Abstract
To date, 24 Legionella pneumophila genes (icm and dot genes) have been shown to be required for intercellular growth and host cell killing. A previous report indicated that the regulation of these genes is complicated and probably involves several regulatory proteins. In this study, a genetic screen performed in Escherichia coli identified the CpxR response regulator as an activator of the L. pneumophila icmR gene. Construction of an L. pneumophila cpxR insertion mutant showed that the expression of icmR is regulated by CpxR. In addition, a conserved CpxR binding site (GTAAA) was identified in the icmR regulatory region and L. pneumophila His-tagged CpxR protein was shown to bind to the icmR regulatory region using a mobility shift assay. Besides its dramatic effect on the icmR level of expression, the CpxR regulator was also found to affect the expression of the icmV-dotA and icmW-icmX operons, but to a lesser extent. The role of CpxA, the cognate sensor kinase of CpxR, was also examined and its effect on the icmR level of expression was found to be less pronounced than the effect of CpxR. The RpoE sigma factor, which was shown to coregulate genes together with CpxR, was examined as well, but it did not influence icm and dot gene expression. In addition, when the cpxR mutant strain, in which the expression of the icmR gene was dramatically reduced, and the cpxA and rpoE mutant strains were examined for their ability to grow inside Acanthamoeba castellanii and HL-60-derived human macrophages, no intracellular growth defect was observed. This study presents the first evidence for a direct regulator (CpxR) of an icm-dot virulence gene (icmR). The CpxR regulator together with other regulatory factors probably concerts with the expression of icm and dot genes to result in successful infection.
Legionella pneumophila, the causative agent of Legionnaires' disease, is a broad-host-range facultative intracellular pathogen. L. pneumophila is able to infect, multiply within, and kill human macrophages as well as the free-living amoebae that serve as their environmental reservoir (19, 37). Two regions of icm and dot genes required for human macrophage killing and intercellular multiplication have been discovered in L. pneumophila (reviewed in references 41 and 46). Most of these genes were also shown to be required for intracellular growth in the protozoan host Acanthamoeba castellanii (42). Complementation and primer extension analysis indicated that these genes are probably organized in nine transcriptional units (icmTS, icmR, icmQ, icmPO, icmMLKEGCD, icmJB, icmF-tphA, icmWX, and icmV-dotA) (2, 4, 34, 39, 40, 45). The icm-dot system encodes a type IV secretion system that translocates effector molecules into the host cell and, in this way, modulates the properties of the phagosome containing bacteria (7, 30, 41, 46). The specific function of most of the Icm proteins is not known, but the IcmS and IcmW proteins as well as the IcmQ and IcmR proteins were shown to interact with one another (6). In addition, the IcmR protein was shown to exhibit chaperone activity for IcmQ, which was shown to form homopolymers (13).
Recently, 12 regulatory sites were identified in the upstream region of eight icm and dot genes. Seven of these sites were shown to constitute the −10 promoter elements of these genes, whereas the other five are expected to serve as binding sites for regulatory factors (16). To date, several regulatory factors have been shown to be involved in the regulation of icm and dot genes in L. pneumophila, including the stationary-phase sigma factor RpoS, the ppGpp synthetase RelA (50), and the response regulator LetA (15, 26) but none of them was shown to directly regulate any of the icm and dot genes.
CpxR and CpxA are known to comprise a two-component system, which constitutes a typical signal transduction pathway (reviewed in references 1, 17, and 43). In this system, cpxR encodes the cytoplasmic response regulator (12), while cpxA encodes an inner membrane sensor histidine kinase (36). In E. coli the Cpx system is activated by envelope stress signals, such as accumulation of misfolded proteins in the periplasm. When the signal is received, CpxA is being activated by autophosphorylation on a conserved histidine residue and then acts as a kinase to phosphorylate a conserved aspartate residue of the CpxR regulator. Phosphorylation of CpxR by CpxA-p enhances its binding upstream from target genes, leading to transcriptional activation (reviewed in reference 35). Recent studies of several pathogenic bacteria determined the role of the Cpx system in the regulation of virulence genes. In uropathogenic Escherichia coli, Cpx was found to be involved in P-pilus assembly (21, 22). A Salmonella enterica serovar Typhi cpxA mutant does not invade intestinal epithelial cells (24). In Shigella sonnei, the virF gene (a master regulator of genes required for host cell invasion) was found to be under the control of the Cpx pathway (31, 32).
The goal of the study presented here was to identify the regulator of the icmR gene that was shown to have different levels of expression in L. pneumophila and E. coli. By applying a genetic complementation screen, the L. pneumophila CpxR homolog was found to directly regulate the expression of the icmR gene. In addition, besides its dramatic effect on icmR gene expression, the CpxR regulator was found to positively regulate the expression of two icm-dot operons (icmV-dotA and icmW-icmX), but to a lesser extent. These results indicate that the CpxR regulator plays a major role in the regulation of the icm and dot genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and media.
L. pneumophila strains used in this work were JR32, a streptomycin resistance, restriction-negative mutant of L. pneumophila Philadelphia-1, which is a wild-type strain in terms of intracellular growth (38), and a mutant that cannot grow intracellularly, 25D (19). L. pneumophila strains constructed in this study are all JR32 derivatives, including a cpxR insertion mutant, OG2002, and cpxA and rpoE deletion substitution mutants OG2004 and OG2003, respectively. The E. coli strains used were MC1061 (5) and BL21(DE3) (44). Plasmids and primers used in this work are described in Tables 1 and 2, respectively. Bacterial media, plates, and antibiotic concentrations were used as described before (40).
TABLE 1.
Plasmids used in this study
| Plasmid | Feature(s) | Reference or Source |
|---|---|---|
| pBR322 | rep (pMB1) rop Apr Tcr | 3 |
| pET-15b | oriR (ColE1) Apr, pT7, N-terminal His6 tag | Novagen |
| pGS-lac-02 | pAB-1 with a promoterless lacZ gene | 16 |
| pGS-reg-RP1 | pOG-R125 containing a mutation in the promoter of icmR | This study |
| pGS-reg-RP2 | pOG-R125 containing a mutation in the promoter of icmR | This study |
| pLAW344 | sacB MCSaoriT (RK2) CmroriR (ColE1) Apr | 47 |
| pMMB207 | RSF1010 derivative, IncQ CmroriT MCS | 29 |
| pOG-BCC2 | E. coli cpxR in pOG-BRR3 | This study |
| pOG-BRM1 | pOG-R125 containing a mutation in the BRM1 site of icmR | This study |
| pOG-BRM2 | pOG-R125 containing a mutation in the BRM2 site of icmR | This study |
| pOG-BRR1 | icmR::lacZ fusion from pOG-R-125 in pBR322 | This study |
| pOG-BRR2 | pOG-BRR1 containing a single EcoRI site | This study |
| pOG-BRR3 | Ptac promoter in pOG-BRR2 | This study |
| pOG-cpxA-Km1 | pOG-U-cpxRA with the Kmr cassette in cpxA | This study |
| pOG-cpxA-Km2 | Insert of pOG-cpxA-Km1 in pLAW344 | This study |
| pOG-cpx-C2 | E. coli cpxR in pUC18 | This study |
| pOG-cpxR-1 | L. pneumophila cpxR in pUC18 | This study |
| pOG-cpxR-2 | pOG-cpxR-1 with the Kmr cassette in cpxR | This study |
| pOG-cpxR-3 | Insert of pOG-cpxR-2 in pLAW344 | This study |
| pOG-ECP2 | L. pneumophila cpxR in pET-15b | This study |
| pOG-R-125 | icmR::lacZ fusion in pGS-lac-01 | 50 |
| pOG-R-cpx3 | L. pneumophila cpxR in pOG-R125 | This study |
| pOG-RCPR3 | L. pneumophila cpxR in pOG-BRR3 | This study |
| pOG-R-cpxRA | L. pneumophila cpxRA in pOG-R125 | This study |
| pOG-rpoE-1 | L. pneumophila rpoE in pUC18 | This study |
| pOG-rpoE-2 | pOG-rpoE-1 with the Kmr cassette in rpoE | This study |
| pOG-rpoE-3 | Insert of pOG-rpoE-2 in pLAW344 | This study |
| pOG-RR16 | L. pneumophila genomic insert in pOG-BRR3 | This study |
| pOG-U-cpxRA | L. pneumophila cpxRA in pUC18 | This study |
| pOG-U-ECP | L. pneumophila cpxR in pUC18 | This study |
| pSS-R27 | Regulatory region of icmR in pMC1403 | 50 |
| pUC18 | oriR (ColE1) MCS Apr | 49 |
MCS, multiple cloning site.
