Abstract
The Bacillus subtilis extracytoplasmic function sigma factor σY is of unknown function. We demonstrate that the sigY operon is expressed from an autoregulatory promoter site, PY. We selected for transposon-induced mutations that upregulate PY transcription in an attempt to identify genes involved in σY regulation. The resulting insertions disrupted yxlC, the gene immediately downstream of sigY. However, the phenotype of the yxlC::Tn10 insertion was due to polarity on the downstream genes of the sigY operon; a nonpolar insertion in yxlC did not lead to derepression of PY. Further analyses revealed that both yxlD and yxlE encoded proteins important for the negative regulation of σY activity. A comparison of the transcriptomes of wild-type and yxlC::Tn10 mutant strains revealed elevated expression of several operons. However, only one additional gene, ybgB, was unambiguously identified as a direct target for σY. This was supported by analysis of direct targets for σY transcription with whole-genome runoff transcription followed by macroarray analysis.
The extracytoplasmic function (ECF) σ factors function as global regulators of a variety of stress responses often triggered by changes in the cell envelope (12). In some organisms, this particular family of regulators has expanded to include large numbers of paralogues. The Bacillus subtilis genome encodes seven ECF σ factors, Mycobacterium tuberculosis encodes 10, and, remarkably, Streptomyces coelicolor encodes at least 50. In most cases, the function of these σ factors is not yet known.
In B. subtilis, most studies to date have concentrated on three of the ECF σ factors, σX, σW, and σM. The roles of these factors have been investigated by phenotypic analysis of mutant strains altered in σ activity (14, 15), identification of target operons (3, 4, 6, 17, 18, 30), and identification of signals that function to induce the various regulons (7, 28, 30, 32). The results indicate that σX controls functions associated with modification of the cell envelope, while σW and σM control overlapping regulons that are induced by antibiotics that target the cell envelope (12). The σW regulon is strongly induced by alkali shock (30), although this may be due to effects of high pH on cell wall synthesis.
Despite this progress, the roles of the other four ECF σ factors, σY, σYlaC, σV, and σZ, are still mysteries. As one approach to defining the roles of these regulators, we generated mini-Tn10 transposon libraries to identify mutants with increased expression of σY, σYlaC, σV, or σZ. In principle, selection for upregulation might identify proteins that interact directly with the operon control region (e.g., repressors) or genes that affect cell physiology in ways that trigger operon expression. In addition, since most of these operons are thought to be autoregulated by the encoded σ factor, insertions might identify negative regulators of σ factor activity (e.g., anti-σ factors).
Here we report the characterization of mutants that are derepressed for expression of sigY. We identified an insertion mutation, yxlC::Tn10, that activated expression of σY-dependent genes, including the autoregulated sigYyxlCDEFG operon. Using a combination of molecular genetic and genomic approaches, we identified genes within the sigY operon that regulate the activity of σY and characterized two σY target promoters.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All B. subtilis and Escherichia coli strains used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) medium at 37°C with vigorous shaking. Antibiotics were added to the growth medium when appropriate to 100 μg/ml for ampicillin or 200 μg/ml for spectinomycin for E. coli and 100 μg/ml for spectinomycin, 10 μg/ml for kanamycin, 20 μg/ml for tetracycline, 8 μg/ml for neomycin, and 1 μg/ml for erythromycin plus 25 μg/ml for lincomycin (for macrolide-lincomycin-streptogramin B resistance) for B. subtilis.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| B. subtilis | ||
| CU1065 | W168 trpC2 attSPβ | Lab stock |
| ZB307A | W168 SPβc2Δ2::Tn917::pSK10Δ6 | 33 |
| HB4245 | JH642 but sigY::mls | 16 |
| HB0009 | CU1065 but sigY::mls | This work |
| HB0060 | ZB307A SPβ(PY-cat-lacZ) | This work |
| HB0065 | CU1065 SPβ(PY-cat-lacZ) | This work |
| HB0061 | ZB307A SPβ(PV-cat-lacZ) | This work |
| HB0067 | CU1065 SPβ(PV-cat-lacZ) | This work |
| HB0063 | ZB307A SPβ(PZ-cat-lacZ) | This work |
| HB0071 | CU1065 SPβ(PZ-cat-lacZ) | This work |
| HB0064 | ZB307A SPβ(PylaA-cat-lacZ) | This work |
| HB0073 | CU1065 SPβ(PylaA-cat-lacZ) | This work |
| HB0120 | HB0065 but yxlC::Tn10 (Spcr) | This work |
| HB0119 | HB0120 cured of SPβ | This work |
| HB0121 | HB0119 but sigY::mls | This work |
| HB0122 | HB0121 SPβ(PY-cat-lacZ) | This work |
| HB0915 | CU1065 but yxlC::mls | This work |
| HB0917 | HB0915 SPβ(PY-cat-lacZ) | This work |
| HB0916 | CU1065 but yxlCDEFG::kan | This work |
| HB0918 | HB0916 SPβ(PY-cat-lacZ) | This work |
| HB5302 | CU1065 but yxlFG::kan | This work |
| HB5303 | HB5302 SPβ(PY-cat-lacZ) | This work |
| HB5306 | CU1065 but yxlCDE::tet | This work |
| HB5307 | HB5306 SPβ(PY-cat-lacZ) | This work |
| HB5308 | CU1065 but yxlDE::tet | This work |
| HB5309 | HB5308 SPβ(PY-cat-lacZ) | This work |
| HB5310 | CU1065 but yxlD::tet | This work |
| HB5311 | HB5310 SPβ(PY-cat-lacZ) | This work |
| HB5312 | CU1065 but yxlE::tet | This work |
| HB5313 | HB5312 SPβ(PY-cat-lacZ) | This work |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Lab stock |
| Jm2r− | mcrAB hsdR recA1 Δ(lac-proAB) thi gyrA96 relA1 srl::Tn10 F′ (proAB lacZΔM15) | S. Zahler |
| Plasmids | ||
| pJPM122 | Vector for integration of reporter fusions in SPβ (Apr Neor) | 26 |
| pMC80 | PY-cat-lacZ cloned in pJPM122 | This work |
| pIC333 | Shuttle vector carrying mini-Tn10 (Apr MLSr Spcr) | 27 |
| pXT | Derivative of pDG1731 | 3 |
| pDG646 | Macrolidelincomycin-streptogramin B (MLS) resistance cassette vector | 10 |
| pDG780 | Kanamycin resistance cassette vector | 10 |
| pDG1513 | Tetracycline resistance cassette vector | 10 |
Construction of transcriptional fusions and sigY mutant.
