Abstract
The xeroderma pigmentosum variant (XPV) is a genetic disease involving high levels of solar-induced cancer that has normal excision repair but shows defective DNA replication after UV irradiation because of mutations in the damage-specific polymerase hRAD30. We previously found that the induction of sister chromatid exchanges by UV irradiation was greatly enhanced in transformed XPV cells, indicating the activation of a recombination pathway. We now have identified that XPV cells make use of a homologous recombination pathway involving the hMre11/hRad50/Nbs1 protein complex, but not the Rad51 recombination pathway. The hMre11 complexes form at arrested replication forks, in association with proliferating cell nuclear antigen. In x-ray-damaged cells, in contrast, there is no association between hMre11 and proliferating cell nuclear antigen. This recombination pathway assumes greater importance in transformed XPV cells that lack a functional p53 pathway and can be detected at lower frequencies in excision-defective XPA fibroblasts and normal cells. DNA replication arrest after UV damage, and the associated S phase checkpoint, is therefore a complex process that can recruit a recombination pathway that has a primary role in repair of double-strand breaks from x-rays. The symptoms of elevated solar carcinogenesis in XPV patients therefore may be associated with increased genomic rearrangements that result from double-strand breakage and rejoining in cells of the skin in which p53 is inactivated by UV-induced mutations.
Keywords: UV light, postreplication repair, recombination, sister chromatid exchange, skin cancer
Xeroderma pigmentosum (XP) is a rare autosomal recessive disorder characterized by sun sensitivity, cutaneous and ocular deterioration, and premature malignant skin neoplasms upon sun exposure (1). The mechanism of cancer development in XP remains a matter of conjecture, despite the appealing association of defective excision repair of UV damage to DNA in the cells of the skin with increased risk of actinic cancer (2, 3). One of the difficulties with this notion is that about a quarter of XP patients with characteristic symptoms of sunlight-induced cancer do not have deficiencies in DNA excision repair (1, 4). These patients have been classified with a different abnormality in the response to UV irradiation involving semiconservative DNA replication (postreplication repair) and are known as XP variants (XPVs) (5, 6). This defect is associated with increased rates of UV-induced mutagenesis (7–11), with cells being less likely to incorporate adenine opposite pyrimidine dimers (10).
The XPV gene was recently cloned by homology with a corresponding gene from Saccharomyces cerevisiae (12) and by development of a cell-free DNA replication system (13). They showed that the gene hRAD30 corresponded to an error-free DNA polymerase, pol η, homologous to a member of the S. cerevisiae RAD6 epistasis group, which encompasses many genes involved in postreplication repair. The fact that this polymerase seems to be part of an error-free pathway for processing UV damage may be attributable to a fortuitous combination of factors. The polymerase shows a relaxed substrate specificity and replicates damaged and undamaged DNA equally, with a high error rate (14). However, operation of the “A” rule on photoproducts that are predominantly thymines will result in accurate replication of damaged DNA. The definition of error-free and error-prone pathways therefore may be restricted to UV damage, and this classification may not hold for all forms of damage.
Postreplication repair in many prokaryotes and lower eukaryotes employs both replication bypass and recombinational modes of recovery from DNA damage (15). A recombination pathway for postreplication repair has been difficult to detect in human and other mammalian cells, in part, because of the complexity of chromosomal DNA replication from multiple origins (16–19). We recently showed that one form of recombination, sister chromatid exchange (SCE), that occurs during the S phase was greatly enhanced in the XPV cells after transformation (20).
In this paper, we report a further investigation of the mechanism of UV-induced recombination in XPV cells as well as a role for the recombination pathway involving the hMre11/hRad50/Nbs complex (the hMre11 complex) (21, 22).
Materials and Methods
Cell Cultures.
Primary and simian virus 40 (SV40)-transformed human fibroblasts included repair-proficient (normal), nucleotide excision repair-deficient (XPA), and replication bypass-deficient (XPV) genetic backgrounds (20). Cells were grown in Eagle's MEM, supplemented with 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FBS (Life Technologies, Gaithersburg, MD). The normal primary fibroblast FS was obtained from a local foreskin biopsy. The XPA fibroblasts (XP19BR and GB) were gifts from C. Arlett (University of Sussex, England) and D. Busch (Armed Forces Institute of Pathology, Washington, DC), respectively, and were homozygous for a splice site mutation or a 2-nt deletion in the XPA gene that impairs nucleotide excision repair. XP12R0 is an SV40-transformed group A cell line that is homozygous for a stop codon in the XPA gene. The XPV (GM3617) fibroblasts were obtained from The Human Genetic Mutant Cell Repository, Camden, NJ. The normal fibroblast cell line (GM037) was permanently transformed by SV40 and maintained for many generations as GM637. The XPV fibroblast GM3617 (also designated XP30R0) was transformed with SV40 to produce the permanently transformed line XP30R0 (23). The primary human fibroblast GM3617 contains a mutation in the XPV gene, which results in a chain termination of DNA polymerase η.