TABLE 2.
Primers used in this study
| Primer name | Sequence (5′-3′) |
|---|---|
| cpxA-R-2 | GGATGTTTCAATTTCCATGCG |
| cpxA-R long | GTGGTTTACAAG TACGAGAGG |
| cpx-box-1F | GTTGTTTTGAAAGAATTAGAAAGTTTTATTGG |
| cpx-box-1R | CTAATTCTTTCAAAACAACTAATCATACATTAAC |
| cpx-box-2F | GATATATTGAAAGTAAGAGATTTAGCTCAGG |
| cpx-box-2R | CTCTTACTTTCAATATATCAAAATATATCTTTCAA |
| CpxR-Coli-F | ACATGCTGCTCAATCATCAGCCCC |
| CpxR-Coli R2 | GAATTCATGAATAAAATCCTGTTAGTTG |
| cpxR-F-2 | TAAAACAC ATGAAGGACACTGC |
| cpxR-pET-F | GGCATATGAGCAGCTCTATTCTCATTATTG |
| cpxR-pET-R | CCGGATCCTACAGACTACGCATTAAACATGTACCC |
| icmR-Down | ATGGGAACCAAGAATTAGGG |
| icmR-PRO-1F | TATATTTTGATAGATGTAAAGTAAGAGATTTAGCTC |
| icmR-PRO-1R | ACTTTACATCTATCAAAATATATCTTTCAATATATC |
| icmR-PRO-2F | TATATTTTGATCTATGTAAAGTAAGAGATTTAGCTC |
| icmR-PRO-2R | ACTTTACATAGATCAAAATATATCTTTCAATATATC |
| icmR-reg-up | CCCTGGATGAGTTAATGTATG |
| icmR-Up | GAATTCAGGAGTGGTAATAATGGGT |
| pMC-lac | TAAGTTGGGTAACGCCAGGG |
| rpoE-F | TTACTTCAATTAAATCTTGGGGC |
| rpoE-R | CCATTGAAGTGTTCTATCACCG |
Genetic screen to identify an icmR::lacZ activator.
To construct the vector for the genetic screen, the plasmid pBR322 was digested with HindIII and EcoRI and the icmR::lacZ fusion from pOG-R125 digested with the same enzymes was cloned into it, resulting in the plasmid pOG-BRR1. This plasmid was then partially digested with EcoRI, filled in and religated. A plasmid containing a single EcoRI site was isolated and designated pOG-BRR2. Subsequently, pOG-BRR2 was digested with HindIII and NruI, and a HindIII-Eco47III fragment containing the Ptac promoter from pMMB207 was cloned into it to generate pOG-BRR3. In this plasmid the unique EcoRI site is located immediately downstream from the Ptac promoter. To construct an L. pneumophila genomic library in pOG-BRR3, chromosomal DNA from the L. pneumophila JR32 wild-type strain was partly digested with Tsp509I. The digested DNA was separated on 1% agarose gel and the 2.5- to 3.5-kb fragments were purified from the gel. These fragments were cloned into the EcoRI site (Tsp509I has compatible cohesive ends with EcoRI) of pOG-BRR3 and electroporated into E. coli MC1061. The resulting transformants were screened on MacConkey plates containing ampicillin and IPTG (isopropyl-β-d-thiogalactopyranoside). The background clones gave pale pink colonies, while the suspected clones, which had higher levels of β-galactosidase, gave dark-red colonies.
Construction of plasmids for allelic exchange.
All the primers used for the allelic exchange procedure were designed according to the L. pneumophila genome sequence information (http://genome3.cpmc.columbia.edu/∼legion/index.html). In order to construct an L. pneumophila cpxR insertion mutant, the primers cpxR-F-2 and cpxA-R-2 (Table 2) were used to amplify a 1,784-bp DNA fragment containing the complete cpxR gene and part of the cpxA gene. This fragment was cloned into pUC18 digested with HincII to generate pOG-cpxR-1. To knock out the cpxR gene, the kanamycin resistance cassette (Pharmacia) was cloned into the EcoRV site in cpxR to generate pOG-cpxR-2. The plasmid pOG-cpxR-2 was digested with EcoRI and XmnI and filled in, and the insert was cloned into the EcoRV site of the allelic exchange vector pLAW344, resulting in pOG-cpxR-3. This plasmid was used for allelic exchange, as was previously described (42), to result in the L. pneumophila cpxR insertion mutant, OG2002. Several isolates of OG2002 were analyzed by PCR to confirm that the right change occurred on the chromosome (data not shown).
To construct an L. pneumophila cpxA deletion substitution mutant, the primers cpxR-F-2 and cpxA-R-long (Table 2) were used to amplify a 3,294-bp DNA fragment containing the entire cpxR and cpxA genes. This fragment was cloned into pUC18 digested with SmaI to generate pOG-U-cpxRA. To knock out the cpxA gene, the kanamycin resistance cassette was cloned instead of an internal 85-bp SmaI-Eco47III fragment to generate pOG-cpxA-Km1. This plasmid was digested with EcoRV and the insert was cloned into the EcoRV site of the vector pLAW344 to generate pOG-cpxA-Km2. This plasmid was used for allelic exchange, which resulted in an L. pneumophila cpxA deletion substitution mutant, OG2004. Several isolates of OG2004 were analyzed by PCR and Southern hybridization to confirm that the right change occurred on the chromosome (data not shown).
An L. pneumophila rpoE deletion substitution mutant was constructed by amplification of the rpoE gene using the primers rpoE-F and rpoE-R (Table 2). The resulting 2,704-bp PCR fragment was cloned into pUC18 digested with SmaI to generate pOG-rpoE-1. The kanamycin resistance cassette was cloned instead of an internal 474-bp NsiI fragment (after being filled in) to result in pOG-rpoE-2. This plasmid was then digested with EcoRI and filled in, and the insert was cloned into the EcoRV site of the allelic exchange vector pLAW344, resulting in the plasmid pOG-rpoE-3. This plasmid was used to construct the L. pneumophila rpoE deletion substitution mutant OG2003. Several isolates of OG2003 were analyzed by PCR to confirm that the correct change occurred on the chromosome (data not shown).
Construction of plasmids for CpxR and CpxA complementation.
To complement the expression of the icmR::lacZ fusion in E. coli, the L. pneumophila cpxR gene was isolated by digesting the plasmid pOG-cpxR-1 with EcoRI and PvuII. The insert (1,135 bp) containing the L. pneumophila cpxR gene was filled in and cloned into the vector pOG-BRR3 (after digestion with EcoRI and fill in) to generate the plasmid pOG-RCPR3. To complement the expression of the icmR::lacZ fusion in the L. pneumophila cpxR mutant (OG2002), the later insert containing the cpxR gene was cloned into pOG-R125 digested with XmnI to generate pOG-R-cpx3. In order to complement the expression of the icmR::lacZ fusion in the L. pneumophila cpxA mutant (OG2004), the plasmid pOG-U-cpxRA was digested with PstI and HindIII and the insert (2,865 bp) was cloned into the plasmid pOG-R-cpx3 digested with the same enzymes to generate pOG-R-cpxRA.
Cloning of the E. coli cpxR gene.
In order to determine whether the E. coli CpxR protein can activate the L. pneumophila icmR::lacZ fusion, the intact E. coli cpxR gene was amplified by PCR from the E. coli MC1061 chromosome, using the primers cpxR-coli-F and cpxR-coli R2 (Table 2). The resulting 858-bp PCR product was cloned into pUC18 digested with HincII to generate pOG-cpx-C2, which was verified by sequencing. This plasmid was subsequently digested with EcoRI and HindIII and the resulting fragment was cloned into pOG-BRR3 digested with the same enzymes to generate pOG-BCC2, containing the complete E. coli cpxR gene under the regulation of the Ptac promoter.
Site-directed mutagenesis.