The sigY promoter region was amplified from B. subtilis chromosomal DNA by PCR with primers 339 and 340 (Table 2). The resulting fragment was digested with HindIII and BamHI and cloned into pJPM122 (26) to generate plasmid pMC80 (PY-cat-lacZ). The sequence of the promoter region was verified by DNA sequencing (Cornell DNA sequencing facility). The promoter fusion was introduced into the SPβ prophage by a double-crossover event, in which plasmid pMC80 was linearized with ScaI and transformed into B. subtilis strain ZB307A (33) with selection for neomycin resistance. SPβ lysates were prepared by heat induction as described (9) and used to transduce CU1065 to generate strain HB0065 (PY-cat-lacZ). Reporter fusions for PX, PW, and PM have been described (4, 15, 16). Reporter fusions for putative sigV, ylaC, and sigZ promoters were constructed with a similar strategy (Tables 1 and 2). The sigY mutant HB0009 was constructed by transforming chromosomal DNA from HB4245 (sigY::MLS) (16) into CU1065.
TABLE 2.
Oligonucleotides used in this studya
| No. | Name | Sequence |
|---|---|---|
| 339 | sigY fwd (pJPM122) | 5′-GGCCCAAGCTTCGCCTTTCTACTTTCAATGC-3′ |
| 340 | sigY rev (pJPM122) | 5′-CGCGGATCCGCCGCTGTTCTTCTTGTGTAT-3′ |
| 335 | sigV fwd (pJPM122) | 5′-CCCCAAGCTTGGAATAATGTTTGTCATC-3′ |
| 437 | sigV rev (pJPM122) | 5′-CGGGATCCTTGCTTATGGTCAGTTATGCA-3′ |
| 438 | ylaA fwd (pJPM122) | 5′-CCCAAGCTTGATAGTATTGTCCTGTGT-3′ |
| 439 | ylaA rev (pJPM122) | 5′-CGGGATCCATTGAATCAGCAGGGTGCTTT-3′ |
| 440 | sigZ fwd (pJPM122) | 5′-CCCAAGCTTTTGTCGCCAGAACAA-3′ |
| 441 | sigZ rev (pJPM122) | 5′-CGGGATCCAACGGCTGATGAAATTGATCC-3′ |
| 50 | mini-Tn10 (left) | 5′-GCCGATTCATTAATGCAG-3′ |
| 51 | mini-Tn10 (right) | 5′-CCCACTTATAAACAAAG-3′ |
| 776 | ybgB fwd | 5′-GGAAGCTTAAGGACAAAATACAA-3′ |
| 777 | ybgB rev | 5′-AAGGATCCGGCAGAAAAGGGTAAA-3′ |
| 141 | sigY-f (pET16x) | 5′-GGGGTACCATGGATACACAAGAAGAACAG-3′ |
| 142 | sigY-r (pET16x) | 5′-CGGGATCCTTATTCATCATCCCACTCCT-3′ |
| 1295 | kan fwd | 5′-CAGCGAACCATTTGAGGTGATAGG-3′ |
| 1296 | kan rev | 5′-CGATACAAATTCCTCGTAGGCGCTCGG-3′ |
| 1297 | mls fwd | 5′-GATCCTTTAACTCTGGCAACCCTC-3′ |
| 1298 | mls rev | 5′-GCCGACTGCGCAAAAGACATAATCG-3′ |
| 941 | tet fwd1 | 5′-TCTTGCAATGGTGCAGGTTGTTCTC-3′ |
| 942 | tet fwd2 | 5′-GCTTATCAACGTAGTAAGCGTGG-3′ |
| 940 | tet rev | 5′-GAACTCTCTCCCAAAGTTGATCCC-3′ |
| 1301 | yxlC-up fwd | 5′-GGCTTTGAATCATTTGCGGGATGCCTAGC-3′ |
| 1302 | yxlC-up rev (mls) | 5′-CATTCAATTTTGAGGGTTGCCAGGATTCGGTATAGAGGGATTGGC-3′ |
| 1304 | yxlC-dw fwd (mls) | 5′-CGATTATGTCTTTTGCGCAGTCGGCGGCATCTCGGCGAATGCGAG-3′ |
| 1305 | yxlC-dw rev | 5′-CACACACCTGTTCTGCATCGTGC-3′ |
| 1303 | yxlC-up rev (kan) | 5′-CCTATCACCTCAAATGGTTCGCTGGTGACAGCTGGTGAAGCAG-3′ |
| 1306 | yxlG-dw fwd (kan) | 5′-CGAGCGCCTACGAGGAATTTGTATCGTGCCAGAGAACCGGCTCC |
| 1307 | yxlG-dw rev | 5′-CGGCATCATTCTCGGCAGCTACGG-3′ |
| 936 | yxlF-up fwd | 5′-GCATCGCGGCTCTATCTCGATCACC-3′ |
| 937 | yxlF-up rev (kan) | 5′-CACCTCAAATGGTTCGCTGGTTTACAGCTTCATGGTGCCTGTACG-3′ |
| 939 | yxlG-up rev (kan) | 5′-CACCTCAAATGGTTCGCTGGTCCAGCCACTCCTTCTGCAATAGCG-3′ |
| 943 | yxlC-up rev (tet1) | 5′-GAACAACCTGCACCATTGCAAGATTCGGTATAGAGGGATTGGC-3′ |
| 945 | yxlE-dw fwd (tet) | 5′-GGGATCAACTTTGGGAGAGAGTTCAGCAAAGGTAAGCCGATATGC-3′ |
| 935 | yxlE-dw rev | 5′-CAGGAAGGTTCCCTCCATGTGCG-3′ |
| 1445 | yxlD-up rev (tet1) | 5′-GAACAACCTGCACCATTGCAAGACCAATACGATGAGACAAGCC-3′ |
| 1447 | yxlD-dw fwd (tet) | 5′-GGGATCAACTTTGGGAGAGAGTTCTCAAGCACCGATGCCTCTG-3′ |
| 1448 | yxlE-up rev (tet2) | 5′-CCACGCTTACTACGTTGATAAGCCTTCCGATCCGGCCTAATGAC-3′ |
| 1446 | yxlD-up rev (tet2) | 5′-CCACGCTTACTACGTTGATAAGCGTCATCCGCGTTTCACCTCGC-3′ |
The underlined sequences correspond to the 5′ and 3′ ends of the drug resistance cassette used in each construction.