Immunofluorescence.
One day before irradiation, 1 × 105 cells were seeded in dual-chambered slides (Nalge Nunc, Naperville, IL) and allowed to anchor. Exponentially growing cells were exposed to either UV light (254 nm at a fluence of 1.3 W⋅m−2) or x-rays (Phillips RT250 x-ray machine; 250-kV peak, 15 mA, half-value layer 1.0 mm copper, at a dose rate of 2.5 Gy/min) and were fixed 4–8 h after irradiation in ice-cold methanol for 20 min, then air-dried and stored at −70°C.
Fixed cells were permeabilized in an ice-cold 50:50 acetone:methanol and allowed to come to room temperature before air drying. Cell preparations were blocked for 1 h in 10% FBS in PBS at 37°C, rinsed, and incubated with primary antibody for 1 h at 37°C. Rabbit polyclonal anti-hMre11 antibody (Novus Biologicals, Littleton, CO) was diluted 1:100. Mouse monoclonal anti-human proliferating cell nuclear antigen (PCNA) antibody (Santa Cruz Biotechnology) was diluted to 5 μg/ml. Secondary IgG (heavy plus light) antibodies (Pierce) were selected to provide the appropriate combinations of species specificity (goat anti-rabbit or -mouse) and color discrimination (conjugated to either fluorescein or rhodamine). These antibodies were diluted 1:200 and incubated with cells for 1 h at 37°C. Cells were counterstained with 0.1 μg/ml of 4′,6-diamidino-2-phenylindole in Vectashield (Vector Laboratories). All incubations were interspersed with three 5-min washes in PBS, multiple primary or secondary antibody combinations were incubated simultaneously, and all antibodies were diluted in 1× PBS in 1% BSA.
Foci Quantification and Digital Image Analysis.
Fluorescently labeled cell preparations were examined using a Zeiss Axioplan microscope equipped with a charge-coupled device digital camera (24). Cells were examined by eye at a magnification of ×600 for the presence of Mre11 foci. Those cells showing at least five or more foci per nucleus were scored positive. Excitation of individual fluorochromes was accomplished by using single and dual bandpass filter sets allowing for the visualization of FITC, rhodamine, and 4′,6-diamidino-2-phenylindole. This filter set made it possible to collect sequential, properly registered images (red, green, blue) with three excitation wavelengths (green, blue, UV). The three-color images were captured and processed with a Sun IPX workstation and processed by using scil-image analysis software (Univ. of Amsterdam, Amsterdam).
Apoptosis.
UV-irradiated SV40-transformed cells detached from the substrate between 10 and 20 h after UV exposure. Floating cells had the hallmarks of apoptosis: They were positive for annexin V immunofluorescence and contained cleaved poly(ADP-ribose) polymerase, whereas attached cells were not apoptotic. Annexin V binding, which detects phosphatidylserine residues externalized to the outer leaflet of the plasma membrane, was measured by using the ApoAlert Annexin V Apoptosis Kit (CLONTECH). Inhibition of caspase 3 with zVAD (Sigma) prevented detachment completely. Apoptosis therefore was recorded in these experiments as the percentage of cells detached at 16–20 h after doses of UV light. A balance sheet for the total number of cells in floating and attached populations showed that loss of cells through complete lysis and degradation occurred only at high doses of 13 J⋅m−2 and above. Caffeine (1 mM) and/or the anti-apoptotic peptide zVAD (1 μg/ml) were added immediately after UV irradiation and remained until cells were assayed for apoptosis or Mre11 foci.
