Abstract
We have characterized the in vivo phenotypes of 17 mutations of Escherichia coli ftsZ. In particular, we determined whether these mutations can complement a null ftsZ phenotype, and we demonstrated that two noncomplementing mutations show partial dominant-negative behavior. We performed immunofluorescence microscopy to determine whether these mutants could assemble into normal or abnormal structures in vivo. The mutants separated into four classes—those that complemented the null and formed normal FtsZ rings, those that complemented the null but formed aberrant FtsZ structures, those that formed aberrant FtsZ structures and did not complement, and those that were unable to form any FtsZ structures. We did not find any mutations that produced nonfunctional Z rings of normal appearance. Surprisingly, some mutants that produced extensively spiraled Z-ring structures divided and grew with a normal doubling time. The analysis was carried out using a complementation system based on an ftsZ deletion strain, a temperature-sensitive rescue plasmid, and a complementation vector that placed mutated ftsZ alleles under the control of the pBAD promoter, which offered several advantages over previous systems.
FtsZ, a prokaryotic homologue of tubulin (14), is the primary component of the bacterial cell division apparatus. In vitro, FtsZ can assemble structures based on straight and curved protofilaments, similar to those formed by tubulin (8, 11, 17, 22). In Escherichia coli cells, FtsZ forms a ring (called the Z ring) at the site of division (6, 19). It is believed that the Z ring, in conjunction with other cell division proteins, acts as a contractile ring that effects cell division.
In a previous study (16), a number of site-directed mutants of ftsZ were constructed. Many of these mutations were made in an attempt to interfere with the assembly of FtsZ structures. We concentrated on conserved acidic residues that are believed to be on the surface of the FtsZ molecule. Mutant proteins were tested in vitro for GTPase activity and assembly, and the mutants were examined in vivo for the ability to complement ftsZ84, a temperature-sensitive ftsZ allele. Although many of the mutated alleles did not complement this temperature-sensitive background, most of the noncomplementing ftsZ alleles were not expressed or were expressed at very low levels, possibly due to second-site reversions selected for by constitutive expression of toxic FtsZ protein. Furthermore, there was always a mixture of FtsZ in the cells from the genomic ftsZ84 and the plasmid-borne mutated ftsZ allele. Growing the cultures at the restrictive temperature inactivates FtsZ84 but does not remove it from the cells. Overexpression of FtsZ84 can also rescue the ftsZ84 phenotype (23), which posed additional complications.
An ideal complementation vector would allow both tight repression of the mutated allele during cloning and controlled expression during testing. We have developed a complementation vector (pJSB2) for E. coli ftsZ that expresses mutated ftsZ alleles under the control of the pBAD promoter. Expression of the mutated alleles can be repressed by the addition of glucose or induced by the addition of arabinose. We have combined our complementation vector with an ftsZ null strain (JKD7-1) to create a complementation system that avoids the problems inherent in earlier work. This null strain is maintained by the addition of a rescue plasmid (pKD3) containing a functional ftsZ allele (10). pKD3 is itself temperature sensitive for replication (4). When strains containing both pKD3 and pJSB2-derived plasmids are grown at the restrictive temperature and in the presence of arabinose, pKD3 fails to replicate and the mutated FtsZ replaces the wild-type protein. This strategy has been used successfully in other complementation studies (20, 24, 29, 31).
This complementation system was used to characterize our collection of ftsZ mutants, including testing them for division competence and dominant-negative behavior, as well as immunofluorescence (IF) microscopy to examine Z-ring structure.
MATERIALS AND METHODS
Construction of the complementation system.
The complementation vector, pJSB2, was constructed from pBAD18 (12) in two steps. The multicloning site of pBAD18 was removed by digestion with NheI-SalI and replaced by two partially complementary oligonucleotides, 5′-CTAGCGGAGTCGACCCGGGTACC-3′ and 5′-TCGAGGTACCCGGGTGCACTCCG-3′. When ligated into the digested vector, these oligonucleotides provided a blunt SmaI site (underlined) in proximity to a Shine-Dalgarno sequence (boldface). The Shine-Dalgarno sequence is altered from the consensus sequence, AGGAGG (26). This reduces its efficiency so that maximal induction generates only moderate levels of expression. This vector was named pJSB1, and the alteration was confirmed by sequencing.
pJSB2 was constructed from pJSB1 by disrupting the bla gene with a cat cassette. The cat cassette was constructed by PCR amplification of the cat gene and upstream sequence from pBCSK+ (Stratagene) using primers (5′-ACTCATAGCGTAAGAGGTTCCAACTTTCA-3′ and (5′-AACGAAGCCAGAAGGGCCACCAATAACTGCCTTAAAAAAA-3′) that appended half of a ScaI site and a stop codon to the 5′ end of the PCR product and a BglI site to the 3′ end (the restriction enzyme sites are underlined). This PCR product was digested with BglI and ligated into ScaI-BglI-digested pJSB1, replacing the middle of the bla gene with the cat cassette and truncating the bla gene.
Transferring ftsZ mutants to pJSB2.