Site-directed mutagenesis was performed by using the overlap extension PCR method (18), as described before (16). The PCR template for all the mutations constructed in the icmR regulatory region was pSS-R27. The resulting PCR products were digested with BamHI and cloned into the vector pGS-lac-02. All of the mutations were confirmed by sequencing of the whole regulatory region. The changes made were always A to C, T to G, C to A, and G to T. The primers used to mutate the BRM1 (for binding of regulator mutation 1) site (pOG-BRM1) were cpx-box-1F and cpx-box-1R; the BRM2 (for binding of regulator mutation 2) site (pOG-BRM2), cpx-box-2F and cpx-box-2R; the second nucleotide of the promoter (pGS-reg-RP1), icmR-PRO-1F and icmR-PRO-1R; and the third nucleotide of the promoter (pGS-reg-RP2), icmR-PRO-2F and icmR-PRO-2R (Table 2).
Northern hybridization analysis.
RNA preparation was performed as described before (16). RNA samples (10 μg) were separated on 1.5% agarose gel containing formaldehyde and transferred to a nitrocellulose membrane (Schleicher & Schuell) by capillary transfer in 10× SSC (0.15 M sodium chloride, 15 mM sodium citrate). To prepare a probe for the icmR gene, a 452-bp PCR fragment was amplified using the primers icmR-Up and icmR-Down (Table 2). The icmR probe was purified from agarose gel and radiolabeled with [α-32P]dCTP by the random prime labeling kit (Roche). Hybridization was performed at 65°C for 16 h in a solution containing 5× SSPE (0.18 M sodium chloride, 10 mM sodium phosphate [pH 7.7], 1 mM EDTA), 2.5× Denhardt's solution (0.02% Ficoll, 0.02% polyvinylpyrrolidone, 0.02% bovine serum albumin [BSA]), 0.25% sodium dodecyl sulfate (SDS), and 150 mg of denatured herring sperm DNA per ml. After the hybridization, the filter was washed three times (briefly) in 2× SSPE-0.1% SDS at room temperature, once in 1× SSPE-0.1% SDS at 65°C for 20 min, and then in 0.1× SSPE-0.1% SDS at 65°C for 20 min. Then, the membrane was air dried and exposed to X-ray film (Fuji).
Construction of plasmids for L. pneumophila His6-CpxR protein purification.
A 700-bp fragment containing the L. pneumophila cpxR gene was amplified by PCR using the primers cpxR-pET-F and cpxR-pET-R containing an NdeI and a BamHI site, respectively (Table 2). The 700-bp insert was cloned into pUC18 digested with HincII to generate pOG-U-ECP. This plasmid was sequenced and digested with NdeI and BamHI and the resulting insert was cloned into the pET-15b vector (Novagen) digested with the same enzymes to generate pOG-ECP2. The resulting plasmid expresses a full-length L. pneumophila CpxR protein fused to a His6 tag on its N terminus, with a predicted molecular mass of ∼28 kDa, under the T7 promoter.
Protein purification.
The L. pneumophila His6-CpxR protein was purified from E. coli BL-21(DE3) transformed with pOG-ECP-2. The protein was purified by nickel-affinity chromatography using Ni-nitrilotriacetic acid (Ni-NTA) resin (Qiagen). Protein purification was performed at room temperature under native conditions, according to adjusted protocols described in standard protocol 7 and 9 in the QIAexpressionist manual (available from Qiagen). Briefly, strain BL21(DE3) containing pOG-ECP-2 was grown overnight in NZCYM medium (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl, 1 g of Casamino Acids, 20.28 ml of 0.4 M MgSO4 · 7H2O in 1 liter of water) containing ampicillin (50 μg/ml) at 37°C with shaking. The culture was then diluted 1/100, and after 2 h, IPTG was added to 1 mM to induce expression of the fusion protein. The bacteria were grown for an additional 4 h at 37°C under vigorous aeration to an optical density at 600 nm (OD600) of approximately 2.0 and harvested by centrifugation at 7,600 × g for 10 min. The pellet was resuspended in 1 ml of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, and lysozyme [1 mg/ml]). Cells were disrupted by sonication and then centrifuged at 10,000 × g for 20 min. The soluble fraction was collected and 1 ml of 50% Ni-NTA resin was added to it, and the mixture was gently shaken on ice for 60 min and then loaded on a column. The column was washed twice with 4 ml of wash buffer (50 mM NaH2PO4, 600 mM NaCl, 20 mM imidazole, 0.1% Triton). The protein was eluted in four fractions with 0.2, 0.8, 0.5 and 0.5 ml of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole). Fractions 2 to 4 were then collected and dialyzed (at 4°C) initially against 0.1 M sodium phosphate (pH 8.0), 1 mM EDTA, 150 mM NaCl, 10 mM β-mercaptoethanol, 10% glycerol, and 0.5 mM phenylmethylsulfonyl fluoride and then with the same buffer containing 25% glycerol. Following dialysis the protein was concentrated to ∼0.37 mg/ml in a centrifugal filter device (Centricon YM-10; Amicon) and stored at −20°C. Protein concentration was estimated by using the Bio-Rad protein assay reagent (Bio-Rad), using BSA as a standard.
Mobility shift assay.
A 200-bp DNA fragment containing the icmR regulatory region (consisting of nucleotides −98 to + 102 from the icmR transcription start site), was amplified by PCR from the plasmid pOG-R125 using the primers pMC-lac and icmR-reg-up (Table 2). This fragment was labeled with digoxigenin (DIG)-dUTP by the PCR DIG probe synthesis kit (Roche) according to the manufacturer's instructions (but with modified PCR DIG mix that was diluted 1:2.5 with unlabeled deoxynucleoside triphosphates). In each assay 10 fmol of the DIG-labeled probe was used. The His6-CpxR protein (at the concentration indicated) was incubated in the presence or absence of 50 mM acetyl phosphate (Ac-P) at 37°C for 30 min in a 15-μl reaction volume containing 10 mM Tris (pH 7.4), 50 mM KCl, 1 mM EDTA, 5% glycerol, BSA (50 μg/ml), 1 mM dithiothreitol, 20 mM potassium glutamate, and 10 mM MgSO4. The DIG-labeled probe (10 fmol) was added, and the reaction mixture was incubated for another 30 min at 37°C. Reactions were stopped by the addition of 3 μl of loading dye, and products were electroporated on 6% nondenaturing polyacrylamide gels (29:1) in 0.5× TAE running buffer (0.04 M Tris-acetate, 1 mM EDTA). Subsequently, the gels were electroblotted onto a positively charged nylon membrane (Schleicher & Schuell) and fixed by UV cross-linking. Detection of the DIG-labeled DNA probe by anti-DIG Fab fragment-alkaline phosphatase conjugate (Roche) and substrate CSPD was performed as directed by the manual from Roche.
β-Galactosidase assays.
β-Galactosidase assays were performed as described elsewhere (28). L. pneumophila strains were grown on ACES (N-[2-acetamido]-2-aminoethane-sulfonic acid) buffered charcoal yeast extract (ABCYE) plates containing chloramphenicol for 48 h. The bacteria were scraped off the plate and suspended in ACES yeast extract (AYE) medium, and the bacterial OD600 was calibrated to 0.1 in AYE. The resulting cultures were grown on a roller drum for 17 to 18 h until reaching an OD600 of about 3.2 (stationary phase), and the assays were performed with 50 μl of culture. E. coli strains were grown in LB broth containing ampicillin for 16 h. The cultures were then diluted 1:100 into fresh LB broth with or without IPTG (1 mM) and grown on a roller drum for 2 h until reaching an OD600 of about 0.6 (exponential phase); 100 μl from this culture was taken to the assays. The substrate for β-galactosidase hydrolysis was o-nitrophenyl-β-d-galactopyranoside (Sigma).
Intracellular growth in A. castellanii.
Intracellular growth assays were performed in a similar way to what was previously described (42). A. castellanii (ATCC 30234) cells (1.5 × 105) in proteose yeast glucose broth were added to wells of a 24-well microtiter plate and the amoebae were incubated for 1 h at 37°C to let the amoebae adhere. Then the proteose yeast glucose was aspirated, and the wells were washed once with 0.5 ml of warm (37°C) Acanthamoeba buffer (Ac-buffer), and 0.5 ml of warm Ac-buffer was added to the wells. Then, L. pneumophila, in Ac-buffer, was added to the wells at a multiplicity of infection of approximately 0.1. The plate was incubated for 30 min at 37°C, the Ac-buffer was aspirated, the wells were washed three times with 0.5 ml of warm Ac-buffer, and 0.6 ml of warm Ac-buffer was added to the wells. The supernatant of each well was sampled at intervals of about 24 h and CFU were determined by plating multiple dilutions on ABCYE plates.