Results
We previously reported that in SV40- and HPV16 (E6 or E7 genes)-transformed XPV cells, UV damage induced a much higher frequency of SCEs than it did in normal cells (20). Because SCEs represent recombination events occurring in the S phase (25), this finding could represent the activation of a recombination pathway for postreplication repair that has been previously difficult to demonstrate (26). Our initial experiments concentrated on hRad51/Brca1, because these have been shown to require p53 (27), which is inactivated by both methods of transformation. However, we were unable to demonstrate any differences, by using immunofluorescence, between normal and XPV cells in the response of hRad51/Brca1 to UV damage (R. Scully and J.E.C., unpublished observations). We therefore turned to the hMre11 complex that has been associated with NHEJ of DNA double-strand breaks from ionizing radiation (21). The hMre11 complex undergoes a redistribution after x-irradiation, becoming associated in large nuclear structures at sites of DNA breaks, referred to as foci (21). We compared the formation of these foci after x- and UV irradiation in primary fibroblasts and SV40-transformed normal and XP cells (group A and variant).
X-Ray-Induced hMre11 Foci in Primary and SV40-Transformed Human Fibroblasts.
We first established that normal and XP cells showed hMre11 foci after x-ray damage, as seen in other human cell types (21). Primary human fibroblasts were x-irradiated, and each primary cell line exhibited a similar capacity to form foci after a 6-Gy dose of x-rays (Table 1). X-ray-induced foci formed in XP30R0 cells 4 h after 6 Gy showed the typical scattered nuclear distribution previously reported in other cell types (Fig. 1 a, d, and g). GM037 (normal), XP19BR (XPA), and GM3617 (XPV) cells showed 33, 33, and 52% foci-positive cells when examined 8 h after x-irradiation (Table 1).
Table 1.
hMre11 foci in x-irradiated human cells
| Cell line | Genotype | Dose, Gy | Time, h | Foci-positive cells/total cells | % Foci-positive |
|---|---|---|---|---|---|
| GM037 | Normal | 0 | 8 | 0/500 | 0 |
| GM037 | Normal | 6 | 8 | 176/540 | 33 |
| GM637 | Normal(SV40) | 0 | 8 | 0/500 | 0 |
| GM637 | Normal(SV40) | 6 | 4 | 262/681 | 38 |
| GM637 | Normal(SV40) | 6 | 8 | 309/1020 | 30 |
| XP19BR | XPA | 0 | 8 | 0/500 | 0 |
| XP19BR | XPA | 6 | 8 | 165/500 | 33 |
| XP12R0 | XPA(SV40) | 0 | 8 | 0/500 | 0 |
| XP12R0 | XPA(SV40) | 6 | 4 | 272/500 | 54 |
| XP12R0 | XPA(SV40) | 6 | 8 | 573/1228 | 47 |
| GM3617 | XPV | 0 | 8 | 1/500 | 0.2 |
| GM3617 | XPV | 6 | 8 | 259/500 | 52 |
| XP30R0 | XPV(SV40) | 0 | 8 | 0/500 | 0 |
| XP30R0 | XPV(SV40) | 6 | 4 | 585/1573 | 37 |
| XP30R0 | XPV(SV40) | 6 | 8 | 419/1190 | 35 |
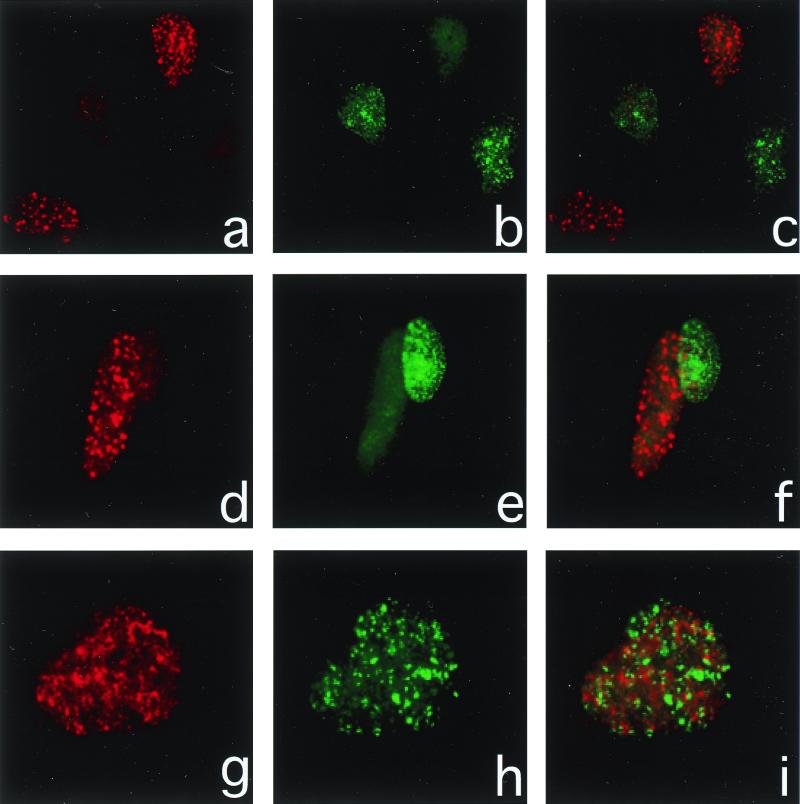
Figure 1.