ftsZ mutant alleles were originally constructed by QuikChange site-directed mutagenesis (Stratagene) in pET11b-ftsZ (16). The mutant alleles were transferred to pJSB2 by excising them from pET11b-ftsZ mutant plasmids with NdeI-BamHI, filling in the restriction site overhangs with Klenow fragment (New England Biolabs), and ligating them into SmaI-digested pJSB2. The promoter region and ftsZ gene of each plasmid was sequenced to confirm proper mutation and orientation. These plasmids were called pJSB100 to -117, and were transformed into JKD7-1/pKD3 to form strains JSN100 to -117 (Table 1).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| JKD7-1 | ftsZ::Kanr (genomic ftsZ null) recA | J. Lutkenhaus (10) |
| JFL101 | ftsZ84(Ts) recA | J. Lutkenhaus (18) |
| JSN2 | JKD7-1/pKD3/pJSB2 | This study |
| JSN100 | JKD7-1/pKD3/pJSB100 | This study |
| JSN101 | JKD7-1/pKD3/pJSB101 | This study |
| JSN102 | JKD7-1/pKD3/pJSB102 | This study |
| JSN103 | JKD7-1/pKD3/pJSB103 | This study |
| JSN104 | JKD7-1/pKD3/pJSB104 | This study |
| JSN105 | JKD7-1/pKD3/pJSB105 | This study |
| JSN106 | JKD7-1/pKD3/pJSB106 | This study |
| JSN107 | JKD7-1/pKD3/pJSB107 | This study |
| JSN108 | JKD7-1/pKD3/pJSB108 | This study |
| JSN109 | JKD7-1/pKD3/pJSB109 | This study |
| JSN110 | JKD7-1/pKD3/pJSB110 | This study |
| JSN111 | JKD7-1/pKD3/pJSB111 | This study |
| JSN112 | JKD7-1/pKD3/pJSB112 | This study |
| JSN113 | JKD7-1/pKD3/pJSB113 | This study |
| JSN114 | JKD7-1/pKD3/pJSB114 | This study |
| JSN115 | JKD7-1/pKD3/pJSB115 | This study |
| JSN116 | JKD7-1/pKD3/pJSB116 | This study |
| JSN117 | JKD7-1/pKD3/pJSB117 | This study |
| pKD3 | pSC101 origin; repA(Ts) ftsZ Ampr | J. Lutkenhaus (10) |
| pBAD18 | pBR322 origin; arabinose regulation; Ampr | D. Weiss (12) |
| pJSB1 | pBAD18 with MCSa for blunt insertion | This study |
| pJSB2 | pJSB1-bla::cat (Amps Cmr) | This study |
| pJSB100 | pJSB2-ftsZ | This study |
| pJSB101 | pJSB2-ftsZ-D45A | This study |
| pJSB102 | pJSB2-ftsZ-A70T | This study |
| pJSB103 | pJSB2-ftsZ-D86K | This study |
| pJSB104 | pJSB2-ftsZ-D96A | This study |
| pJSB105 | pJSB2-ftsZ-G105S | This study |
| pJSB106 | pJSB2-ftsZ-D158A | This study |
| pJSB107 | pJSB2-ftsZ-D166K/F268V | This study |
| pJSB108 | pJSB2-ftsZ-D187A | This study |
| pJSB109 | pJSB2-ftsZ-D209A | This study |
| pJSB110 | pJSB2-ftsZ-D212A | This study |
| pJSB111 | pJSB2-ftsZ-D212G | This study |
| pJSB112 | pJSB2-ftsZ-E238A | This study |
| pJSB113 | pJSB2-ftsZ-S245F | This study |
| pJSB114 | pJSB2-ftsZ-E250A | This study |
| pJSB115 | pJSB2-ftsZ-E250K/D253K | This study |
| pJSB116 | pJSB2-ftsZ-D269A | This study |
| pJSB117 | pJSB2-ftsZ-D299A | This study |
MCS, multicloning site.
Growth of JSN strains.
Cells were grown in a variety of Luria broth (LB)-based media. Repression medium was LB containing 34 μg of chloramphenicol/ml (selecting for the pJSB2-derived plasmid containing the mutant ftsZ), 100 μg of ampicillin/ml (selecting for the pKD3 rescue plasmid containing wild-type ftsZ), and 0.2% (wt/vol) glucose (suppressing expression of the mutant ftsZ allele). Cells were usually grown at 30°C in repression medium to retain pKD3. To test the effects of mutated ftsZ alleles, cells were grown on agar plates or in liquid culture in induction medium (LB containing 34 μg of chloramphenicol/ml and various concentrations of arabinose) at 42°C. This eliminated the pKD3 rescue plasmid, making arabinose-induced expression of the pJSB-based ftsZ allele necessary for cell division.
Certain JSN strains were also grown under pseudochemostat conditions, in which the cells were grown in induction medium containing 0.05% arabinose at 42°C and repeatedly diluted 1:10 in fresh medium at 42°C whenever the cultures grew to an optical density at 600 nm (OD600) of >0.3 to 0.5. This maintained the cells in log phase under complementation conditions for extended periods.
Complementation tests.
Plate complementation assays were performed as follows. JSN strains containing mutant ftsZ alleles were grown in repression medium overnight at 30°C. Equal volumes of diluted overnight culture were then plated on either induction plates containing a range of arabinose from 0 to 0.5% or repression plates containing 0.5% glucose. Repression plates were grown at 30°C to determine the number of CFU in the plated volume, and induction plates were grown at 42°C. Colonies on induction plates were counted, and these values were normalized to the number of colonies on the repression plate. A mutation was considered complementing when induction plates containing 0.05 to 0.5% arabinose grew at least 80% as many colonies as the repression plate. Noncomplementing mutations produced no colonies on any of the induction plates. There were no intermediate cases.
Dominant-negative assays were performed similarly, except that induction plates were grown at 30°C to retain pKD3 and its wild-type ftsZ expression. Two mutants were designated partially dominant negative on the basis of very small colonies or colonies with abnormal morphologies.
Liquid complementation assays were performed by growing JSN strains at 30°C to an OD600 of 0.5 in repression medium and then diluting these cultures 1:5 × 106 in induction medium containing 0.05% arabinose and growing them at 42°C for 24 h. Complementation was defined as the ability of the culture to grow to an OD600 of >0.5; these cells had normal morphology. Noncomplementing strains did not produce measurable turbidity.
Quantitation of ftsZ expression.
JSN100 (expressing wild-type FtsZ from pJSB100) was used to quantitate the levels of FtsZ expressed at different arabinose concentrations. Cells were grown at 30°C overnight in repression medium and were then diluted 1:1,000 in induction medium containing various amounts of arabinose and grown at 30°C to an OD600 of 0.5 to 0.7. The cultures were treated with NaN3 to 0.02% and harvested by centrifugation. The cell pellet was frozen and thawed, resuspended in sodium dodecyl sulfate sample buffer without β-mercaptoethanol, sonicated at low power three times for 10 s each time, heated at 95°C for 10 min, sonicated again as previously, and then split into two aliquots. β-Mercaptoethanol was added to one aliquot to 5% in preparation for quantitative Western blotting, and an equal volume of H2O was added to the other aliquot in preparation for protein quantitation. Quantitative Western blotting and protein quantitation by the bicinchoninic acid assay (Pierce) were performed as described previously (15, 16). Western blots were alternately prepared with a secondary antibody of goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (BioSource International, Camarillo, Calif.) and developed using 3,3′-diaminobenzidine (100 mM Tris-HCl, pH 7.4, 2 mM 3,3′-diaminobenzidine, 4 mM NiCl2, 1% H2O2). These showed levels of expression equivalent to those of quantitative Western blots. To verify expression of mutated ftsZ alleles, JSN cultures were grown and diluted as described above and then induced with 0.05% arabinose and grown at 30°C to midlog phase. Extracts were prepared, and FtsZ expression was quantitated as described above.
IF microscopy.