Intracellular growth in HL-60-derived human macrophages.
Intracellular growth assays were performed in a similar way to what was previously described (42). Wells of a 24-well microtiter plate containing 6 × 106 differentiated HL-60-derived macrophages were used for infection. L. pneumophila was added to the wells at a multiplicity of infection of approximately 0.1 and the infected HL-60-derived macrophages were incubated for 1 h at 37°C under CO2 (5%). Then, the wells were washed three times, and 0.6 ml of RPMI 1640 containing 2 mM glutamine and 10% normal human serum was added to the wells. The supernatant of each well was sampled at intervals of about 24 h and CFU were determined by plating multiple dilutions on ABCYE plates.
Nucleotide sequence accession numbers.
Sequence data of the L. pneumophila cpxRA and rpoE genes have been assigned GenBank accession numbers AY295086 and AY295087, respectively.
RESULTS
The levels of expression of nine translational fusions of L. pneumophila icm and dot genes (icmT::lacZ, icmR::lacZ, icmQ::lacZ, icmP::lacZ, icmM::lacZ, icmJ::lacZ, icmF::lacZ, icmW::lacZ, and icmV::lacZ) have been previously compared between E. coli and L. pneumophila (16). It was found that the level of expression of the icmR::lacZ fusion in E. coli in comparison to L. pneumophila was fourfold lower at exponential phase (295 ± 16 and 1,157 ± 159 Miller units [MU], respectively) and sevenfold lower at stationary phase (369 ± 19 and 2,435 ± 339 MU, respectively) (16). (Unless otherwise noted, results are presented as means ± standard deviations.) These results led us to assume that L. pneumophila possesses a regulatory factor(s) required for optimal expression of the icmR gene that is missing or functions differently in E. coli. These differences in the level of expression of the icmR::lacZ fusion served as the basis for a genetic screen aimed at the identification of the L. pneumophila icmR regulator.
Genetic screen for the identification of an icmR regulator.
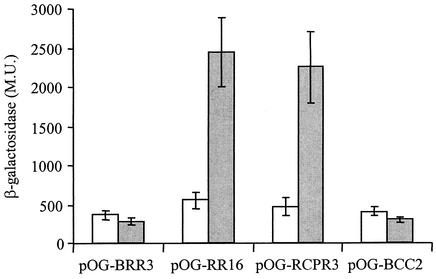
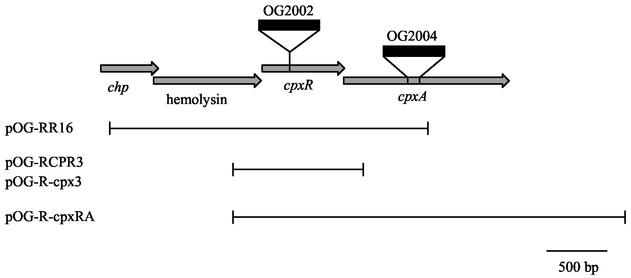
To identify the activator that participates in the regulation of icmR, a genetic screen of the L. pneumophila library in E. coli was performed. The L. pneumophila library was constructed using a plasmid (pOG-BRR3) that contains the icmR::lacZ fusion and at a different position a unique EcoRI site located immediately downstream from a Ptac inducible promoter (see Materials and Methods). All together, about 13,700 E. coli transformants were screened on MacConkey plates containing IPTG for higher expression of the icmR::lacZ fusion, and 24 suspected (dark-red) clones were identified. One clone (pOG-RR16) was found to have a particularly high level of β-galactosidase activity (2,452 ± 446 MU) in comparison to the vector (pOG-BRR3) β-galactosidase background level (283 ± 37 MU). This high level of expression of the icmR::lacZ fusion was found to be dependent on IPTG induction (Fig. 1). Most of the other 23 clones isolated showed β-galactosidase levels of expression of about 300 to 350 MU. The positive clone that had a high level of expression (pOG-RR16) was partly sequenced and examined by restriction analysis. pOG-RR16 was found to contain two joined inserts from different regions of the L. pneumophila chromosome, 2.6 and 6.4 kb in size. The sequence data from pOG-RR16 was compared to the L. pneumophila genome database, which showed that the 2.6-kb fragment (located immediately downstream from the Ptac promoter) contains the following genes: (i) an open reading frame (ORF) homologous to a conserved hypothetical protein (chp), (ii) an ORF homologous to hemolysin, (iii) an ORF homologous to cpxR, and (iv) a portion (217 of 455 amino acids) from an ORF homologous to cpxA (Fig. 2). The L. pneumophila CpxR ortholog was found to be 49% identical and 68% similar to the E. coli CpxR, whereas the L. pneumophila CpxA ortholog was found to be 27% identical and 46% similar to the E. coli CpxA. The L. pneumophila cpxR and cpxA genes were found to overlap each other by 8 bp, an organization that was also found in other bacteria (10).
FIG. 1.
Level of expression of the icmR::lacZ fusion in E. coli. E. coli containing different plasmids was grown to exponential phase, and the level of expression of the icmR::lacZ fusion was determined without (white) and with (gray) IPTG induction. The plasmids examined were the vector (pOG-BRR3), the clone isolated from the genetic screen (pOG-RR16), the vector harboring the L. pneumophila cpxR gene by itself (pOG-RCPR3), and the vector harboring the E. coli cpxR gene (pOG-BCC2). The β-galactosidase specific activity is presented as an average of the results of at least three different experiments. The error bars represent standard deviations.
FIG. 2.
Linkage map of the L. pneumophila cpxRA locus. The arrows indicate ORFs (conserved hypothetical protein, chp; hemolysin; cpxR and cpxA). The position of the kanamycin resistance cassette in the cpxR insertion mutant (OG2002) and cpxA deletion substitution mutant (OG2004) is indicated. The thin lines indicate the regions covered by the plasmids listed on the left.
L. pneumophila CpxR enhances the expression of the icmR::lacZ fusion in E. coli.
The most reasonable candidate for being responsible for the elevated expression of the icmR::lacZ fusion in the clone described above (pOG-RR16) was the response regulator CpxR (see introduction). In order to test this assumption, the L. pneumophila cpxR gene by itself was cloned into the library vector (pOG-BRR3) and the resulting plasmid (pOG-RCPR3, Fig. 2) was examined for the level of expression of the icmR::lacZ fusion, in the presence and absence of IPTG. As can be seen in Fig. 1, in the presence of IPTG, the level of expression of the icmR::lacZ fusion from pOG-RCPR3 (2,255 ± 459 MU) was similar to the level of expression obtained with the original clone isolated by the genetic screen (pOG-RR16, 2,452 ± 446 MU) and was eightfold higher than the level of expression of the library vector (pOG-BRR3, 283 ± 37 MU). These data indicate that supplying the L. pneumophila cpxR gene on a plasmid significantly elevates the level of expression of the icmR::lacZ fusion in E. coli.
CpxR controls the expression of three L. pneumophila icm genes.
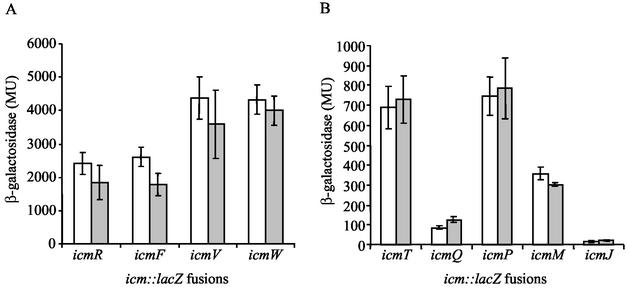
To examine, whether CpxR also plays a role in the regulation of icmR in L. pneumophila, an insertion mutant was constructed in the cpxR gene. The resulting L. pneumophila strain (OG2002, Fig. 2) grows well on bacteriological media with a growth rate similar to that of the wild-type strain (data not shown). To determine the involvement of CpxR in icm and dot gene expression, the levels of expression of the nine icm::lacZ fusions described above were compared between the cpxR mutant strain (OG2002) and the wild-type strain (JR32). As can be seen in Fig. 3, the level of expression of three icm::lacZ fusions was found to be significantly lower in OG2002. The level of expression of the icmV::lacZ and icmW::lacZ fusions decreased more than threefold (from 4,384 ± 634 to 1,278 ± 213 MU for icmV::lacZ and from 4324 ± 425 to 1418 ± 130 MU for icmW::lacZ in JR32 and OG2002, respectively) in the cpxR mutant strain (OG2002). However, the most profound change was the reduction in the level of expression of the icmR::lacZ fusion, which decreased 13-fold from 2,410 ± 329 MU in the wild-type strain (JR32) to 187 ± 26 MU in the cpxR mutant strain (OG2002). These results indicate that the response regulator CpxR is involved in the regulation of icmV, icmW, and especially icmR in L. pneumophila.
