Abstract
Regulated expression of the Escherichia coli dam gene has been achieved with the araBAD promoter lacking a ribosome binding site. Cultures of dam mutants containing plasmid pMQ430 show no detectable methylation in the absence of arabinose and complete methylation in its presence. Dam methyltransferase is a substrate for the Lon protease.
In Escherichia coli K-12, about 2% of adenines are modified to N6-methyladenine by the action of the Dam methyltransferase (8). This modification occurs at GATC sequences in transiently hemimethylated double-stranded DNA trailing the replication fork. Hemimethylated DNA has one parental chain which is methylated, but its newly synthesized complementary strand is not methylated. Methylation of hemimethylated DNA is delayed due to the low concentration of Dam methyltransferase, which is present at about 130 molecules per cell (2). Increasing Dam methyltransferase levels reduce the amount of hemimethylated DNA (3).
The large number of phenotypic differences associated with Dam deficiency in E. coli is consistent with multiple roles for dam methylation (8, 11). These roles can broadly be defined as affecting (i) DNA mismatch repair (12), (ii) alterations in gene expression (15), (iii) the initiation of chromosome replication (1), and (iv) the chromosome structure (6). The phenotypes of dam mutants have been explored using presumed missense (e.g., dam-3) and insertion and deletion (e.g., dam-13::Tn9 and dam-16::Kan, respectively) mutations. In spite of numerous attempts, no temperature-sensitive dam alleles have been isolated from E. coli, although such alleles would be very useful in functional studies to turn Dam methylation on and off. These alleles would also allow testing of the effects of partially methylated or hemimethylated DNA on cell functions.
In an alternative approach, we placed the dam gene borne on a chromosomal XbaI-HindIII fragment from pYIN2 (13) under the control of the araBAD promoter in vector pBAD18 (5), creating pMQ400. In the absence of inducer, however, the cells still showed a Dam+ phenotype, as was determined by the action of DpnI (digests methylated dam DNA), DpnII (digests unmethylated dam DNA), and Sau3AI (digests both methylated and unmethylated dam DNA) on total DNA extracted from cultures using a MasterPure DNA purification kit (Epicentre). To reduce the amount of Dam produced from the plasmid, we removed the ribosome binding site downstream of the araBAD promoter in pMQ400. This was achieved by removing the native DNA upstream of the ATG of the dam gene and fusing the translation initiation codon (an NlaIII site, ↓CATG) directly to the SphI site (G↓CATG) in the polylinker sequence of pBAD18. The resulting construct was designated pMQ430.
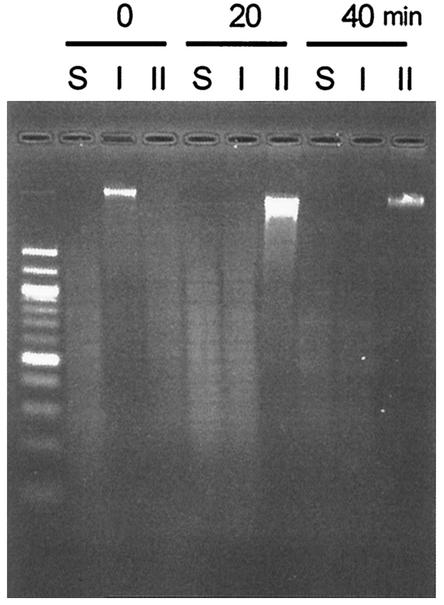
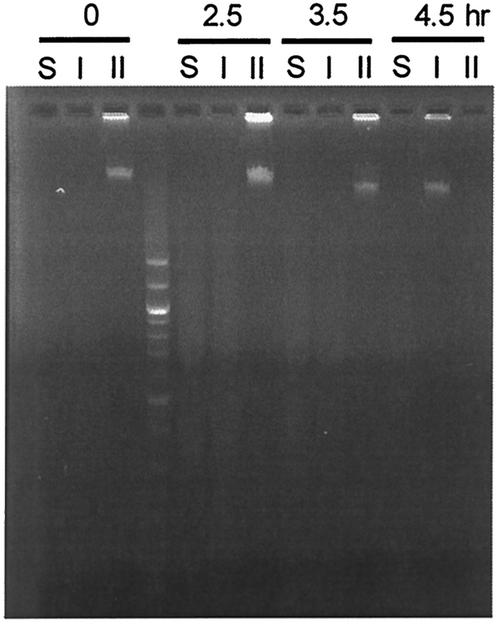
Log-phase cultures of strain GM3819 (dam-16::Kan [16]) (Table 1) bearing pMQ430 growing in L broth show no detectable methylation of chromosomal DNA in the absence of inducer (Fig. 1). Upon exposure to 0.2% arabinose, however, chromosomal DNA was completely methylated within one generation (40 min), as was determined with the restriction enzymes DpnI, DpnII, and Sau3AI (Fig. 1). Removal of the inducer after a 40-min exposure and its replacement with 0.2% glucose led to a slow reduction in chromosomal methylation requiring 4.5 h or nine generations to effect complete loss of detectable methylation (Fig. 2). It should be noted that neither DpnI nor DpnII digests hemimethylated DNA. The cells showed no decrease in growth rate (30-min generation time) or cell number during this period. The time required to demethylate chromosomal DNA was longer than expected, and we suspect that this may be due to the slow degradation of the inducer. Consequently, we have not quantitated it further by the use of Southern or Western blots.