The fixation method used was adapted from several sources (1, 13, 25). JSN strains were grown in repression medium at 30°C overnight and then diluted 1:1,000 to 1:2,000 in induction medium containing 0.05% arabinose and grown at 42°C to an OD600 of 0.4 to 0.6 (∼2 to 3 h). These samples were fixed as described by Addinall et al. (1), washed three times with PBS plus 0.05% Tween 20 (PBST), and resuspended in 25 mM Tris-HCl-50 mM glucose-10 mM EDTA, pH 8.0. Twenty microliters of cell suspension was dropped onto a polylysine-coated 12-well multiwell slide (ICN). The cells were allowed to adhere to the slide for 10 min and were permeabilized with 100 μl of 2-mg/ml lysozyme/well (freshly dissolved in 25 mM Tris HCl-50 mM glucose-10 mM EDTA, pH 8.0) for ∼5 min at room temperature. The slide was then washed three times in PBST.
Samples were blocked and stained as described by Addinall et al. (1) using 3.1 μg of affinity-purified rabbit anti-EcFtsZ 5435/ml (15) as the primary antibody and 2 μg of goat anti-rabbit immunoglobulin G conjugated to Alexa 488 (Molecular Probes)/ml as the secondary antibody. The slide was then washed three times in PBST, mounted in SlowFade Light (Molecular Probes) according to the manufacturer's directions, and sealed with nail polish. Samples were viewed on a Zeiss Axiophot microscope using plan-Neofluar oil immersion lenses (numerical aperture = 1.30) at 40× and 100×. Images were captured using a cooled charge-coupled device camera and manipulated using Adobe Photoshop. Measurements of cells were performed using NIH Image software.
RESULTS
Rescue of ftsZ null by pJSB100.
We constructed pJSB2, a pBAD-based complementation vector inducible by arabinose, and inserted mutated alleles of ftsZ into the expression site. The resulting pJSB plasmids were transformed into JKD7-1/pKD3 cells, forming a collection of JSN strains. Each JSN strain had three ftsZ alleles—a genomic null allele; a wild-type allele under native promoters in pKD3, which is temperature sensitive for replication; and a mutant allele under the pBAD promoter. In repression medium at 30°C, the mutant allele would not be expressed and the ftsZ allele on pKD3 would rescue the null allele; in induction medium at 42°C, pKD3 would be lost in the majority of cells and growth would require complementation by the arabinose-induced mutant ftsZ allele on the pJSB plasmid.
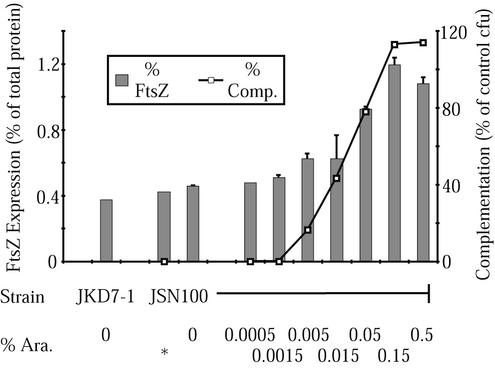
To test pJSB2 as a complementation vector, we inserted wild-type E. coli ftsZ into the expression site to create pJSB100 and transformed this into JKD7-1/pKD3 cells to form JSN100. We then tested JSN100 for the ability to grow at 42°C, given a range of levels of arabinose to induce wild-type ftsZ expression from pJSB100. While JKD7-1/pKD3 (without a complementation plasmid) and JKD7-1/pKD3/pJSB2 (without ftsZ) did not grow at 42°C, JSN100 (wild-type ftsZ) was able to grow when arabinose was present at levels of >0.01% (Fig. 1). We also quantitated the levels of FtsZ present in JSN100 grown at 30°C by quantitative Western blotting (Fig. 1). At arabinose concentrations of 0.05% and above, which gave full complementation, the FtsZ concentration was doubled at 30°C. Since there would be no pKD3-based FtsZ expression at 42°C, we conclude that complementation occurs when pJSB-based arabinose-induced expression is sufficient to supply normal levels of FtsZ. We did not directly assay expression at 42°C because we could not extend those measurements to induction levels insufficient to complement or to noncomplementing mutants. However, the successful complementation of wild-type and several mutant alleles means that ftsZ alleles on pJSB must be expressed at the appropriate level at 42°C. It was a somewhat fortuitous result that our construct, with its altered Shine-Dalgarno sequence, produced approximately the same level of FtsZ as the pKD3 plasmid when fully induced.
FIG. 1.
Comparison of wild-type FtsZ induction with rescue of a null ftsZ allele. The left axis (% FtsZ) indicates the level of FtsZ in the cells grown at 30°C, while the right axis (% Comp.) indicates the level of complementation in cells grown at 42°C. The FtsZ measured includes that expressed from both pKD3 and pJSB100. At 100% complementation, there were as many colonies on the induction plate as on the repression control. Arabinose levels (% Ara.) are given on the horizontal axis; ∗ indicates addition of 0.2% glucose in lieu of arabinose to repress plasmid-borne FtsZ production. The error bars indicate standard errors of the mean.
Testing ftsZ mutants for complementation.
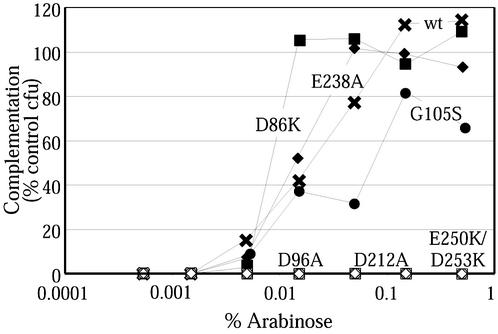
We inserted 17 ftsZ mutant alleles into pJSB2 and transformed JKD7-1/pKD3 cells with these plasmids. Each was then tested for its ability to complement the null allele of JKD7-1 at 42°C in our plate complementation assay. The results of these experiments are summarized in Table 2. Each of the complementing mutants gave a complementation curve similar to that of wild-type ftsZ in that colonies started to appear at 0.005% arabinose and the number of colonies plateaued around 0.05% arabinose (Fig. 2). An exception was ftsZ-G105S (the 105 codon encoding a Ser rather than a Gly), discussed below. None of the noncomplementing mutants showed any ability to form colonies at 42°C at any level of arabinose induction. Liquid complementation assays gave identical results.
TABLE 2.