FIG. 3.
CpxR affects the level of expression of three icm::lacZ fusions. The level of expression of nine icm::lacZ fusions (icmT, icmR, icmQ, icmP, icmM, icmJ, icmF, icmV, and icmW) was examined in the L. pneumophila wild-type strain JR32 (white) and the cpxR insertion mutant OG2002 (gray). The β-galactosidase activity was measured as described in Materials and Methods. Four icm::lacZ fusions that had high β-galactosidase activities (A) and five that had low activities (B) are shown. The data are presented as averages of results of at least three different experiments. The error bars represent standard deviations. The level of expression of the vector was 7.3 ± 2.4 MU.
CpxR activates the expression of icmR in L. pneumophila.
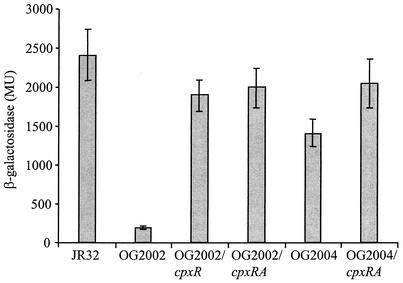
In order to give further support to the assumption that the reduction in the level of expression of icmR in OG2002 was due to the insertion in the cpxR gene, a complementation experiment was performed. The L. pneumophila cpxR gene was cloned into a plasmid containing the icmR::lacZ fusion (pOG-R125) and the resulting plasmid (pOG-R-cpx3) (Fig. 2) was introduced into the cpxR insertion mutant (OG2002). When the plasmid containing the cpxR gene (pOG-R-cpx3) was introduced into the cpxR insertion mutant strain (OG2002), the level of expression of icmR::lacZ was restored (1,901 ± 202 MU) to a level similar to that in the wild-type strain (2,410 ± 329 MU) (Fig. 4).
FIG. 4.
The expression of the icmR::lacZ fusion in different L. pneumophila strains. Strains of L. pneumophila containing different plasmids were grow to stationary phase and analyzed for the level of expression of the icmR::lacZ fusion. All the strains examined contain an icmR::lacZ fusion located on a plasmid. The following were used: wild-type strain (JR32), the cpxR insertion mutant (OG2002), the cpxR insertion mutant containing the L. pneumophila cpxR gene (OG2002/cpxR), the cpxR insertion mutant containing the L. pneumophila cpxRA genes (OG2002/cpxRA), the cpxA deletion substitution mutant (OG2004), and the cpxA deletion substitution mutant containing the L. pneumophila cpxRA genes (OG2004/cpxRA). The β-galactosidase activity was measured as described in Materials and Methods. The data are presented as an average of at least three different experiments. The error bars represent standard deviations. The level of expression of the vector was 7.3 ± 2.4 MU.
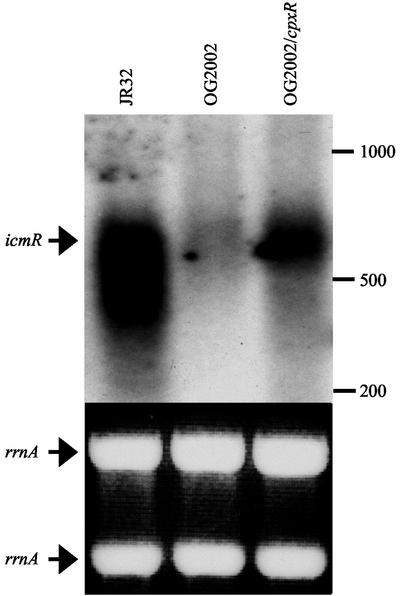
The differences in the levels of expression of the icmR gene between wild-type L. pneumophila (JR32), the cpxR mutant strain (OG2002) and the cpxR mutant strain (OG2002) containing a complementing plasmid (pOG-R-cpx3) were also determined at the mRNA level. Total RNA was purified from these strains and analyzed by Northern hybridization using the icmR gene as a probe (Fig. 5). Quantification of the hybridization signals indicated that the level of icmR mRNA in the L. pneumophila wild-type strain (JR32) was about 10-fold higher than its level in the cpxR mutant strain (OG2002) and introduction of the cpxR gene resulted in partial complementation. However, the cpxR mutant strain (OG2002) was found to hybridize normally with other probes such as the flaA gene (data not shown). Therefore, the Northern hybridization results are in agreement with the results obtained with the icmR::lacZ fusion (compare Fig. 4 and 5), showing that icmR gene expression is controlled by the CpxR regulator.
FIG. 5.
CpxR affects icmR mRNA level. RNA was purified from stationary-phase cultures of the L. pneumophila wild-type strain (JR32), the cpxR insertion mutant (OG2002), and the cpxR insertion mutant containing the complementing plasmid pOG-R-cpx3 (OG2002/cpxR). The upper part of the figure shows the icmR mRNA levels that were analyzed by Northern hybridization, using the icmR gene as a probe. The arrow indicates the icmR mRNA signal (about 620 bp); the size maker is indicated to the right. The lower part of the figure shows equal loadings of samples (10 μg) demonstrated by ethidium bromide staining of rrnA (23S and 16S) (indicated by arrows) in the agarose gel prior to transfer.
Identification of the CpxR binding site in the icmR regulatory region.
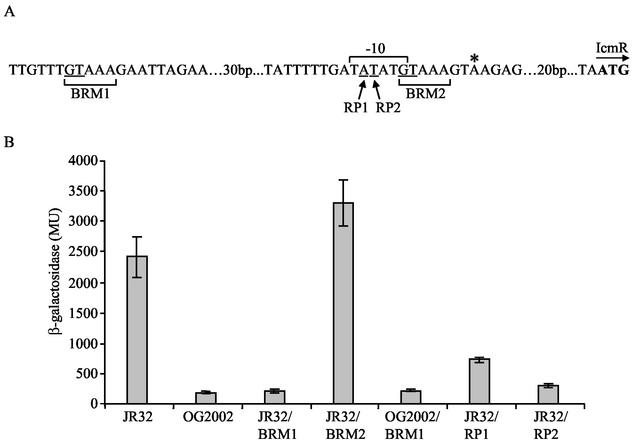
The binding site of the response regulator CpxR in the upstream regulatory region of its target genes has been identified before in E. coli (21, 33) and in S. sonnei (31). According to these reports, CpxR has a conserved recognition site that contains the sequence GTAAA (11). This sequence was found twice in the icmR regulatory region (Fig. 6A). The first GTAAA site (which will be referred to as BRM1) is located 87 bp upstream from the first IcmR methionine and 60 bp upstream from the icmR transcription start site (16). The second GTAAA site (which will be referred to as BRM2) is located 29 bp upstream from the first IcmR methionine and only 2 bp upstream from the transcription start site. To find which of these two potential sites (if any) serves as a binding site for CpxR, each of them was changed separately using site-directed mutagenesis (changing the first two nucleotides from GT to TG). The plasmids containing these mutations (pOG-BRM1 and pOG-BRM2) were introduced into L. pneumophila JR32 and the levels of expression of the icmR::lacZ fusion were determined (Fig. 6B). The expression of the icmR::lacZ fusion containing the mutation at the BRM1 site (pOG-BRM1) was found to be 11-fold lower than that of the icmR::lacZ fusion that contains a wild-type regulatory region (209 ± 37 and 2,411 ± 329 MU, respectively), a result expected from a mutation in an activator binding site. In contrast, the mutation constructed at the BRM2 site resulted in a higher level of expression of the icmR::lacZ fusion than that of the fusion that contains the wild-type regulatory region (3,291 ± 382 and 2,411 ± 329 MU, respectively) (Fig. 6B). This result might be explained by the fact that the mutation at the BRM2 site altered the −10 promoter element of icmR from TATATG to TATATT (Fig. 6A). The latter sequence was shown to constitute the −10 promoter element of the icmV gene, and our previous studies showed that the icmV::lacZ fusion has a higher level of expression than the icmR::lacZ fusion (16). Hence, changing the −10 promoter element of icmR to the sequence TATATT might be the cause for the higher level of expression of icmR::lacZ fusion containing the mutation at the BRM2 site.