Construction of mini-Tn10 libraries and identification of mutants upregulated in σY activity.
B. subtilis strain HB0065 was transformed with pIC333 (27) to generate random mini-Tn10 libraries as described previously (28). Nine libraries were generated and plated onto LB containing spectinomycin, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and growth-inhibitory levels (6, 8, or 10 μg/ml) of chloramphenicol (Cm). Cmr mutants with elevated β-galactosidase activity were isolated following 2 days of incubation at 37°C. Chromosomal DNA was extracted from each mutant and used to transform HB0065, with selection on LB plates with spectinomycin (Spc), neomycin, and X-Gal. Only mutants that had a high level of linkage between the mini-Tn10(spc) and elevated expression of β-galactosidase expression were characterized further. Plasmids containing the mini-Tn10 element with a ColE1 origin and flanking B. subtilis chromosomal DNA were recovered by transformation into E. coli. DNA sequences upstream and downstream of the transposon were obtained with two primers (50 and 51) corresponding to the left and right ends of the mini-Tn10, respectively. We generated a sigY yxlC double mutant (HB0121) by transformation of chromosomal DNA from HB4245 (sigY::mls) into HB0119 (HB0120 cured of SPβ; Table 1). The PY-cat-lacZ fusion was then introduced into this strain by transduction, and β-galactosidase was measured.
Construction of null mutants of yxlC, yxlCDEFG, yxlFG, yxlCDE, yxlDE, yxlD, and yxlE.
Long-flanking homology PCR was used as described (29) to generate allelic replacement mutants for each gene or group of genes. In brief, approximately 1,000-bp genomic regions flanking the gene(s) to be deleted were amplified from CU1065 chromosomal DNA by PCR. The primers used are summarized in Table 2. Drug resistance cassettes were amplified by PCR from pDG646 (macrolide-lincomycin-streptogramin B, mls), pDG780 (kanamycin, kan), or pDG1513 (tetracycline, tet) (10).
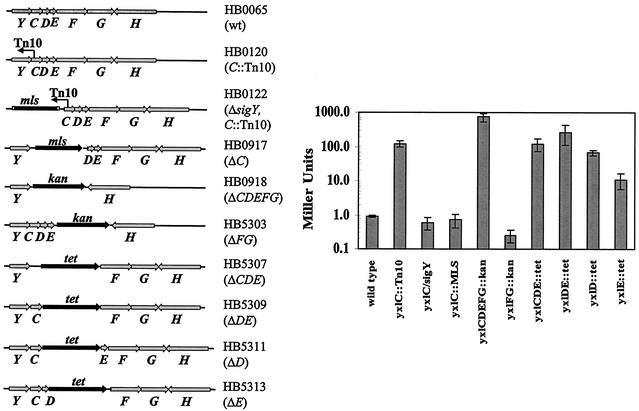
For each mutant construction, equal amounts (approximately 200 to 300 ng) of purified upstream flanking fragment, downstream flanking fragment, and the corresponding drug resistance cassette were used in a joint PCR procedure as described (29), with either the Expand polymerase (Roche) or the HotStarTaq Master Mix kit (Qiagen). The resulting PCR products were purified and then directly transformed into B. subtilis wild-type strain CU1065, selecting for the corresponding antibiotic resistance. The generated mutant strains are listed in Table 1 and shown in Fig. 1.
FIG. 1.
Genetic analysis of sigY operon. The genetic organization of the sigY-yxlCDEFG mutations used in these studies is illustrated. The corresponding level of β-galactosidase synthesis (mean ± standard deviation; n = 3) for the PY-cat-lacZ reporter fusion is shown to the right. wt, wild type.
β-Galactosidase assay.
In preliminary studies, overnight cultures were diluted 1:100 into 15 ml of LB medium. Samples were taken when the optical density at 600 nm (OD600) reached 0.1, 0.2, 0.4, 0.6, 0.8, and 1.0 and 1 h after it reached 1.0. The β-galactosidase activity of each sample was measured according to Miller (20). At an OD600 of 0.8 (late log phase), PY-cat-lacZ expression reached its maximum level.
To compare different strains, three individual colonies were inoculated in LB medium with corresponding antibiotics and incubated at 37°C overnight. Then 50 μl of each overnight culture was used to inoculate 5 ml of warm LB medium. Samples were taken at an OD600 of 0.8, and β-galactosidase activity was assayed. Averages and standard deviations were calculated for each strain.
Primer extension assays.