TABLE 1.
Description of E. coli K-12 strainsa
| Strain | Relevant characteristics | Source |
|---|---|---|
| GM2807 | Hfr dam-16::Kan (PO68) thi-1 relA1 | Lab stock |
| AB1157 | F−thr-1 leuB6 thi-1 argE3 hisG4 proA2 lacY1 galK2 mtl-1 xyl-5 ara-14 rpsL31 tsx-33 supE44 rfbD1 kdgK51 | Lab stock |
| GM3819 | Like AB1157 but dam-16::Kan | Lab stock |
| GM7428 | pMQ430/N4454 | Lab Stock |
| GM7481 | pMQ430/EC18 lon::Tn10 dam-13::Tn9 | R. Woodgate |
| GM7482 | pMQ430/EC22 clpP::Kan dam-13::Tn9 | R. Woodgate |
| GM7483 | pMQ430/EC28 clpX::Kan dam-13::Tn9 | R. Woodgate |
| GM7484 | pMQ430/EC210 rcsA166::Kan dam-13::Tn9 | R. Woodgate |
| GM7485 | pMQ430/SG22099 clpA319::Kan dam-13::Tn9 | S. Gottesman |
| GM7486 | pMQ430/SG2210 clpB::Kan dam-13::Tn9 | S. Gottesman |
| N4454 | Like AB1157 but DE(ruvABC)::Cam | R. Lloyd |
The EC derivatives also carry the following markers: DE(umuDC)::Erm sulA211 thi-1 DE(lac-gpt)35 ilv(Ts) mtl-1 rpsL31. Full descriptions of dam mutant strains can be found at http://users.umassmed.edu/martin.marinus/dstrains.html.
FIG. 1.
Effect of adding inducer to log-phase cultures. Arabinose was added to a culture of pMQ430/GM3819 growing in L broth to a final concentration of 0.2% for 20 and 40 min. Cells were harvested, and total DNA was extracted and digested with Sau3A (lanes S) (cuts independently of methylation status), DpnI (lanes I) (cuts methylated DNA), and DpnII (lanes II) (cuts unmethylated DNA). First lane, 100-bp ladder.
FIG. 2.
Loss of methylation after removal of inducer. Log-phase cells of pMQ430/GM3819 were exposed to arabinose, harvested, and resuspended in L medium with glucose. The cells were kept in the logarithmic phase of growth by dilution into fresh medium. Samples were removed at 0, 2.5, 3.5, and 4.5 h. For each time point the order of digestion was Sau3A (lanes S), DpnI (lanes I), and DpnII (lanes II). Fourth lane, 100-bp ladder.
We have also tested the effect of sequentially turning Dam production on and off during cell growth. Cultures were exposed to 0.2% arabinose for 60 min, diluted, and then allowed to grow logarithmically in L broth with 0.2% glucose for 14 generations. The culture was again exposed to inducer, and the cycle was repeated two more times. We found that there was no change in growth rate or numbers of CFU during these cycling experiments. We conclude that alternating cycles of methylation and demethylation have no deleterious effects on cell growth.
Several proteases have been described for E. coli (4), and we wanted to determine if any were active in degrading Dam methyltransferase. We constructed pMQ430/dam-13::Tn9 derivatives of a series of protease-deficient strains and determined the state of DNA methylation 14 generations after a 60-min exposure to inducer. The results in Table 2 show that under these conditions, methylated DNA was found only in a Lon-deficient strain, thus implicating it in Dam methyltransferase stability. This is the first direct evidence for proteolytic degradation of Dam and confirms previous indirect data to this effect (7).
TABLE 2.
Dam methylation in protease-deficient strainsa
| Genotype | Dam methylation |
|---|---|
| Wild type | − |
| lon::Tn10 | + |
| clpA319::Kan | − |
| clpB::Kan | − |
| clpP::Kan | − |
| clpX::Kan | − |
| rcsA166::Kan | − |
Log-phase dam-13::Tn9 bacteria containing pMQ430, with or without a protease defect, were exposed to arabinose and after growth in the absence of inducer were assayed for Dam methylation in total chromosomal DNA.