FIG. 6.
Identification of the CpxR binding site in the icmR regulatory region. (A) Sequence of icmR regulatory region. The first ATG codon is shown in boldface type. The position of the transcription start site is indicated by an asterisk. The two GTAAA sites (BRM1 and BRM2) are indicated by their designations. The two nucleotides that were changed (GT to TG) within the BRM1 and BRM2 sites are underlined. The icmR −10 promoter element (TATATG) is indicated and the pair of nucleotides mutated in this site (A to C and T to G) are underlined and indicated by their designations (RP1 and RP2). (B) Effect of different mutations in the regulatory region of icmR on the expression of the icmR::lacZ fusion. The following were used: the wild-type strain containing the icmR::lacZ fusion (JR32), the cpxR insertion mutant containing the icmR::lacZ fusion (OG2002), the wild-type strain containing the icmR::lacZ fusion with a mutation at the BRM1 site (JR32/BRM1), the wild-type strain containing the icmR::lacZ fusion with a mutation at the BRM2 site (JR32/BRM2), the cpxR insertion mutant containing the icmR::lacZ fusion with a mutation at the BRM1 site (OG2002/BRM1), the wild-type strain containing the icmR::lacZ fusion with a mutation at the second nucleotide of the −10 promoter element (JR32/RP1), and the wild-type strain containing the icmR::lacZ fusion with a mutation at the third nucleotide of the −10 promoter element (JR32/RP2). The β-galactosidase activity was measured as described in Materials and Methods. The data are presented as averages of results of at least three different experiments. The error bars represent standard deviations. The expression level of the vector was 7.3 ± 2.4 MU.
To give further support to the hypothesis that the BRM1 site functions as the binding site for CpxR (and not a binding site of another unrelated icmR activator), the plasmid containing the mutation at the BRM1 site (pOG-BRM1) was examined in the L. pneumophila cpxR mutant strain (OG2002). As can be seen in Fig. 6B, no additive effect was found when the plasmid containing the mutation at the BRM1 site (pOG-BRM1) was examined in the cpxR mutant strain (OG2002), indicating that the sequence GTAAA at the BRM1 site constitutes the binding site for the CpxR activator. It was expected that if the BRM1 site was a recognition site for another regulator, and not CpxR, the effect of introducing the icmR::lacZ fusion containing the mutation at the BRM1 site into the cpxR mutant strain would have resulted in a lower level of expression of this fusion than that of the wild-type strain containing the same fusion (additive effect). In agreement with this observation, the level of expression from pOG-BRM1 (209 ± 37 MU) was very similar to the level of expression of the wild-type icmR::lacZ fusion in the cpxR mutant strain (188 ± 26 MU). These results show that the same level of expression could be achieved when either the binding site of CpxR was mutated or when the regulator itself was absent.
In order to learn more about the importance of the BRM1 site in icmR gene expression, the effect of the mutation in this site was compared to the effects of two mutations (RP1 and RP2) constructed in the −10 promoter element of icmR (Fig. 6A). Similar mutations that were previously constructed in the same two positions in the −10 promoter element of several icm genes resulted in dramatic reductions in the levels of expression of the genes located downstream from them (16). As can be seen in Fig. 6B, the level of expression from pOG-BRM1 (209 ± 37 MU) was found to be lower than the level of expression of the icmR::lacZ fusion carrying the mutations at the −10 promoter element (727 ± 42 MU for RP1 and 293 ± 31 MU for RP2). These results indicate that the CpxR binding site plays a critical role in the expression of icmR, and its contribution to the icmR gene expression is at least as significant as that of the −10 promoter element.
The E. coli CpxR protein does not activate the L. pneumophila icmR gene.
The finding that the L. pneumophila CpxR regulator recognizes a regulatory element similar to that of E. coli CpxR is not surprising; however, this finding makes it hard to understand the initial ability to identify the L. pneumophila CpxR regulator in the screen performed, as the E. coli strain used in the screen contained an intact cpxR gene. In order to clarify this issue, we cloned the E. coli cpxR gene into the vector that was used for the screen (pOG-BRR3) in the same way in which the L. pneumophila cpxR regulator was cloned. The resulting plasmid (pOG-BCC2) was examined for the level of expression of the icmR::lacZ fusion with or without the addition of IPTG. The expression of the icmR::lacZ fusion was found to be equal to that obtained with the vector (pOG-BRR3) (Fig. 1), indicating that the E. coli CpxR regulator cannot activate the L. pneumophila icmR gene. The analysis was performed during exponential as well as stationary phase, and the same result was obtained (Fig. 1 and data not shown).
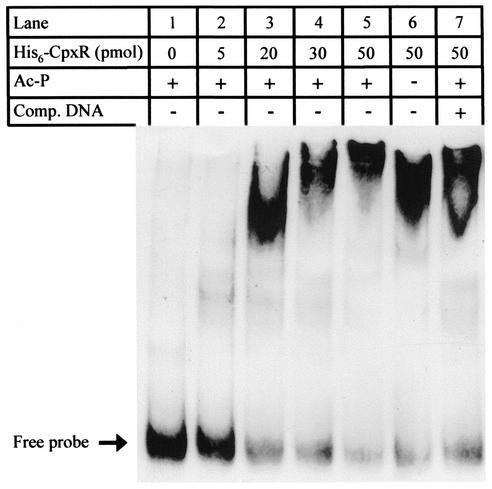
The L. pneumophila His6-CpxR protein binds to the icmR regulatory region.
To determine whether L. pneumophila CpxR binds to the icmR regulatory region, L. pneumophila CpxR was His tagged, overexpressed, and purified as described in Materials and Methods. We utilized a 200-bp fragment that covers the icmR regulatory region as a probe in a mobility shift assay with the His6-CpxR protein. The L. pneumophila His6-CpxR was found to bind to the regulatory region of icmR, as evidenced by a shift in the migration of the DNA probe after electrophoresis (Fig. 7, lanes 2 to 5), and no effect was observed when BSA (50 pmol) was added (Fig. 7, lane 1). The degree of the band shift as well as the amount of the shifted probe was correlated with increasing amounts of the His6-CpxR protein (Fig. 7, compare lanes 2 to 5). In addition, the amount of the shifted probe was found to be affected by the elimination of Ac-P from the reaction. This result indicates that phosphorylation of the His6-CpxR protein is required for maximum binding (Fig. 7, compare lanes 5 and 6). Finally, a competition with unlabeled probe reduced the band shift (Fig. 7, compare lanes 5 and 7), and no effect was observed when the same amount of Herring sperm DNA was added (data not shown). These data indicate that the L. pneumophila His6-CpxR protein binds to the icmR regulatory region.
FIG. 7.
L. pneumophila His6-CpxR binds to the icmR regulatory region. Mobility shift assays were performed with pure His6-CpxR and the DIG-labeled icmR regulatory region in the presence (+) and absence (−) of 100 fmol of the unlabeled icmR regulatory region (Comp. DNA) or acetyl phosphate (Ac-P). Each lane contains 10 fmol of the DIG-labeled probe and the amount of His6-CpxR (pmol) is indicated above each lane. Lane 1 contains 50 pmol of BSA.
CpxA has moderate effect on the expression of icmR.
Phosphorylation of CpxR, which usually occurs by its cognate sensor kinase CpxA, allows the upregulation of its target genes. Therefore, it was interesting to examine whether the expression of the icmR::lacZ fusion is dependent on the presence of a functional CpxA. To illuminate this issue, a deletion substitution was constructed in the cpxA gene. The resulting strain (OG2004) (Fig. 2), grows well on bacteriological media with a growth rate similar to that of the wild-type strain (data not shown).