Summary of results
| Mutant Class | Mutation | Complements the nulla | IF | GTPase activityb | Assembly in:
|
|
|---|---|---|---|---|---|---|
| Mgc | DEAE-dextrand | |||||
| Complementing; normal FtsZ structures | ||||||
| G105S (ftsZ84)e | Y | Rings | 10 | ND | More sheets | |
| D158A | Y | Rings | 155 | PF | Normal | |
| D187A | Y | Rings | ND | PF | Normal | |
| E238A | Y | Rings | 145 | PF | Normal | |
| D269A | Y | Rings | 10 | PF | Normal | |
| Complementing; aberrant FtsZ structures | ||||||
| A70T (ftsZ1) | Y | Mild spirals | 14 | PF | Normal | |
| D86K | Y | Spirals | 49 | twPF + tubes | More tubes | |
| E250A | Y | Spirals | 67 | PF | Fewer tubes | |
| D299A | Y | Mild spirals | 200 | PF | Normal | |
| Noncomplementing; aberrant FtsZ structures | ||||||
| D212A | N | Severe spirals | 7 | None | More tubes | |
| D212G (ftsZ2) | N | Severe spirals | 0.5 | ND | ND | |
| E250K/D253K | D | Longitudinal filaments | 23 | PF | No tubes | |
| Noncomplementing; no FtsZ structures | ||||||
| D45A | D | No FtsZ structure; blebs | 5 | None | All tubes | |
| D96A | N | No structure | ND | PF | No tubes | |
| D209A | N | No structure | 7 | PF | All tubes | |
| Not expressed | ||||||
| D166K/F268V | ? | Dim; punctate | 15 | PF | No tubes | |
| S245F | ? | Dim; punctate | 76 | PF + tubes | Normal | |
Y, complements the null allele; N, does not complement the null allele; ?, does not complement but was poorly expressed; D, does not complement the null allele and is at least partially dominant negative.
As percentage of wild-type FtsZ. See reference 16 for details and references.
PF, protofilaments (wild-type FtsZ forms protofilaments); twPF, twinned protofilaments; ND, not determined. See reference 16 for details.
ND, not determined. See reference 16 for details.
ftsZ84 does not complement when expressed from the genomic locus; however, it can complement if overexpressed (23), as in this study.
FIG. 2.
Complementation curves for a number of ftsZ mutants. Complementation curves are shown as in Fig. 1. Three noncomplementing mutants (ftsZ-D96A, ftsZ-D212A, and ftsZ-E250K/D253K) are at the bottom of the graph. wt, wild type.
Many of these mutations were previously tested for the ability to complement ftsZ84, a temperature-sensitive allele of ftsZ (16). The majority of the mutations behaved similarly in both complementation tests, but there were three exceptions. Plasmids expressing ftsZ-D86K, ftsZ-E238A, and ftsZ-E250A all failed to complement ftsZ84 in the previous study but complemented the null ftsZ in this study. In the previous study, it was found that these noncomplementing alleles were poorly expressed, so we wanted to determine whether they could complement the ftsZ84 allele from pJSB2 when they were fully expressed. We tested this by transforming JFL101 cells (which contain the ftsZ84 mutation at their genomic locus) with pJSB plasmids containing the three mutants and testing the resulting strains in our plate complementation assay. Low-NaCl medium (1% tryptone, 0.5% yeast extract, 0.05% NaCl, 1.5% agar) was used to avoid salt rescue of ftsZ84 (23). ftsZ-E238A and ftsZ-E250A rescued JFL101, confirming that they are not lethal mutations. However, ftsZ-D86K did not rescue JFL101, despite its ability to complement the null allele. Since D86 is located on the left side of the FtsZ molecule, while G105 is located in the GTP binding pocket, it is unlikely that the mutant residues interact directly. The interaction causing the failure of complementation is unknown.
Two mutants in our collection had previously been shown to have temperature-sensitive phenotypes. FtsZ-G105S (FtsZ84) is functional at 30°C but does not form a Z ring at 42°C (2). In our study, ftsZ-G105S complemented the null ftsZ allele at 42°C, but it required higher levels of arabinose to achieve complementation (Fig. 2). This is consistent with the observation that the defect can be compensated for by overexpression of FtsZ-G105S (23). ftsZ-D212G (ftsZ2) did not complement the null ftsZ allele in our study. However, FtsZ-D212G is temperature sensitive (7), meaning that ftsZ-D212G should complement the null ftsZ allele at 30°C in our system. We confirmed this by curing JSN110 of the pKD3 plasmid under induction conditions at 30°C. We obtained strains in which ftsZ-D212G was able to complement the null allele at 30°C, although they appeared to be unstable and were lost.
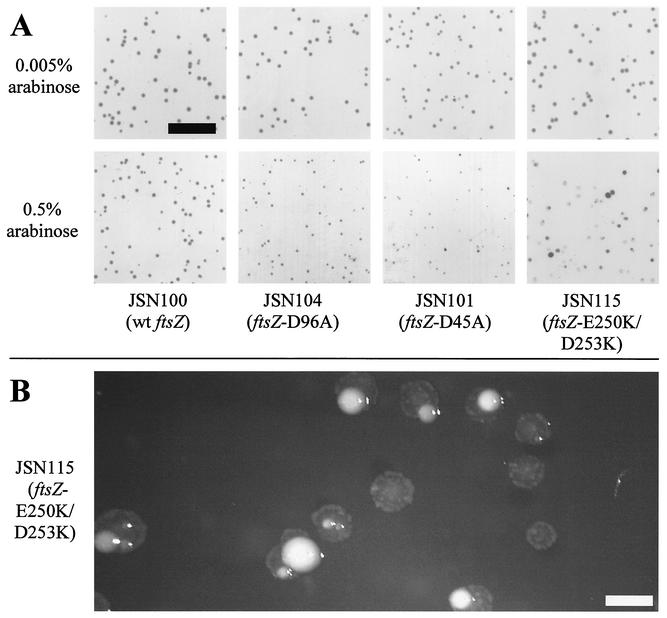
Since ftsZ is an essential gene and its product presumably acts in a multimeric fashion, it is possible that mutants might have dominant-negative phenotypes. Indeed, the lack of expression of several ftsZ mutants in previous work (16) suggested that these mutants might have been toxic. We tested each noncomplementing mutant for dominant-negative behavior by growing the corresponding JSN strain at 30°C, retaining the wild-type ftsZ allele from pKD3. These mutants were tested on induction plates containing the complete range of concentrations of arabinose used for complementation testing. Most of the alleles tested did not interfere with colony formation, even at high levels of induction, indicating that they do not have dominant-negative behavior (Table 2). However, two strains grew into abnormal colonies under high levels of induction. JSN101 (ftsZ-D45A) colonies were very small at high levels of induction compared to JSN101 colonies grown at lower levels of induction (Fig. 3A). JSN115 (ftsZ-E250K/D253K) colonies grown on high levels of arabinose (sufficient to result in complementation of the null allele by wild-type ftsZ) were often translucent and flat, sometimes with a high ridge around the perimeter or with normal-appearing subcolonies in the middle of a flat colony (Fig. 3B). Both of these mutants have partial dominant-negative phenotypes.