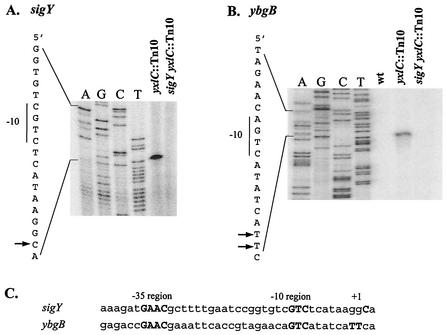
RNA was prepared from mid-logarithmic-phase cells (OD600 ≈ 0.5) with the Qiagen RNeasy mini kit; 100 μg of total RNA (from CU1065 or the sigY yxlC double mutant strain) or 10 μg of total RNA (from the yxlC::Tn10 mutant) and 2 pmol of end-labeled reverse primer were mixed for each primer extension experiment following the procedures described previously (4). For mapping the sigY transcriptional start site, the end-labeled reverse primer 340 was used. The PCR-amplified sigY promoter region (with primers 339 and 340) was sequenced with the same primer, and the reaction products were electrophoresed adjacent to the primer extension products. For ybgB, primer 777 was used, and primers 776 and 777 were used for amplification of the ybgB promoter region for the sequence ladder.
Microarray analysis.
Total RNA was prepared from B. subtilis CU1065 and the yxlC::Tn10 mutant grown aerobically in LB medium. The cell cultures were grown to an OD600 of 0.4, and the cells were harvested immediately. The protocol for RNA isolation, cDNA synthesis, and slide hybridization was described previously (31). Each RNA preparation was used to make both indocarbocyanine- and indodicarbocyanine-labeled cDNA, and all hybridizations were done twice, once with each cDNA preparation, to control for differences in labeling between the two fluorophores. Since all PCR products were spotted twice on each slide, all signal intensities and calculated ratios are the averages of four values. Two microarray experiments (yxlC mutant versus wild type) were performed with RNA prepared from two independent cell cultures. Signal intensities were quantified with ArrayVision software (Molecular Dynamics) and assembled into Excel spreadsheets. Mean values and standard deviations were calculated with Excel. Genes with a standard deviation in expression values (fluorescence intensity) greater than the mean value were ignored. Complete datasets are available as supplementary material at http://www.micro.cornell.edu/faculty.JHelmann.html.
Overproduction and purification of σY protein.
The sigY gene was PCR amplified from B. subtilis chromosomal DNA with oligonucleotides 141 and 142, designed to engineer an NcoI site upstream and a BamHI site downstream of the sigY gene. The PCR product was cloned into pET16x (Novagen) via the NcoI and BamHI sites to generate pKF85. The sequence of sigY in pKF85 was verified by DNA sequencing (Cornell DNA sequencing facility). σY was purified from E. coli strain BL21/DE3(pLysS) transformed with pKF85. Expression was induced by addition of 20 μM isopropylthiogalactopyranoside (IPTG) for 3 h, resulting in the formation of inclusion bodies. σY was purified from the inclusion bodies as follows: 2 ml of disruption buffer (50 mM Tris-HCl [pH 8.0], 2 mM EDTA, 0.1 mM dithiothreitol, 1 mM β-mercaptoethanol, 233 mM NaCl, 10% glycerol) was added to a frozen pellet generated from 50 ml of induced culture. Then 0.4 ml of the resuspended cells was sonicated for 5-s pulses, 12 pulses total.
Inclusion bodies were collected by centrifugation at 13,000 rpm for 20 min at 4°C and resuspended twice in 10 ml of TEDG buffer(50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 0.5% [vol/vol] Triton X-100). The washed pellet was resuspended in 2 ml of TEDG with 0.4% Sarkosyl and gradually diluted to 20 ml with TEDG buffer to allow refolding of σY. The sample was then dialyzed against 200 ml of TEDG for 8 h at 4°C. A 1-ml Hi-trap heparin column was equilibrated with 3 ml of TEDG, and 1 ml of the dialyzed sample was loaded onto the column. The column was washed five times with 500 μl of TEDG. σY was eluted from the column with washes of increasing NaCl concentrations (50 to 500 mM NaCl) in TEDG buffer. Each eluate was tested for the presence of protein with the Bio-Rad protein detection assay, and peak fractions were collected. The renatured σY eluted with ≈0.5 M NaCl and was analyzed by polyacrylamide gel electrophoresis (PAGE) and confirmed to migrate at approximately 21.2 kDa, which is the predicted molecular mass for σY.
In vitro runoff transcription assay and microarray analysis.
The runoff transcription/macroarray analysis (ROMA) experiment was performed as described previously (6). Purified σY was added in 17-fold molar excess relative to the core RNA polymerase. With the σY autoregulated promoter as a template, we determined that the specificity of σY-dependent transcription was optimal between 100 and 150 mM KCl (data not shown). For the ROMA experiment, 100 mM KCl (final concentration) was used.
RESULTS
Characterization of mini-Tn10 mutants with elevated expression of sigY.
We used the presumptive sigY regulatory region to generate a cat-lacZ operon fusion (sigY′-cat-lacZ) integrated ectopically into the SPβ prophage. The resulting fusion was expressed weakly if at all under a variety of laboratory growth conditions, suggesting that the signals that normally activate expression of sigY were not present. We selected for Tn10(spc) insertions that led to chloramphenicol resistance. We identified five Cmr mutants from three independent libraries (approximately 10,000 transposants) that had dramatically increased β-galactosidase activity. In each case, these phenotypes were tightly linked to the spc marker associated with the transposon. DNA linked to each transposon was recovered by transformation into E. coli, and the sites of insertion were determined by DNA sequencing. All five transposants had the Tn10(spc) insertion at the same position and in the same direction within yxlC, the gene immediately downstream of sigY (HB00120, Fig. 1). Expression of sigY′-cat-lacZ in the yxlC::Tn10 mutant was increased more than 100-fold compared to the wild type, as measured during late logarithmic growth (Fig. 1).
σY is positively autoregulated.
Most ECF σ factors are positively autoregulated, often with an adjacent anti-σ factor gene that regulates activity (11, 12, 19). To determine whether sigY is autoregulated, we took advantage of the high level of expression from the sigY′-cat-lacZ reporter fusion in the yxlC::Tn10 mutant. When a sigY mutation was introduced into this genetic background, expression was reduced to the background level (Fig. 1, strain HB0122). Thus, the sigY′-cat-lacZ reporter fusion is also a reporter of σY-dependent transcriptional activity, PY-cat-lacZ.