We tested the utility of pMQ430 to produce Dam+ or Dam− conditions in conjugal crosses between a dam Hfr strain (GM2807 Kanr Strs [Table 1]) and a recipient strain (GM7428 Strr [Table 1]) bearing a ruvABC::Cam deletion. Logarithmic-phase cells were mixed in a ratio of 1 donor to 10 recipients (at about 108 per ml), mated for 60 min, and plated on selective media with or without arabinose. As shown in Table 3, no dam ruvABC recombinants (Kanr Strr) were recovered in the absence of arabinose, confirming previous results indicating that this combination of mutations is lethal (10). A few recombinant colonies did appear on the selective plates, but although these were dam by their kanamycin-resistant phenotype, they were also chloramphenicol sensitive, indicating the loss of the ruvABC allele and its replacement by the wild-type genes. The same low frequency of colonies was obtained when the mating mixture was plated in the presence of 0.002 and 0.02% arabinose. At a concentration of 0.2% arabinose, however, we observed a high frequency of recombinants which were sensitive to UV light and chloramphenicol resistant, indicating a ruvABC defect. These recombinants were resistant to ampicillin, indicating retention of pMQ430. They were unable to grow when they were patched onto the same selective medium without arabinose, thereby confirming that the dam ruvABC combination is inviable.
TABLE 3.
Recombinant formation in crosses between Hfr GM2807 (dam-16::Kan) and pMQ430/GM7323 (ΔruvABC::Cam Strr)a
| % Arabinose | Recombination frequency |
|---|---|
| None | 5 (Cams UVr) |
| 0.2 | 1,250 (Camr UVs) |
| 0.02 | 4 (Cams UVr) |
| 0.002 | 2 (Cams UVr) |
Bacteria were mated for 60 min and then plated for Kanr Strr recombinants on plates with various concentrations of arabinose. The numbers of recombinants in 50 μl of mating mixture are shown. The Cams UVr recombinants arise from the transfer of the wild-type ruvA, -B, and -C genes from the donor.
The viability of E. coli strains carrying the dam-16::Kan deletion mutation indicated that the loss of Dam methyltransferase is not a lethal event provided that recombination proficiency is not impaired (9). The present study with pMQ430 indicates that cells can tolerate the progressive loss of Dam methylation (and the formation of partially methylated DNA) and subsequent remethylation without any effect on growth. These findings imply that it is highly unlikely that any essential genes have their transcription coupled to hemimethylated DNA in a fashion similar to that of the transposase gene of Tn10 (17). Our results also make it highly unlikely that hemi- or partial methylation of the overabundant GATC sites in the oriC region (14) is essential for the initiation of chromosome replication. Finally, regulating transcription of the dam gene on plasmid pMQ430 is a simple way to manipulate the level of Dam methyltransferase in the cell in the absence of any temperature-sensitive mutations in the gene.
Acknowledgments
We thank Patricia Foster, S. Gottesman, R. Lloyd, and R. Woodgate for providing us with E. coli K-12 strains and Johnny Park for constructing pMQ430.
This work was supported by Public Health Service grant GM63790 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Boye, E., and A. Lobner-Olesen. 1990. The role of dam methyltransferase in the control of DNA replication in E. coli. Cell 62:981-989. [DOI] [PubMed] [Google Scholar]
- 2.Boye, E., M. G. Marinus, and A. Lobner-Olesen. 1992. Quantitation of Dam methyltransferase in Escherichia coli. J. Bacteriol. 174:1682-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, J. L., and N. Kleckner. 1990. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967-979. [DOI] [PubMed] [Google Scholar]
- 4.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 5.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lobner-Olesen, A., M. G. Marinus, and F. G. Hansen. 2003. Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proc. Natl. Acad. Sci. USA 100:4676-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maas, R. 2001. Change of plasmid DNA structure, hypermethylation, and Lon-proteolysis as steps in a replicative cascade. Cell 105:945-955. [DOI] [PubMed] [Google Scholar]
- 8.Marinus, M. G. 1996. Methylation of DNA, p. 782-791. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 9.Marinus, M. G. 2000. Recombination is essential for viability of an Escherichia coli dam (DNA adenine methyltransferase) mutant. J. Bacteriol. 182:463-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Messer, W., and M. Noyer-Weidner. 1988. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell 54:735-737. [DOI] [PubMed] [Google Scholar]
- 11.Modrich, P., and R. Lahue. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65:101-133. [DOI] [PubMed] [Google Scholar]
- 12.Nwosu, V. U. 1992. Overexpression of the wild-type gene coding for Escherichia coli DNA adenine methylase (dam). Biochem. J. 283:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oka, A., K. Sugimoto, M. Takanami, and Y. Hirota. 1980. Replication origin of the Escherichia coli K-12 chromosome: the size and structure of the minimum DNA segment carrying the information for autonomous replication. Mol. Gen. Genet. 178:9-20. [DOI] [PubMed] [Google Scholar]
- 14.Oshima, T., C. Wada, Y. Kawagoe, T. Ara, M. Maeda, Y. Masuda, S. Hiraga, and H. Mori. 2002. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol. 45:673-695. [DOI] [PubMed] [Google Scholar]
- 15.Parker, B., and M. G. Marinus. 1988. A simple and rapid method to obtain substitution mutations in Escherichia coli: isolation of a dam deletion/insertion mutation. Gene 73:531-535. [DOI] [PubMed] [Google Scholar]
- 16.Roberts, D., B. C. Hoopes, W. R. McClure, and N. Kleckner. 1985. IS10 transposition is regulated by DNA adenine methylation. Cell 43:117-130. [DOI] [PubMed] [Google Scholar]