FIG. 3.
Abnormal colony morphology in partially dominant-negative ftsZ mutants. (A) Colonies at approximately normal magnification (bar = 1 cm). The colonies in the top row were grown on 0.005% arabinose, which is insufficient to complement or cause dominant-negative effects. The colonies in the bottom row were grown on 0.5% arabinose, sufficient to cause dominant-negative effects. All plates were grown for 24 h at 30°C, except JSN101 at 0.5% arabinose, which was grown for 48 h due to its extremely low rate of growth. (B) JSN115 colonies at greater magnification (bar = 1 mm). Note the “fried-egg” appearance of the colonies and the normal sectors in some colonies.
Quantitation of mutant ftsZ expression.
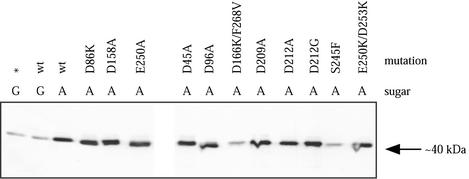
To test whether each of the mutant ftsZ alleles was being expressed, we prepared cell lysates of JSN strains containing noncomplementing mutations, as well as several strains containing complementing mutations, that were grown at 30°C in induction medium containing 0.05% arabinose. This level of expression resulted in complementation of the null allele by all complementing mutants (Fig. 2). Western blotting of the lysates showed that all of the mutant alleles were expressed at levels similar to that of wild-type ftsZ under the pBAD promoter, except for ftsZ-D166K/F268V and ftsZ-S245F (Fig. 4). This confirmed that most of the mutant alleles were being expressed as expected. Repeated transformations of the two underexpressing strains showed identical phenotypes. We cannot explain why these two exceptions were not expressed normally.
FIG. 4.
Expression of mutated ftsZ alleles. JSN mutant strains were grown at 30°C and lysed, and Western blotting was performed using anti-FtsZ antibodies and secondary antibodies conjugated to horseradish peroxidase. A, 0.05% arabinose was added to the culture to induce FtsZ expression; G, 0.2% glucose was added to repress FtsZ expression; ∗, JSN2 without a pJSB plasmid.
Characterization of the complementation system under pseudochemostat conditions.
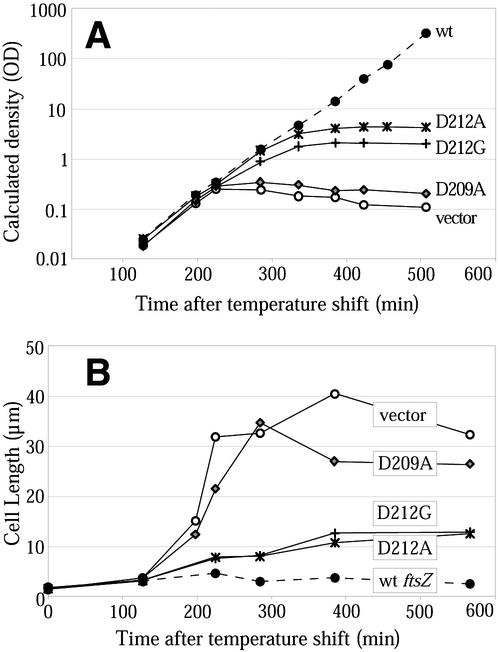
This complementation system cannot effect an immediate removal of wild-type FtsZ. When cultures are shifted to 42°C, the pKD3 vector stops replicating, and its very low copy number (9) means that the majority of the cells will not contain pKD3 plasmid after a small number of division periods. However, even these cells will have a pool of FtsZ protein that will be diluted only by a factor of 2 with each division. Eventually, most cells will have no pKD3 and a substantially reduced pool of wild-type FtsZ. We investigated the time course of the complementation system by growing several strains under pseudochemostat conditions (see Materials and Methods for details) to maintain cultures in log phase for longer periods.
Cells containing wild-type ftsZ continued to grow (as measured by increasing turbidity) over a long period at 42°C (Fig. 5A). These cells were not elongated after prolonged growth (Fig. 5B), indicating that the complementation system was able to support division over many division cycles. Cells containing a representative complementing mutation (ftsZ-D269A) were also grown under these conditions and continued to grow and divide normally. Cells that contained the complementation vector without an ftsZ allele, or contained the noncomplementing ftsZ allele ftsZ-D209A, reached maximum cell density at 240 min (Fig. 5A). This increase in density was accompanied by cell division until ∼180 min, followed by filamentation from 200 to 240 min. At 240 min, the culture was at an OD600 of ∼0.4 (Fig. 5A), and the cells were 6 to 12 times longer than wild-type cells on average (Fig. 5B). Further incubation resulted in slightly greater filamentation, and the OD of the culture then decreased, perhaps due to the lysis of filamentous cells.
FIG. 5.
Pseudochemostat growth of various strains. (A) Growth curves of five strains (measured as the OD of the culture multiplied by the dilution factor due to pseudochemostat dilutions). (B) Average cell lengths of the strains in panel A. Each average is for 30 to 60 cells; error bars are not given due to the lack of normal distribution of cell lengths. wt, wild type.
Cells containing ftsZ-D212A and ftsZ-D212G had an intermediate phenotype under pseudochemostat conditions (Fig. 5). They reached a maximum density higher than that of cells without FtsZ, although they did not continue to grow indefinitely. The cells did eventually elongate, although they did so later and were only to three to six times longer than wild-type cells when they ceased to grow. As the cells elongated, they grew into progressively more abnormal forms, with twists, blebs, and thickenings of the cell becoming more common as the culture incubation time increased, especially in cells containing ftsZ-D212A (see below). Importantly, at 240 min, these cells were not appreciably longer than wild-type cells. At this time point, cell division is apparently still occurring. We conclude that this division must be achieved through a combination of the wild-type FtsZ, diluted but still present, and the mutant D212A/G protein. Division and growth both cease at later time points when the wild-type FtsZ is further depleted, indicating that the D212A/G mutant proteins by themselves are not able to support division at 42°C.
Characterization of mutant ftsZ strains.
We wanted to examine the Z-ring formation of these mutants to see if abnormal rings or morphologically normal but nonfunctional rings were formed. Initial studies were performed by diluting an overnight culture 1:1,000 and growing it to an OD600 of 0.4. Pseudochemostat experiments (see above) showed that this was just after the point where cells without a complementing FtsZ started to filament. This is thus the earliest point where a mutant phenotype should be clearly visible.