The upstream region of sigY contains a candidate promoter similar to other promoters recognized by ECF σ factors (13). We mapped the transcription start site of sigY by primer extension, taking advantage of the high level of expression in the yxlC::Tn10 mutant. Transcription started from a C residue 9 bases downstream from the −10 region CGTC motif (Fig. 2A). Consistent with the β-galactosidase result, this transcript was not detectable in the sigY yxlC double mutant even when 10 times more total RNA was used as the template. No other start sites were observed within the sigY regulatory region (≈250 bp upstream from the start codon). We conclude that σY positively autoregulates its own expression.
FIG. 2.
Primer extension mapping of sigY (A) and ybgB (B) transcription start sites. RNA was extracted from mid-log-phase cells of strains CU1065 (wild type, wt), HB0119 (yxlC::Tn10), and HB0121 (sigY yxlC double mutant) in LB medium. The putative −10 regions are indicated, and the transcription start sites are shown by arrows. (C) Alignment of the σY autoregulated promoter sequence with the ybgB promoter region. Conserved bases and transcription start sites (+1) are in bold uppercase type.
The σY autoregulatory promoter PY has consensus elements of TGAAC (−35) and CGTC (−10) with a 17-bp spacer (Fig. 2C). This is very similar to the consensus sequences recognized by other B. subtilis ECF σ factors, including σW and σX (Fig. 3). Although both σX and σW can recognize promoters with a CGTC motif in the −10 region (25), sigY has not been identified as part of either the σX or σW regulon (5, 6, 18), nor is sigY upregulated by the induction of other ECF σ factors (2; our unpublished results). These results suggest that other sequence features, in addition to those highlighted (Fig. 3), are important for promoter discrimination.
FIG. 3.
Known autoregulated promoter regions of σY (PY), σX (PX), σW (PW), and σM (PM) were compared with the putative promoter sequences just upstream of the genes for σV (sigV), YlaA (ylaA) (the first gene in the ylaABCD operon; ylaC encodes σYlaC), and σZ (sigZ). The −35 and −10 regions are in uppercase, with conserved bases in bold. Mapped transcription start sites (+1) are in bold uppercase.
σY does not activate transcription of other ECF σ factors.
Next, we tested the effect of the yxlC::Tn10 insertion (leading to high in vivo σY activity) on the expression of autoregulatory promoters recognized by various ECF σ factors. We replaced the PY-cat-lacZ fusion in strain HB0120 with reporter fusions containing the known or putative autoregulatory regions for each of the other six ECF σ factors (Fig. 3 and data not shown). The results indicate that the yxlC::Tn10 insertion and consequent upregulation of σY activity do not lead to elevated expression of any of the other ECF σ factors. We conclude that, in general, σY does not regulate other ECF σ factors.
Effect of yxlC::Tn10 insertion is due to polarity.
In several well-characterized examples, the gene immediately downstream of an ECF σ factor gene encodes an anti-σ factor (12). Therefore, we hypothesized that yxlC might encode an anti-σ factor. However, when we engineered a yxlC::mls allelic replacement mutant (HB0917), we failed to observe an increase in σY activity (Fig. 1). Note that in this mutant the mls cassette was oriented to allow expression of downstream genes from the mls promoter. In light of this result, we hypothesized that the original Tn10 insertion was polar on downstream genes. sigY is the first of six codirectional genes, many with overlapping start and stop codons (Table 3), that likely constitute an operon.
TABLE 3.
The σY regulon
| Protein | No. of amino acids | Loca- tiona | Positions on genome | Known or putative function |
|---|---|---|---|---|
| SigY | 178 | C | 3969867-3969334 | RNA polymerase ECF-type sigma factor |
| YxlC | 106 | M | 3969338-3969021 | Unknown |
| YxlD | 68 | M | 3969021-3968818 | Unknown |
| YxlE | 62 | M | 3968818-3968633 | Unknown |
| YxlF | 295 | C | 3968623-3967739 | Similar to ABC transporter (ATP-binding protein) |
| YxlG | 259 | M | 3967739-3966963 | Unknown |
| YbgB | 91 | M | 258520-258792 | Unknown |
C, predicted cytoplasmic localization; M, predicted membrane protein.
σY is negatively regulated by both YxlD and YxlE.
To determine which of the four downstream genes might have been affected by the yxlC::Tn10 insertion mutation, we constructed a series of allelic replacement mutants (Fig. 1). When the whole yxlCDEFG region was deleted, PY was derepressed (HB0918), indicating that σY is negatively regulated by one or more proteins encoded by the downstream genes. High-level expression from PY was also observed in the ΔyxlCDE but not the ΔyxlFG mutant. We conclude that YxlF and YxlG are not essential for PY regulation but might function as accessory factors, because expression was always lower in the ΔyxlCDE than in the ΔyxlCDEFG mutant.
To investigate the role of individual gene products of the yxlCDE region, three additional deletions (ΔyxlDE, ΔyxlD, and ΔyxlE) were constructed (Fig. 1). The results indicate that both YxlD and YxlE are important for negative regulation of σY activity, with YxlD being the major negative regulator. Note that both YxlD and YxlE are small proteins predicted to associate with the cell membrane (Table 3). Thus, the signaling complex likely to regulate σY activity may be membrane localized.
σY regulon includes sigY operon and ybgB gene.
