3). Binding of successively greater numbers of NanR molecules might then involve recognition of the tandem repeats, with the added potential for protein-protein interactions stabilizing the complex. This mechanism would be a hybrid of models 2 and 3. The binding of NanR to artificial operators, especially those with more than three GGTATA motifs, is consistent with this hypothesis (Tables 2 and 3). The regularly spaced ladder of NanR-DNA complexes shown in Fig. 5 was observed with crude wild-type extract or an extract from a strain overproducing NanR, as well as purified NanR-His6 or native NanR. This should exclude possible artifacts due to the peptide tag or extra components supplied by the extract and indicates that the affinity of NanR for its target and possible NanR-NanR interactions are the only factors involved in DNA binding. Although we cannot exclude the possibility of positional or configurational isomers to explain the banding pattern, studies in systems with known stoichiometries indicate that the primary factor affecting DNA mobility during electrophoresis is the molecular weight of the bound protein (12). Moreover, the DNA footprint analysis (Fig. 2B) would seem to require binding of at least two NanR molecules to explain the protection pattern. Binding of multiple regulatory units to transcriptional control sites is a mechanism for fine tuning gene expression in response to environmental conditions (15) and for increasing the range of induction, as seen in the lacZYA system. Our results demonstrate that the high level of nanA induction which is achieved through loss of NanR by mutation or titration or the presence of Neu5Ac is comparable to that produced by LacI binding to its three separated operators (22). However, binding of multiple LacI molecules requires a DNA looping mechanism, while NanR achieves a similar regulatory range by binding to adjacent sites.
Although both our present and previous results (30, 38, 39) clearly implicate Neu5Ac as the nan inducer, we were initially surprised by the failure to duplicate the in vitro equivalent of induction by incubating NanR with Neu5Ac. However, the temporal dependency of the in vitro induction suggests that the thermodynamically minor α isomer of Neu5Ac might be the true inducer. Because we have not detected orthologues of NanR in organisms with putative nan systems that are not closely related to E. coli, nan expression in different organisms may involve alternative regulators and inducing signals. In other words, the coordination between Neu5Ac synthesis or acquisition and its catabolism in different species may require distinct regulatory circuitry and involve diverse effector molecules. The relative ease of genetic manipulation provided by E. coli offers an attractive system to further explore the role of sialic acid metabolism in host-microbe interactions and provides the basis for similar studies in other pathogens or commensals.
Acknowledgments
We are indebted to James Murray for his expert molecular modeling of NanR, Annie Kolb for the gift of CAP and comments on the manuscript, Kurt Braun of Miltenyi Biotec for advice and encouragement in using microbead technology for the purification of native NanR from crude extracts, and Kerry Helms and Eric Deszo for expert graphical assistance. We are grateful to one anonymous reviewer for helpful suggestions.
K.A.K. was supported in part by a College of Agricultural, Consumer and Environmental Sciences Value-Added Predoctoral Fellowship and an American Heart Association (Midwest Affiliate) Predoctoral Fellowship. This work was supported by National Institutes of Health grant R01 AI42015 (E.R.V.) and by the CNRS and the Université Paris 7 to UPR9073 (J.P.).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Angata, T., and A. Varki. 2002. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102:439-469. [DOI] [PubMed] [Google Scholar]
- 3.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco, A. G., M. Sola, F. X. Gomis-Ruth, and M. Coll. 2002. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10:701-713. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, R. G. 1993. The winged-helix DNA-binding motif: another helix-turn-helix takeoff. Cell 74:773-776. [DOI] [PubMed] [Google Scholar]
- 6.Buning, H., U. Gartner, D. von Schack, P. A. Baeuerle, and H. Zorbas. 1996. The histidine tail of recombinant DNA binding proteins may influence the quality of interaction with DNA. Anal. Biochem. 234:227-230. [DOI] [PubMed] [Google Scholar]
- 7.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 8.Carey, J. 1991. Gel retardation. Methods Enzymol. 208:103-112. [DOI] [PubMed] [Google Scholar]
- 9.Davis, R. W., D. Botstein, and J. R. Roth. 1980. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Deijl, C. M., and J. F. Vliegenthart. 1983. Configuration of substrate and products of N-acetylneuraminate pyruvate-lyase from Clostridium perfringens. Biochem. Biophys. Res. Commun. 111:668-674. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson, K. 1964. Starch-gel electrophoresis—application to the classification of pituitary proteins and polypeptides. Metabolism 13:985-1002. [DOI] [PubMed] [Google Scholar]