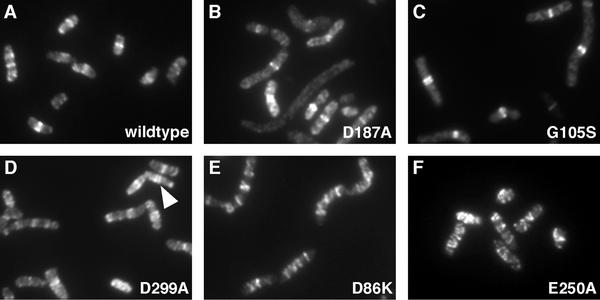
JSN100 (wild-type ftsZ) grew normally at 42°C, and IF microscopy showed cells with normal morphology. Most of the cells had normal Z rings that appeared as a single bright band, located at the median of the cell (Fig. 6A). A small percentage (<5%) of the Z rings at any time point appeared as double bands. Focusing though the specimen plane showed that these double bands were actually helices of 0.5- to 1-μm pitch running one circumference around the cell. Cells containing these helical Z rings were not elongated and occasionally showed division furrows, suggesting that the helical Z rings could carry out cell division. We hypothesize that these helices are due to overproduction of FtsZ, as this effect has been seen in other strains with higher-than-normal FtsZ levels (J. Stricker, personal observation, and reference 19).
FIG. 6.
FtsZ structures in complementing strains. All panels show IF microscopy of FtsZ. All strains were grown under complementing conditions at 42°C with 0.05% arabinose to mid-log phase (see Materials and Methods). The arrowhead in panel D points to a spiral that resolves as a double ring. Magnification, ×2,550.
A number of mutations produced Z rings that were very similar to or indistinguishable from the wild type under the conditions used (Fig. 6B and C). All of these mutations complemented the null ftsZ allele (Table 2 shows a summary of the results). Cells containing a representative mutation (ftsZ-D269A) were grown under pseudochemostat conditions and contained normal Z rings after long periods of growth (data not shown). These structures must have been formed from mutant FtsZ protein without a significant proportion of wild-type FtsZ. Except for JSN105 (ftsZ-G105S), all of these strains had normal cell morphology. JSN105 cells were longer than wild-type cells, but the rings were bright, well-formed bands, and most cells had the correct number of rings at the correct locations (Fig. 6C).
A second group of mutants formed spiral Z rings at high frequency. These further separated into two groups—those that complemented the null allele (ftsZ-D299A, ftsZ-A70T, ftsZ-D86K, and ftsZ-E250A) and those that did not (ftsZ-D212A and ftsZ-D212G). In the first group, there was a range of severities of the defect in Z-ring formation. JSN117 (ftsZ-D299A) formed two-turn FtsZ spirals at the middle of the cells that were similar to those seen in wild-type cells (Fig. 6D), though at higher frequency (∼20% of the cells). JSN102 (ftsZ-A70T) formed similar spirals in 30 to 40% of the cells. JSN103 (ftsZ-D86K) formed more extensive spirals that were found in the majority of the cells. These spirals were often three to four turns long and could extend the length of the cell, although they appeared to have a similar ∼0.5- to 1-μm pitch (Fig. 6E). JSN114 (ftsZ-E250A) had extensive FtsZ spirals in nearly every cell (Fig. 6F). These often extended the length of the cell, and occasional cells with separated spirals could be seen. These spirals also appeared to have a regular pitch of 0.5 to 1 μm. Extensive FtsZ spirals were still seen in JSN114 cells grown for longer times under pseudochemostat conditions, implying that FtsZ-E250A assembles spirals without any contribution from wild-type FtsZ. All four of these mutants were able to complement the null allele and had normal cell morphologies, despite these defects in Z-ring formation. All four strains had doubling times close to that of wild-type E. coli under induction conditions at 42°C. Also, all four strains localized their Z rings (or FtsZ spirals) to the median of the cell, even if they subsequently extended away from this site.
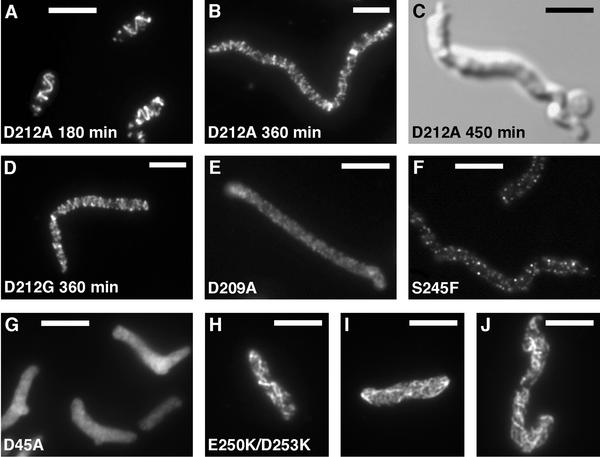
The noncomplementing mutants ftsZ-D212A and ftsZ-D212G also formed spirals, but these were qualitatively different than those of the complementing mutants. These spirals formed after short growth periods while the cells had not yet elongated and often appeared brighter and of wider pitch than functional spirals (Fig. 7A). Some spirals also appeared to be tilted and may have partially released from the cell membrane (not shown). Occasionally, spiral structures would double back upon themselves. As deduced from the pseudochemostat experiment, these spirals are functional for cell division while the cells still contain some wild-type FtsZ. When the cells were grown under pseudochemostat conditions for longer periods to deplete wild-type FtsZ levels, the cells elongated and became more misshapen, and the spirals became nonfunctional and grew more extensive as elongation progressed (Fig. 7B to D).
FIG. 7.
FtsZ structures in noncomplementing strains. All panels except C show IF microscopy of FtsZ; panel C shows a differential interference contrast image. All strains were grown at 42°C with 0.05% arabinose to mid-log phase (see Materials and Methods) except panels B to D, where the cells were grown under pseudochemostat conditions (see the text) for the indicated times. Bars = 5 μm. (Panels B and D are at a slightly different magnification.)