To identify other genes transcribed by σY, we used DNA microarray analysis to compare RNA populations from wild-type and yxlC::Tn10 mutant cells in two independent experiments. Overall, ≈99.5% of the expressed genes varied less than twofold in expression level, despite the nearly 100-fold effect of the yxlC insertion on expression of sigY itself. Only seven genes were significantly and reproducibly upregulated (>3.5-fold) in the yxlC mutant (Table 4). The most dramatic changes were the sigY (71-fold) and the yxlC (14-fold) genes. The interpretation of this finding is complicated by the fact that the mutant strain has a Tn10 insertion in the yxlC gene and the upregulation noted in the microarray study could, in principle, be due to countertranscription from the promoter of the spectinomycin resistance cassette in the Tn10 insertion (Fig. 1). Nevertheless, it is clear from the reporter fusion studies that the yxlC::Tn10 insertion leads to upregulation of PY. Therefore, we prefer a model in which the Tn10 insertion leads to upregulation of PY, which leads to elevated levels of sigY and the 5′-proximal part of yxlC.
TABLE 4.
Genes that were induced >2-fold in the yxlC::Tn10 mutanta
| Gene | Induction (fold ± SD)
|
Avg induction (fold) | |
|---|---|---|---|
| Set 1 | Set 2 | ||
| sigY | 95.4 ± 21.0 | 46.8 ± 3.7 | 71.1 |
| yxlC | 17.0 ± 4.6 | 11.0 ± 2.8 | 14.0 |
| ybgB | 9.0 ± 4.6 | 6.6 ± 0.7 | 7.8 |
| ybgE | 6.1 ± 1.8 | 5.9 ± 1.7 | 6.0 |
| yvdF | 9.0 ± 2.5 | 5.9 ± 1.9 | 7.4 |
| des | 6.0 ± 1.6 | 4.4 ± 0.4 | 5.2 |
| tyrZ | 4.1 ± 1.0 | 3.5 ± 0.7 | 3.8 |
Complete datasets are available as supplementary material at http://www.micro.cornell.edu/faculty.JHelmann.html. Set 1 and set 2 are the results from two microarray experiments (yxlC versus wild type) with RNA prepared from two independent cell cultures.
Five other genes that were significantly upregulated in the yxlC mutant were ybgB, ybgE, yvdF, des and tyrZ. YbgB is a small hydrophobic protein (91 amino acids) of unknown function, while YbgE is similar to a branched-chain amino acid aminotransferase. Using primer extension, we mapped the ybgB transcription start site and confirmed that this gene is σY dependent (Fig. 2B). It is not yet known if the upregulation of ybgE is physiologically relevant, since it is separated from ybgB by a 212-bp intergenic region. This region has recently been shown to bind CodY, and ybgE is derepressed in a codY mutant (22). We suggest that the upregulation of ybgE is due to readthrough of the ybgB transcript into ybgE, which is otherwise repressed under our growth conditions.
Apart from ybgB, none of the other upregulated genes appeared to be direct targets for σY-directed transcription. The des gene encodes a cold-inducible membrane phospholipid desaturase, and its transcription is controlled by σA (1). tyrZ is a monocistronic gene encoding a minor tyrosyl-tRNA synthetase. No obvious σY-dependent promoter was found upstream of tyrZ, and we failed to detect its transcription start site even in the yxlC mutant. The yvdF gene encodes a putative maltogenic amylase (98% identical to the BbmA sugar hydrolase from B. subtilis SUH4-2 [8]) and is the second gene of a large cluster. Most genes in this cluster seem to be involved in maltose or maltodextrin utilization. However, no obvious σY-dependent promoter was found upstream of either yvdF or the larger gene cluster, and we were unable to detect any transcription start site for yvdF in primer extension experiments. Since only the second gene in this region was induced in the yxlC mutant, this could be a false-positive, the result of cross-hybridization, or an indirect effect.
Analysis of σY regulon by ROMA.
With the microarray approach, we identified two operons (sigY-yxlCDEFG and ybgB) as direct targets for σY-directed transcription. As a complementary approach, we used reconstituted σY holoenzyme to identify in vitro targets in a whole-genome transcription study. As described previously (6), in the runoff transcription/macroarray analysis (ROMA) experiment, we generated 33P-labeled runoff transcripts with total genomic DNA and used these to probe a DNA macroarray (Sigma/GenoSys) containing 4,107 B. subtilis open reading frames.
We detected only three strong hybridization signals by ROMA: sigY, ybgB, and yabE (Fig. 4). In contrast, dozens of strong signals were detected when the σW or σX holoenzyme was used (5, 6). Hybridization to yabE is apparently due to an RNA generated by a σ factor contaminating the core preparation (E), since the signal appeared in both E and EσY experiments. Additional weak signals generated in the EσY experiment but lacking in the E alone experiment were also identified. These signals correspond to ybgE, the downstream gene of ybgB, and the genes downstream of sigY. These signals were reduced in intensity due to the use of restriction enzyme-digested DNA in the in vitro transcription reaction, which served to limit transcription to promoter-proximal genes. Significantly, the other three genes (yvdF, des, and tyrZ) that were induced in the yxlC::Tn10 mutant (Table 4) were not detected in ROMA, consistent with our conclusion that these are not likely to be direct targets for σY.
FIG. 4.
Total B. subtilis chromosomal DNA was digested with EcoRI and transcribed in vitro with either the core alone (E) or the core with an excess of σY (EσY). Signals generated specifically by the σY holoenzyme belong to either the sigYyxlCDEFG or ybgB operon (ovals). The yabE gene (rectangle) also gave a strong signal in the control experiment (E). The positions of the identified genes (on the panorama macroarray) are listed beside each gene. Similar results were obtained in a replicate experiment with HindIII-digested B. subtilis chromosomal DNA as the transcription template.
In summary, both transcriptional profiling and ROMA experiments suggest that σY directs transcription of a small regulon including only the sigY-yxlCDEFG and the ybgB genes (Table 3). Note that most of these gene products are small, hydrophobic proteins, suggestive of a role in transport or other membrane-associated functions.