The level of expression of the icmR::lacZ fusion in the L. pneumophila cpxA mutant strain (OG2004) was found to be mildly lower than that of the wild-type strain (1,416 ± 186 and 2,410 ± 329 MU, respectively) (Fig. 4). The lower level of expression of the icmR::lacZ fusion in OG2004 was complemented by introducing the cpxRA genes on a plasmid (pOG-R-cpxRA, 2,045 ± 307 MU) (Fig. 4). When this plasmid was introduced into the cpxR mutant strain (OG2002), the level of expression of the icmR::lacZ fusion (1,991 ± 250 MU) was similar to the level obtained by complementation with the cpxR gene by itself (pOG-R-cpx3, 1,901 ± 202 MU) and similar to the level of expression of the icmR::lacZ fusion in the wild-type strain (2411 ± 329 MU) (Fig. 4). These results indicate that in the conditions used, CpxA has only a moderate effect on the expression of icmR and that CpxR may be phosphorylated by factors other than CpxA (see Discussion).
RpoE is not involved in the regulation of the icm and dot genes.
RpoE is an alternative sigma factor that regulates extracytoplasmic functions (reviewed in reference 48). It was shown before that the Cpx pathway and RpoE share a functionally similar mechanism of signal transduction (9, 35) and that both are involved in the regulation of protein turnover and protein-folding activities in the bacterial envelope. In addition, in some cases the RpoE sigma factor and the Cpx pathways overlap in their target genes, as was demonstrated in E. coli for the regulation of the periplasmic protease DegP (9) and probably the heat shock sigma factor RpoH (33). Considering this, the role of the RpoE sigma factor in icm and dot gene expression was examined. Using a BLAST search against the L. pneumophila genome database, the sequence coding for the L. pneumophila RpoE was identified (the L. pneumophila RpoE was found to be 37% identical and 55% similar to the E. coli RpoE), and a deletion substitution was constructed in it (OG2003). The nine icm::lacZ fusions described above were introduced into this strain (OG2003) and their level of expression was determined. As can be seen in Fig. 8, no significant change was found in the levels of expression of all nine icm::lacZ fusions in OG2003 in comparison to those of the wild-type strain. These results indicate that the icm and dot genes (including the three genes that were shown to be regulated by CpxR) probably do not contain RpoE-dependent promoters. Analysis of the upstream regulatory region of these genes also revealed that promoter consensus sequences recognized by the E. coli RpoE sigma factor (48) are not present there.
FIG. 8.
RpoE does not affect the levels of expression of the icm::lacZ fusions. The levels of expression of nine icm::lacZ fusions (icmT, icmR, icmQ, icmP, icmM, icmJ, icmF, icmV, and icmW) were examined in L. pneumophila wild-type strain JR32 (white) and the rpoE deletion substitution mutant OG2003 (gray). β-Galactosidase activity was measured as described in Materials and Methods. Four icm::lacZ fusions that had high β-galactosidase activities (A) and five that had low activities (B) are shown. The data are presented in Miller units (MU) and are the averages of the results of at least three different experiments. The error bars represent standard deviations. The expression level of the vector was 7.3 ± 2.4 MU.
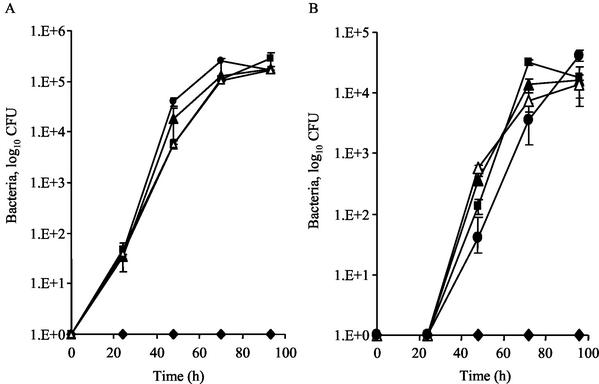
CpxR, CpxA, and RpoE are dispensable for intracellular growth of L. pneumophila.
In the environment, L. pneumophila replicates intracellularly within protozoan hosts, such as A. castellanii (14), while in humans, during infection, these bacteria grow inside alveolar macrophages (20, 37). Given that CpxR was found to affect the level of expression of three icm genes, required for intracellular multiplication, the role of CpxR (as well as of the other two regulators described, CpxA and RpoE) in intracellular growth within A. castellanii and HL-60-derived human macrophages was examined. As can be seen in Fig. 9, the L. pneumophila cpxR, cpxA, and rpoE mutant strains (OG2002, OG2004, and OG2003, respectively) replicated to the same extent as the wild-type strain in the protozoan host A. castellanii (Fig. 9A) and in HL-60-derived human macrophages (Fig. 9B). Therefore, we concluded that the three regulators examined (CpxR, CpxA, and RpoE) are dispensable for intracellular growth of L. pneumophila.
FIG. 9.
CpxR, CpxA and RpoE are dispensable for L. pneumophila intracellular multiplication. The ability of cpxR, cpxA, and rpoE mutant strains to grow intracellularly was examined in A. castellanii (A) and HL-60-derived human macrophages (B). Symbols: squares, L. pneumophila wild-type strain (JR32); diamonds, the mutant strain 25D; closed triangles, cpxR mutant strain (OG2002); open triangles, cpxA mutant strain (OG2004); circles, rpoE mutant strain (OG2003). The experiments were performed as described in Materials and Methods. The experiments were done three times, and the same results were obtained; error bars indicate standard errors.
DISCUSSION
We previously reported the construction of nine icm translational fusions (icmT::lacZ, icmR::lacZ, icmQ::lacZ, icmP::lacZ, icmM::lacZ, icmJ::lacZ, icmF::lacZ, icmW::lacZ, and icmV::lacZ) and the comparison of their levels of expression in E. coli and L. pneumophila (16). Most of these fusions (six out of nine) were found to retain similar levels of expression in both bacteria, suggesting that the same basic regulation of these icm genes is conserved between E. coli and L. pneumophila. However, the icmR::lacZ fusion, was found to have a high level of expression in L. pneumophila and a low level of expression in E. coli (16). These results led to the assumption that optimal expression of icmR requires an L. pneumophila regulatory factor(s) that is absent or functions differently in E. coli. In order to identify this potential regulator, a genetic screen was applied. In this screen, the L. pneumophila response regulator CpxR was identified as an activator of the icmR gene. Construction of an L. pneumophila cpxR insertion mutant demonstrated the critical role that CpxR plays in controlling icmR gene expression. In addition, the binding site (GTAAA) of the CpxR regulator was identified in the icmR regulatory region located 60 bp upstream from its transcription start site. The consensus identified is similar to the one found in other bacteria (11, 21, 33). Mutagenesis of the CpxR binding site was found to reduce the level of expression of the icmR::lacZ fusion to a lower level than that of mutations constructed in the −10 promoter element of icmR, suggesting that the contribution of the CpxR binding site to the expression of icmR is highly significant. Moreover, the L. pneumophila CpxR regulator was shown to bind the icmR regulatory region in vitro by using a mobility shift assay. Furthermore, the presented results indicate that CpxR is also involved in the regulation of icmV and icmW. Considering that the decrease in their level of expression was mild in comparison to the decrease observed with the icmR::lacZ fusion and the similar levels of expression that they had in L. pneumophila and E. coli (16), we speculate that the regulation of icmV and icmW by CpxR might be indirect.
The finding that CpxR regulates the expression of the icmR gene led us to examine the role of its cognate sensor kinase CpxA in the regulation of icmR. The expression of icmR::lacZ fusion in an L. pneumophila cpxA mutant strain was found to be moderately low in comparison to that of the wild-type strain. The differences in the effects of the cpxR and cpxA mutant strains on the level. of expression of the icmR::lacZ fusion might be explained by the possible substitution of CpxA by the low-molecular-weight compound acetyl-phosphate (Ac-P). This compound might phosphorylate the CpxR response regulator in the absence of CpxA as was shown previously for the E. coli CpxR and other response regulators (23, 25, 27) as well as in our gel-shift results. In E. coli, the pta and ackA genes code for a phosphate acyltransferase and an acetate kinase, respectively, and they were shown to be responsible for Ac-P synthesis (27). A BLAST search against the L. pneumophila incomplete genome database identified one ORF homologous to the E. coli pta and ackA genes (23% identity and 41% similarity to pta; 33% identity and 51% similarity to ackA). This homolog could be responsible for the synthesis of Ac-P in L. pneumophila, which might facilitate the phosphorylation of the CpxR regulator in the absence of CpxA.