- 12.Fried, M. G. 1989. Measurement of protein-DNA interaction parameters by electrophoresis mobility shift assay. Electrophoresis 10:366-376. [DOI] [PubMed] [Google Scholar]
- 13.Gajiwala, K. S., and S. K. Burley. 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10:110-116. [DOI] [PubMed] [Google Scholar]
- 14.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harlocker, S. L., L. Bergstrom, and M. Inouye. 1995. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J. Biol. Chem. 270:26849-26856. [DOI] [PubMed] [Google Scholar]
- 16.Harrison-McMonagle, P., N. Denissova, E. Martinez-Hackert, R. H. Ebright, and A. M. Stock. 1999. Orientation of OmpR monomers within an OmpR:DNA complex determined by DNA affinity cleaving. J. Mol. Biol. 285:555-566. [DOI] [PubMed] [Google Scholar]
- 17.Hayden, D. J., and J. R. Guest. 1991. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 79:291-296. [DOI] [PubMed] [Google Scholar]
- 18.Huffman, J. L., and R. G. Brennan. 2002. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 12:98-106. [DOI] [PubMed] [Google Scholar]
- 19.Lavrrar, J. L., C. A. Christoffersen, and M. A. McIntosh. 2002. Fur-DNA interactions at the bidirectional fepDGC-entS promoter region in Escherichia coli. J. Mol. Biol. 322:983-995. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, M., G. Chang, N. C. Horton, M. A. Kercher, H. C. Pace, M. A. Schumacher, R. G. Brennan, and P. Lu. 1996. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science 271:1247-1254. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Oehler, S., M. Amouyal, P. Kolkhof, B. von Wilcken-Bergmann, and B. Muller-Hill. 1994. Quality and position of the three lac operators of E. coli define efficiency of repression. EMBO J. 13:3348-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohta, Y., K. Watanabe, and A. Kimura. 1985. Complete nucleotide sequence of the E. coli N-acetylneuraminate lyase. Nucleic Acids Res. 13:8843-8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen, H., and P. Valentin-Hansen. 1997. Protein-induced fit: the CRP activator protein changes sequence-specific DNA recognition by the CytR repressor, a highly flexible LacI member. EMBO J. 16:2108-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plumbridge, J. 1998. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol. Microbiol. 27:369-380. [DOI] [PubMed] [Google Scholar]
- 26.Plumbridge, J., and E. Vimr. 1999. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J. Bacteriol. 181:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pohl, E., J. C. Haller, A. Mijovilovich, W. Meyer-Klaucke, E. Garman, and M. L. Vasil. 2003. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47:903-915. [DOI] [PubMed] [Google Scholar]
- 28.Pohl, E., R. K. Holmes, and W. G. Hol. 1999. Crystal structure of a cobalt-activated diphtheria toxin repressor-DNA complex reveals a metal-binding SH3-like domain. J. Mol. Biol. 292:653-667. [DOI] [PubMed] [Google Scholar]
- 29.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507-12515. [DOI] [PubMed] [Google Scholar]
- 30.Ringenberg, M., C. Lichtensteiger, and E. Vimr. 2001. Redirection of sialic acid metabolism in genetically engineered Escherichia coli. Glycobiology 11:533-539. [DOI] [PubMed] [Google Scholar]
- 31.Schauer, R. 2000. Achievements and challenges of sialic acid research. Glycoconj. J. 17:485-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultz, S. C., G. C. Shields, and T. A. Steitz. 1991. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science 253:1001-1007. [DOI] [PubMed] [Google Scholar]
- 33.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 34.van Aalten, D. M., C. C. DiRusso, and J. Knudsen. 2001. The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO J. 20:2041-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Aalten, D. M., C. C. DiRusso, J. Knudsen, and R. K. Wierenga. 2000. Crystal structure of FadR, a fatty acid-responsive transcription factor with a novel acyl coenzyme A-binding fold. EMBO J. 19:5167-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vimr, E., and C. Lichtensteiger. 2002. To sialylate, or not to sialylate: that is the question. Trends Microbiol. 10:254-257. [DOI] [PubMed] [Google Scholar]
- 37.Vimr, E., C. Lichtensteiger, and S. Steenbergen. 2000. Sialic acid metabolism's dual function in Haemophilus influenzae. Mol. Microbiol. 36:1113-1123. [DOI] [PubMed] [Google Scholar]
- 38.Vimr, E. R., and F. A. Troy. 1985. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J. Bacteriol. 164:845-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vimr, E. R., and F. A. Troy. 1985. Regulation of sialic acid metabolism in Escherichia coli: role of N-acylneuraminate pyruvate-lyase. J. Bacteriol. 164:854-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White, A., X. Ding, J. C. vanderSpek, J. R. Murphy, and D. Ringe. 1998. Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex. Nature 394:502-506. [DOI] [PubMed] [Google Scholar]
- 41.Xu, Y., R. J. Heath, Z. Li, C. O. Rock, and S. W. White. 2001. The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli. J. Biol. Chem. 276:17373-17379. [DOI] [PubMed] [Google Scholar]