DISCUSSION
The ECF σ factors activate a variety of stress responses that often involve changes in the cell envelope or transport or efflux across the cell membrane (reviewed in references 12, 21, and 24). In B. subtilis, significant progress has been made in defining the regulons controlled by σX, σW, and σM, but the functions of the other four ECF σ factors are unknown (13). Here we demonstrate that σY controls a small regulon, including its own operon, and at least one other target gene, ybgB. However, the function of this regulon is not yet clear.
We have explored several strategies to decipher the regulatory roles of the multiple ECF σ factors in B. subtilis. Mutational analyses revealed that none of the seven σ factors is essential, and even multiply mutant strains often have only subtle phenotypes. Therefore, we focused our efforts on identifying target genes that are recognized by each σ factor with promoter consensus search, DNA microarray, and in vitro transcription-based strategies (3, 4, 6, 7, 15-18). In complementary experiments, we attempted to define the chemical and genetic factors that elicit σ factor activation (28). Together, these studies revealed that σX controls several operons that modulate cell envelope properties, including the d-alanylation of teichoic acids (dltABCDE), phosphatidylethanolamine biosynthesis (pssA psd), and expression of autolysins (lytR) (4, 5, 18). The σW regulon includes at least 30 operons, including several with roles in antibiotic resistance (3, 6). This regulon is induced by antibiotics acting on the cell wall or by alkali stress (7, 30). The regulon controlled by σM has not been well defined, but it appears to overlap the σX and σW regulons and includes at least one gene that functions in antibiotic resistance (bcrC) (4, 23).
Several different strategies have also been explored to define the role of σY. Direct comparison of the transcriptomes of wild-type and sigY null mutant strains did not reveal significant differences (data not shown). One interpretation of this result is that σY may not be active under the conditions of the experiment, and therefore very few genes (if any) were affected by the absence of the σ factor. For both the σX and σW regulons, the rules defining promoter recognition are reasonably well defined (25), and searching the genome for sequences resembling known target sites produced lists of candidate promoters, many of which turned out to be dependent on the expected σ factor (6, 17, 18). Similar search strategies have not been as successful for other ECF σ factors. While it is clear that the two known promoters recognized by σY are similar in sequence (Fig. 2C), searches based on the apparent consensus identified sites already classified as dependent on σX, σW, or both but very few additional candidates (data not shown). It remains possible that the σY regulon may overlap that recognized by σX, σW, or another ECF σ factor. However, this suggestion is not supported by either the in vivo transcriptome analysis or the ROMA studies reported here.
The present work was initiated with the goal of defining the genetic factors that negatively regulate σY activity. In similar studies with the autoregulatory σX- and σW-dependent promoters, we identified transposon insertions in genes for antibiotic biosynthesis, sugar isomerases, and multidrug efflux systems (28). With only one exception, the insertions affected σX or σW, but not both. Of those tested to date, none of these insertions affected σY activity (data not shown). Moreover, σY is not strongly activated by a variety of physical or chemical factors that activate σX, σW, and σM (e.g., antibiotics, high salt concentrations, and extreme pH). Weak activation of sigY expression is observed in cells grown on minimal medium compared to rich medium, and this expression is σY dependent. Thus, the σY regulon appears to respond to different stresses than those known to activate other σ regulons.
In the present study, we only recovered insertions in the sigY operon itself, which focused attention on the regulatory roles of these cotranscribed genes. Our results indicate that the major negative regulators of σY activity, YxlD and YxlE, are two small membrane proteins. It is not yet clear whether these two proteins together form a multisubunit anti-σ factor or whether they act independently. Since their genes are cotranscribed with a predicted component of an ABC transporter (Table 3), we speculate that σY may be regulated by the activity of a membrane transport complex.
The dramatic effect of the yxlC::Tn10 insertion mutation on the activity of the σY autoregulatory promoter encouraged us to pursue global transcriptional profiling to identify other operons upregulated by elevated σY activity. Of the resulting candidate operons, only ybgB was clearly a direct target for σY-dependent transcription, and the function of this gene is not clear. Asai et al. (2) also used transcriptome analysis to define the regulons controlled by ECF σ factors. Their results, based on overexpression of individual ECF σ factors, support the idea that σY positively autoregulates its own expression and that of the downstream genes (yxlCDEFG and yxlH). The apparent upregulation of yxlH may be due to readthrough from the convergent sigY operon. Although these authors reported 10 additional genes as being upregulated by induction of σY, only one (tyrZ) was also identified in our comparison of wild-type and yxlC::Tn10 mutant strains, and our results suggest that the induction of many of these reported target genes may be due to indirect effects. It should be noted, for example, that their studies were done by inducing each σ factor and harvesting cells after 2 h of growth at 37°C, during which time both the control and experimental cultures likely entered the stationary phase.
Transcriptional profiling is a very powerful approach for defining the effects of regulatory proteins on gene expression, but some target operons may be missed. This can occur due to low expression levels (e.g., due to the inactivity of a needed activator), poor hybridization to target probes, and background expression from other promoter sites. Moreover, it is difficult to separate direct from indirect effects. As an independent approach to estimating the size of the σY regulon, we turned to ROMA, whole-genome in vitro transcription to generate σY-dependent transcripts followed by macroarray analysis to identify the corresponding genes. The results confirmed the transcriptional profiling studies; in both cases, the direct targets of σY regulon appeared to include only two operons, sigY-yxlCDEFG and ybgB. Further studies will be needed to define the physiological roles of these genes and their products.
Acknowledgments
We thank Kurt Fredrick for constructing the pKF85 σY overproduction plasmid.
This work was supported by NIH grant GM-47446 (to J.D.H.), NIH MBRS SCORE grant GM-052588 (to L.M.-M.), and NIH MBRS RISE grant GM5-59298 (to C.B.).