Several noncomplementing mutations did not form FtsZ structures. JSN109 (ftsZ-D209A) grew into filaments that were filled with diffuse FtsZ staining when viewed by IF microscopy (Fig. 7E). This strain behaved like JSN2 under pseudochemostat conditions, both in failure to grow to a turbid culture and in rapid elongation of the cells (Fig. 5). JSN104 (ftsZ-D96A) usually failed to form FtsZ structures, although very rare cells with a hint of helical structure of very wide pitch were seen. Both JSN107 (ftsZ-D166K/F268V) and JSN113 (ftsZ-S245F), which showed only low levels of expression from the pBAD-controlled ftsZ allele, grew into long filaments, with occasional cells of normal length in the JSN107 culture. Both strains stained very dimly by IF microscopy, with fluorescence appearing in punctate dots that often associated with the membrane (Fig. 7F). The dim IF correlates with the low level of FtsZ expression.
Two of the mutations we studied had partial dominant-negative phenotypes. Under complementation conditions, JSN101 (ftsZ-D45A) grew into somewhat elongated cells that often had pointed ends and a single bleb in a cell. The bleb was usually in the middle of the cell on one side, and the cell often bent at the bleb. IF microscopy showed a diffuse distribution with no organized FtsZ structures in these cells (Fig. 7G). The blebs were often brighter than the rest of the cell, although this may have been due to their larger volume. The same phenotype was seen in cells grown under dominant-negative conditions (at 30°C with 0.5% arabinose), suggesting that an excess of FtsZ-D45A blocked assembly of wild-type FtsZ. Intermediate levels of arabinose induction at 30°C resulted in intermediate effects on the cells. Z rings appeared as small helices when 0.03% arabinose was added, and further addition of arabinose resulted in increasing proportions of cells with no Z rings and cellular blebs (data not shown). This supports the idea that FtsZ-D45A exerts a dominant-negative effect.
ftsZ-E250K/D253K produced the most aberrant FtsZ structures. IF microscopy of JSN115 cells containing this mutation showed elongated cells that were sometimes swollen and contorted. The swelling was largely due to the fixation and permeabilization performed on these cells. It is noteworthy that identical conditions did not result in swelling in any of the other mutants examined here. Many of the JSN115 cells were filled with a meshwork of FtsZ filaments that ran longitudinally through the cell (Fig. 7H to J). Occasionally, these filaments had some helical character. Similar phenotypes were seen in cells grown under dominant-negative conditions, and intermediate levels of induction under dominant-negative conditions resulted in intermediate phenotypes, as in the case of ftsZ-D45A.
DISCUSSION
In terms of the assembly of Z rings, we might expect ftsZ mutants to fall into four classes: (i) assembly of Z rings with normal morphology and function; (ii) assembly of Z rings with normal morphology that fail to function; (iii) assembly of Z rings with aberrant structure, which might or might not function; and (iv) failure to assemble Z rings. We did not find any mutants that assembled normal Z rings that failed to function, but examples of the other classes were found (Table 2).
Five of our 17 mutants showed apparently normal assembly and function. These mutations are of highly conserved amino acids, so we expected them to have some role in cell division. The only effects seen were slightly longer cells. Some of these mutants showed altered GTPase in vitro, which has been noted previously for functional FtsZ mutants. It is likely that these apparently benign mutations have defects that are too subtle to be seen under our growth conditions.
We did not find any mutants that formed nonfunctional Z rings of normal appearance. This class of mutants would be particularly interesting if they indicated sites essential for the constriction mechanism of the Z ring. Mutation of the conserved C-terminal peptide of FtsZ has been shown to result in the formation of regularly spaced FtsZ concentrations that cannot function. These look like FtsZ rings, but they may represent pools of FtsZ localized by nucleoid exclusion rather than normal Z rings (20, 24). The C-terminal peptide of FtsZ is the binding site for FtsA and ZipA (20, 30), so failure of these mutants to function can be attributed to failure to bind these essential accessory proteins.
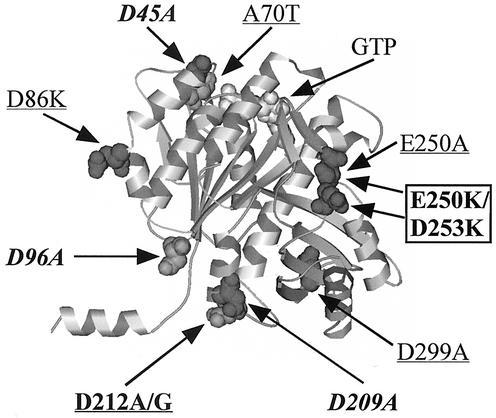
Seven of our mutants fell into the third class, forming aberrant Z rings or FtsZ structures. Most of these mutants formed spiral Z rings, although ftsZ-E250K/D253K formed filaments with a more longitudinal orientation. These spirals had a range of severities, from occasional mild spirals that did not visibly interfere with cell division to wider, more extensive spirals that were not capable of division. It is interesting that mutations that result in FtsZ spirals were found in several positions on the FtsZ molecule, including the upper and lower longitudinal bonding interfaces and lateral positions (Fig. 8). Spiral Z rings have also been seen in the ftsZ26 mutant (3, 7). Overexpression of FtsZ can also cause the formation of spiral Z rings, as can expression of an ftsZ-gfp construct at levels above one-third of the total FtsZ pool (19). E. coli cells deficient in phosphatidylethanolamine, a lipid component of the cell membrane, also form spiral Z rings (21). It would seem that the Z ring can adopt a spiral form under a variety of stresses and that this spiral form can often carry out cell division, even to the point of forming spiral septa (3). In Bacillus subtilis, Z rings are observed in spiral form during sporulation and may be an intermediate in moving the Z ring from central to polar locations (5).
FIG. 8.
Locations of mutations that cause aberrant or no Z-ring formation. Mutations in regular type complement the null allele at 42°C, and boldface mutants do not complement. The underlined mutants formed spirals, the italicized mutants did not form Z rings, and the boxed mutants formed more aberrant FtsZ structures.
A number of the mutants in this study formed discrete linear FtsZ structures, whether rings, spirals, or more severe filaments. The ability to form these linear filamentous structures implies that the various mutated FtsZs retain their basic assembly function. Indeed, all of the mutants that formed spirals showed mostly normal assembly of protofilaments in vitro (16) (Table 2). The generic structure of a linear filament that is constrained to the cell membrane is a spiral, and the primary difference between a spiral and a ring may be that the ring is more tightly constrained to the center of the cell. A number of perturbations of the Z ring or of the division machinery all result in spiral Z rings of a pitch wide enough to be visible in the light microscope. It has recently been concluded that the Z ring is about six protofilaments thick on average (28). The spirals we see are as bright as normal Z rings, implying that they are also only a few protofilaments thick. The normal Z ring may actually be a spiral with a pitch too small to resolve in the light microscope.