Interestingly, despite the sequence similarity between the E. coli and the L. pneumophila CpxRs and their similar recognition sequences, the E. coli CpxR does not activate the expression of the icmR::lacZ fusion when expressed from a plasmid in E. coli. This result might indicate that the E. coli CpxR regulator does not bind to the icmR regulatory region or that it binds but cannot activate the icmR promoter. Currently we cannot distinguish these two possibilities. These data may indicate that these two proteins are not interchangeable. However, this is probably not the only difference between the cpx systems in these two bacteria. In E. coli, CpxP serves as an inhibitor molecule for CpxA (8); however, the cpxP gene is thus far absent in the L. pneumophila genome database (85% completed).
Although CpxR was shown to directly control the expression of the icmR gene and to be involved in the expression of two other icm genes (icmV and icmW), the L. pneumophila CpxR regulator was found to be dispensable for intracellular growth in A. castellanii and HL-60-derived human macrophages. This information indicates that even a 10-fold reduction in the expression of the icmR gene was still sufficient for establishing a successful infection in the eukaryotic hosts examined. It is possible that in other hosts used by L. pneumophila the reduction in the amount of IcmR in the bacterial cell would result in an intracellular growth phenotype. Previously, it was shown that an icmR insertion mutant retains some ability to kill HL-60-derived human macrophages (40) and another mutation in this gene was shown to permit limited intracellular replication in murine bone marrow macrophages (6). These results might also explain the fact that the reduction in the level of expression of the icmR gene in the cpxR mutant strain does not result in an intracellular growth defect, as even a complete null mutation in the icmR gene still allows some ability to replicate within host cells. In addition, the IcmR gene product was suggested to act as the chaperone of IcmQ (13). This information is consistent with the known role of CpxR, in other bacteria, where it was shown to regulate genes involved in protein-folding functions (9, 21, 35). Since IcmR is probably not a structural component of the Icm/Dot complex, it might be that a reduction in its level can result in a no intracellular growth phenotype.
Thus far, several regulatory factors such as RpoS, RelA, and LetA have been shown to be involved in the regulation of the icm and dot genes in L. pneumophila, but not in a direct manner. The data presented here, showing that the regulation of the L. pneumophila icmR virulence gene is mediated by the CpxR response regulator, is the first evidence for a direct regulator of an icm or dot gene. The CpxR regulator together with other regulatory factors probably concerts with the expression of icm and dot genes to result in successful infection.
Acknowledgments
This research was supported by the Charles H. Revson Foundation of the Israel Science Foundation (grant 45/00). G. Segal was supported by the Alon fellowship awarded by the Israeli Ministry of Education.
REFERENCES
- 1.Albright, L. M., E. Huala, and F. M. Ausubel. 1989. Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu. Rev. Genet. 23:311-336. [DOI] [PubMed] [Google Scholar]
- 2.Berger, K. H., J. J. Merriam, and R. R. Isberg. 1994. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol. Microbiol. 14:809-822. [DOI] [PubMed] [Google Scholar]
- 3.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 4.Brand, B. C., A. B. Sadosky, and H. A. Shuman. 1994. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol. Microbiol. 14:797-808. [DOI] [PubMed] [Google Scholar]
- 5.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 6.Coers, J., J. C. Kagan, M. Matthews, H. Nagai, D. M. Zuckman, and C. R. Roy. 2000. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38:719-736. [DOI] [PubMed] [Google Scholar]
- 7.Conover, G. M., I. Derre, J. P. Vogel, and R. R. Isberg. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305-321. [DOI] [PubMed] [Google Scholar]
- 8.Danese, P. N., and T. J. Silhavy. 1998. CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danese, P. N., and T. J. Silhavy. 1997. The sigma(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 11:1183-1193. [DOI] [PubMed] [Google Scholar]
- 10.De Wulf, P., B. J. Akerley, and E. C. Lin. 2000. Presence of the Cpx system in bacteria. Microbiology 146:247-248. [DOI] [PubMed] [Google Scholar]
- 11.De Wulf, P., A. M. McGuire, X. Liu, and E. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277:26652-26661. [DOI] [PubMed] [Google Scholar]
- 12.Dong, J., S. Iuchi, H. S. Kwan, Z. Lu, and E. C. Lin. 1993. The deduced amino-acid sequence of the cloned cpxR gene suggests the protein is the cognate regulator for the membrane sensor, CpxA, in a two-component signal transduction system of Escherichia coli. Gene 136:227-230. [DOI] [PubMed] [Google Scholar]
- 13.Dumenil, G., and R. R. Isberg. 2001. The Legionella pneumophila IcmR protein exhibits chaperone activity for IcmQ by preventing its participation in high-molecular-weight complexes. Mol. Microbiol. 40:1113-1127. [DOI] [PubMed] [Google Scholar]
- 14.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 15.Gal-Mor, O., and G. Segal. 2003. The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb. Pathog. 34:187-194. [DOI] [PubMed] [Google Scholar]
- 16.Gal-Mor, O., T. Zusman, and G. Segal. 2002. Analysis of DNA regulatory elements required for expression of the Legionella pneumophila icm and dot virulence genes. J. Bacteriol. 184:3823-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross, R., B. Arico, and R. Rappuoli. 1989. Families of bacterial signal-transducing proteins. Mol. Microbiol. 3:1661-1667. [DOI] [PubMed] [Google Scholar]
- 18.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 19.Horwitz, M. A. 1987. Characterization of avirulent mutant Legionella pneumophila that survive but do not multiply within human monocytes. J. Exp. Med. 166:1310-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Investig. 66:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung, D. L., T. L. Raivio, C. H. Jones, T. J. Silhavy, and S. J. Hultgren. 2001. Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. EMBO J. 20:1508-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, C. H., P. N. Danese, J. S. Pinkner, T. J. Silhavy, and S. J. Hultgren. 1997. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 16:6394-6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenney, L. J., M. D. Bauer, and T. J. Silhavy. 1995. Phosphorylation-dependent conformational changes in OmpR, an osmoregulatory DNA-binding protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 92:8866-8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leclerc, G. J., C. Tartera, and E. S. Metcalf. 1998. Environmental regulation of Salmonella typhi invasion-defective mutants. Infect. Immun. 66:682-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukat, G. S., W. R. McCleary, A. M. Stock, and J. B. Stock. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. USA 89:718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch, D., N. Fieser, K. Gloggler, V. Forsbach-Birk, and R. Marre. 2003. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol. Lett. 219:241-248. [DOI] [PubMed] [Google Scholar]
- 27.McCleary, W. R., J. B. Stock, and A. J. Ninfa. 1993. Is acetyl phosphate a global signal in Escherichia coli? J. Bacteriol. 175:2793-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular biology. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 30.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama, S., and H. Watanabe. 1998. Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J. Bacteriol. 180:3522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakayama, S., and H. Watanabe. 1995. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J. Bacteriol. 177:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pogliano, J., A. S. Lynch, D. Belin, E. C. Lin, and J. Beckwith. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11:1169-1182. [DOI] [PubMed] [Google Scholar]
- 34.Purcell, M., and H. A. Shuman. 1998. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect. Immun. 66:2245-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 36.Raivio, T. L., and T. J. Silhavy. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 179:7724-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Segal, G., and H. A. Shuman. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 65:5057-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segal, G., and H. A. Shuman. 1998. How is the intracellular fate of the Legionella pneumophila phagosome determined? Trends Microbiol. 6:253-255. [DOI] [PubMed] [Google Scholar]
- 42.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 45.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 46.Vogel, J. P., and R. R. Isberg. 1999. Cell biology of Legionella pneumophila. Curr. Opin. Microbiol. 2:30-34. [DOI] [PubMed] [Google Scholar]
- 47.Wiater, L. A., A. B. Sadosky, and H. A. Shuman. 1994. Mutagenesis of Legionella pneumophila using Tn903dlllacZ: identification of a growth-phase-regulated pigmentation gene. Mol. Microbiol. 11:641-653. [DOI] [PubMed] [Google Scholar]
- 48.Wosten, M. M. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127-150. [DOI] [PubMed] [Google Scholar]
- 49.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 50.Zusman, T., O. Gal-Mor, and G. Segal. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 184:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]