REFERENCES
- 1.Aguilar, P. S., P. Lopez, and D. de Mendoza. 1999. Transcriptional control of the low-temperature-inducible des gene, encoding the delta5 desaturase of Bacillus subtilis. J. Bacteriol. 181:7028-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai, K., H. Yamaguchi, C. M. Kang, K. Yoshida, Y. Fujita, and Y. Sadaie. 2003. DNA microarray analysis of Bacillus subtilis sigma factors of extracytoplasmic function family. FEMS Microbiol. Lett. 220:155-160. [DOI] [PubMed] [Google Scholar]
- 3.Cao, M., B. A. Bernat, Z. Wang, R. N. Armstrong, and J. D. Helmann. 2001. FosB, a cysteine-dependent fosfomycin resistance protein under the control of σW, an extracytoplasmic function sigma factor in Bacillus subtilis. J. Bacteriol. 183:2380-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, M., and J. D. Helmann. 2002. Regulation of the Bacillus subtilis bcrC bacitracin resistance gene by two extracytoplasmic function sigma factors. J. Bacteriol. 184:6123-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, M. 2002. Ph.D. thesis. Cornell University, Ithaca, N.Y.
- 6.Cao, M., P. A. Kobel, M. M. Morshedi, M. F. Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, runoff transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443-457. [DOI] [PubMed] [Google Scholar]
- 7.Cao, M., T. Wang, R. Ye, and J. D. Helmann. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol. Microbiol. 45:1267-1276. [DOI] [PubMed] [Google Scholar]
- 8.Cho, H. Y., Y. W. Kim, T. J. Kim, H. S. Lee, D. Y. Kim, J. W. Kim, Y. W. Lee, S. Leed, and K. H. Park. 2000. Molecular characterization of a dimeric intracellular maltogenic amylase of Bacillus subtilis SUH4-2. Biochim. Biophys. Acta 1478:333-340. [DOI] [PubMed] [Google Scholar]
- 9.Cutting, S. M., and P. B. VanderHorn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for bacillus. John Wiley and Sons, Ltd., Chichester, England.
- 10.Guérout-Fleury, A.-M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 11.Helmann, J. D. 1999. Anti-sigma factors. Curr. Opin. Microbiol. 2:135-141. [DOI] [PubMed] [Google Scholar]
- 12.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 13.Helmann, J. D., and C. P. Moran, Jr. 2002. RNA Polymerase and Sigma Factors, p. 289-312. In A. L. Sonenshein and R. Losick (ed.), Bacillus subtilis and Its Relatives: From Genes to Cells. ASM Press, Washington D. C.
- 14.Horsburgh, M. J., and A. Moir. 1999. Sigma M, an ECF RNA polymerase sigma factor of Bacillus subtilis 168, is essential for growth and survival in high concentrations of salt. Mol. Microbiol. 32:41-50. [DOI] [PubMed] [Google Scholar]
- 15.Huang, X., A. Decatur, A. Sorokin, and J. D. Helmann. 1997. The Bacillus subtilis σX protein is an extracytoplasmic function sigma factor contributing to the survival of high temperature stress. J. Bacteriol. 179:2915-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, X., K. L. Fredrick, and J. D. Helmann. 1998. Promoter recognition by Bacillus subtilis σW: autoregulation and partial overlap with the σX regulon. J. Bacteriol. 180:3765-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, X., A. Gaballa, M. Cao, and J. D. Helmann. 1999. Identification of target promoters for the Bacillus subtilis extracytoplasmic function σ factor, σW. Mol. Microbiol. 31:361-371. [DOI] [PubMed] [Google Scholar]
- 18.Huang, X., and J. D. Helmann. 1998. Identification of target promoters for the Bacillus subtilis σX factor with a consensus-directed search. J. Mol. Biol. 279:165-173. [DOI] [PubMed] [Google Scholar]
- 19.Hughes, K. T., and K. Mathee. 1998. The anti-sigma factors. Annu. Rev. Microbiol. 52:231-286. [DOI] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28:1059-1066. [DOI] [PubMed] [Google Scholar]
- 22.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohki, R., K. Tateno, Y. Okada, H. Okajima, K. Asai, Y. Sadaie, M. Murata, and T. Aiso. 2003. A bacitracin-resistant Bacillus subtilis gene encodes a homologue of the membrane-spanning subunit of the Bacillus licheniformis ABC transporter. J. Bacteriol. 185:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paget, M., H.-J. Hong, M. Bibb, and M. J. Buttner. 2002. The ECF sigma factors of Streptomyces coelicolor A3(2), p. 105-125. In C. M. Thomas and H. F. Jenkinson (ed.), Switches, signals, regulons and cascades: control of bacterial gene expression. Society for General Microbiology Symposium Volume 61. Cambridge University Press, Cambridge, England.
- 25.Qiu, J., and J. D. Helmann. 2001. The −10 region is a key promoter specificity determinant for the Bacillus subtilis extracytoplasmic-function sigma factors σX and σW. J. Bacteriol. 183:1921-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slack, F. J., J. P. Mueller, and A. L. Sonenshein. 1993. Mutations that relieve nutritional repression of the Bacillus subtilis dipeptide permease operon. J. Bacteriol. 175:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinmetz, M., and R. Richter. 1994. Easy cloning of mini-Tn10 insertions from the Bacillus subtilis chromosome. J. Bacteriol. 176:1761-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner, M. S., and J. D. Helmann. 2000. Mutations in multidrug efflux homologs, sugar isomerases, and antimicrobial biosynthesis genes differentially elevate activity of the σX and σW factors in Bacillus subtilis. J. Bacteriol. 182:5202-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 30.Wiegert, T., G. Homuth, S. Versteeg, and W. Schumann. 2001. Alkaline shock induces the Bacillus subtilis σW regulon. Mol. Microbiol. 41:59-71. [DOI] [PubMed] [Google Scholar]
- 31.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zellmeier, S., U. Zuber, W. Schumann, and T. Wiegert. 2003. The absence of FtsH metalloprotease activity causes overexpression of the σW-controlled pbpE gene, resulting in filamentous growth of Bacillus subtilis. J. Bacteriol. 185:973-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuber, P., and R. Losick. 1987. Role of AbrB and Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J. Bacteriol. 169:2223-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]