The ftsZ-E250K/D253K mutation has a novel phenotype. JSN115 colonies somewhat resembled those formed by mycoplasma or E. coli L forms. Both of these organisms lack cell walls and have a consequent defect in cell integrity. JFL101 cells transformed with a plasmid expressing ftsZ-E250K/D253K from the native ftsZ promoters (16) became very weak, often lysing while being centrifuged at 5,000 × g (J. Stricker, unpublished observation). While the JSN115 cells used in the present study did not show this extreme fragility, the abnormal colony morphology and tendency to swell when treated for microscopy were consistent with fragility. This may indicate a link between FtsZ function and formation of the cell wall at the division site.
Three of our mutants could not form Z rings or any filamentous structures and appeared diffusely distributed in the cytoplasm. Each of these mutants was able to assemble in vitro, although they did demonstrate aberrations. It is possible that they also assemble in vivo, but the assembly may be limited to protofilaments or other structures that cannot further assemble into the linear structures we observe as rings or spirals.
The complementation system used in this study, featuring a wild-type ftsZ allele on a rescue plasmid that could not replicate at 42°C, should be generally useful for testing mutations of essential genes. The pseudochemostat experiment allowed us to characterize the timing of the loss of wild-type protein, which occurred only after several hours of growth at 42°C. Complementation and mutant phenotypes are more easily observed at later time points. A limitation of the system is that it needs to be shifted to 42°C; therefore, it will not detect function in mutations that are only functional at lower temperatures. However, temperature-sensitive mutants are rare, even in deliberate screens, so it seems unlikely that many of the targeted mutations of surface amino acids seen here would be temperature sensitive.
A potential disadvantage of using the pBAD promoter to control expression of mutated ftsZ alleles is that the pBAD promoter has been shown to behave in an autocatalytic fashion, with individual cells either fully repressed or fully expressing. Increasing arabinose does not produce a gradation of expression in individual cells but increases the fraction of expressing cells (27). This happens randomly from cell to cell in a population, but it is a largely nonreversible process. This is consistent with our observation that increasing the concentration of arabinose increased the number of colonies, not the colony size. Presumably once a cell has switched on in arabinose, its progeny remain on and form a colony of normal size. This was not a problem for our analysis because complementation was scored at arabinose levels where all cells were fully induced.
Characterization of mutations is an important tool for determining the functions and mechanisms of proteins. The mutations examined here cover a range of surface areas of the FtsZ molecule and may aid in elucidating the mechanism of action of the Z ring. In particular, the frequency of formation of spiral Z rings, both functional and nonfunctional, suggest that the Z ring has a helical basis.
Acknowledgments
We thank Joe Lutkenhaus (University of Kansas) for supplying components of our complementation system, Chunlin Lu for initial construction of many of the mutant ftsZ alleles, and Sambra Redick for her technical assistance and helpful discussion. We also thank our reviewers for helpful comments and suggestions.
This work was supported by NIH grant GM28553.
REFERENCES
- 1.Addinall, S. G., E. F. Bi, and J. Lutkenhaus. 1996. FtsZ ring formation in fts mutants. J. Bacteriol. 178:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. Temperature shift experiments with ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J. Bacteriol. 179:4277-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Addinall, S. G., and J. Lutkenhaus. 1996. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol. 22:231-237. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong, K. A., R. Acosta, E. Ledner, Y. Machida, M. Pancotto, M. McCormick, H. Ohtsubo, and E. Ohtsubo. 1984. A 37 × 103 molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J. Mol. Biol. 175:331-348. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257-266. [DOI] [PubMed] [Google Scholar]
- 6.Bi, E., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 7.Bi, E., and J. Lutkenhaus. 1992. Isolation and characterization of ftsZ alleles that affect septal morphology. J. Bacteriol. 174:5414-5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bramhill, D., and C. M. Thompson. 1994. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc. Natl. Acad. Sci. USA 91:5813-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churchward, G., D. Belin, and Y. Nagamine. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165-171. [DOI] [PubMed] [Google Scholar]
- 10.Dai, K., and J. Lutkenhaus. 1991. ftsZ is an essential cell division gene in Escherichia coli. J. Bacteriol. 173:3500-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson, H. P., D. W. Taylor, K. A. Taylor, and D. Bramhill. 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. USA 93:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harry, E. J., K. Pogliano, and R. Losick. 1995. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 177:3386-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Löwe, J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203-206. [DOI] [PubMed] [Google Scholar]
- 15.Lu, C., J. Stricker, and H. P. Erickson. 1998. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima—quantitation, GTP hydrolysis, and assembly. Cell Motil. Cytoskel. 40:71-86. [DOI] [PubMed] [Google Scholar]
- 16.Lu, C., J. Stricker, and H. P. Erickson. 2001. Site-specific mutations of FtsZ—effects on GTPase and in vitro assembly. BMC Microbiol. 1:7. [DOI] [PMC free article] [PubMed]
- 17.Lu, C. L., M. Reedy, and H. P. Erickson. 2000. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol. 182:164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutkenhaus, J., H. Wolf-Watz, and W. D. Donachie. 1980. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J. Bacteriol. 142:615-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, X., D. W. Ehrhardt, and W. Margolin. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. USA 93:12998-13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, X., and W. Margolin. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli ftsZ. J. Bacteriol. 181:7531-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mileykovskaya, E., Q. Sun, W. Margolin, and W. Dowhan. 1998. Localization and function of early cell division proteins in filamentous Escherichia coli cells lacking phosphatidylethanolamine. J. Bacteriol. 180:4252-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee, A., and J. Lutkenhaus. 1994. Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol. 176:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phoenix, P., and G. R. Drapeau. 1988. Cell division control in Escherichia coli K-12: some properties of the ftsZ84 mutation and suppression of this mutation by the product of a newly identified gene. J. Bacteriol. 170:4338-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pogliano, K., E. J. Harry, and R. Losick. 1995. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol. Microbiol. 18:459-470. [DOI] [PubMed] [Google Scholar]
- 26.Shine, J., and L. Dalgarno. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegele, D. A., and J. C. Hu. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA 94:8168-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stricker, J., P. Maddox, E. D. Salmon, and H. P. Erickson. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. USA 99:3171-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, X. D., P. A. de Boer, and L. I. Rothfield. 1991. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 10:3363-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, X. D., J. A. Huang, A. Mukherjee, C. Cao, and J. Lutkenhaus. 1997. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, Y., B. D. Jones, and Y. V. Brun. 2001. A set of ftsZ mutants blocked at different stages of cell division in Caulobacter. Mol. Microbiol. 40:347-360. [DOI] [PubMed] [Google Scholar]