Immunofluorescence of hMre11 and PCNA after x-irradiation. XP30R0 cells were fixed 4 h after 6 Gy of x-rays and processed for the visualization of hMre11 (red rhodamine signal, a, d, and g) and PCNA (green fluorescein signal, b, e, and h). Merged images (e, f, and i) show the lack colocalization of each protein. The majority of cells show either hMre11 or PCNA foci (a–f), whereas those cells showing both do not show colocalized foci (g–i).
SV40-transformed human fibroblasts were similarly exposed to x-rays, and each cell line showed a similar capacity to form foci after 6 Gy of x-rays (Table 1). GM637 (normal), XP12R0 (XPA), and XP30R0 (XPV) cell lines showed 34, 49, and 36% foci-positive cells when examined 4–8 h after x-irradiation (Table 1). These results show that relocalization of the hMre11 complex within the nuclei after x-irradiation was independent of the XP genotype.
UVC-Induced hMre11 Foci in Primary Human Fibroblasts.
UV exposure has not previously been reported to induce hMre11 foci and we confirmed they were not detectable in primary normal or XPA fibroblasts (XP19BR, GB) (Table 2). In XPV (GM3617) fibroblasts, however, at 4 and 8 h after a fluence of 13 J⋅m−2 of UV light, there was a very low, but non-zero frequency, of hMre11 foci (Table 2). The precise value is uncertain without more extensive investigation, but the level was clearly different from that in normal cells. Occasional foci-positive cells were observed in the XPV cells, whereas none was seen in other cell types.
Table 2.
hMre11 foci in UV-irradiated primary human fibroblasts
| Cell line | Genotype | UV, J⋅m−2 | Time, h | Foci-positive cells/total cells |
|---|---|---|---|---|
| FS | Normal | 0 | 8 | 0/500 |
| FS | Normal | 13 | 4 | 1/500 |
| FS | Normal | 13 | 8 | 0/500 |
| GM037 | Normal | 0 | 8 | 0/500 |
| GM037 | Normal | 13 | 8 | 0/500 |
| GB | XPA | 0 | 8 | 7/1000 |
| GB | XPA | 13 | 4 | 0/500 |
| GB | XPA | 13 | 8 | 2/500 |
| XP19BR | XPA | 0 | 8 | 0/1000 |
| XP19BR | XPA | 13 | 4 | 0/500 |
| XP19BR | XPA | 13 | 8 | 2/1000 |
| GM3617 | XPV | 0 | 8 | 0/500 |
| GM3617 | XPV | 13 | 4 | 8/500 |
| GM3617 | XPV | 13 | 8 | 9/1000 |
UVC-Induced hMre11 Foci in SV40-Transformed Human Fibroblasts.
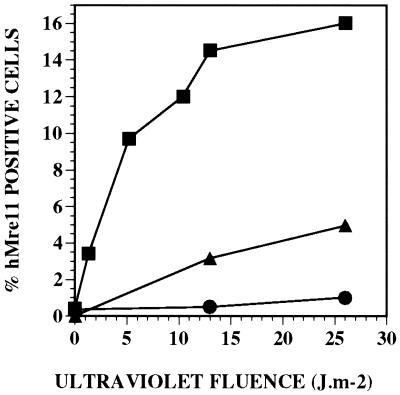
SV40-transformed human fibroblasts were exposed to UV light, fixed, and processed to visualize the formation of nuclear foci, and very different results were obtained. Foci were detected at a low frequency in normal (GM637) cells but were readily observed in both XPA and XPV cell types (Figs. 2 and 3). Foci were especially prominent in XPV cells (Fig. 2 b, e, and h), and in XP12R0 cells, foci were intermediate between those found in normal and XPV cells (Fig. 3). Frequencies were similar at 4 and 8 h after UV irradiation, so these values were combined (Fig. 3), but UV-induced foci could be observed as early as 1 h after irradiation. In XPV cells, the percentage of foci-positive cells increased rapidly up to 10 J⋅m−2, before leveling off at higher fluences (Fig. 3).
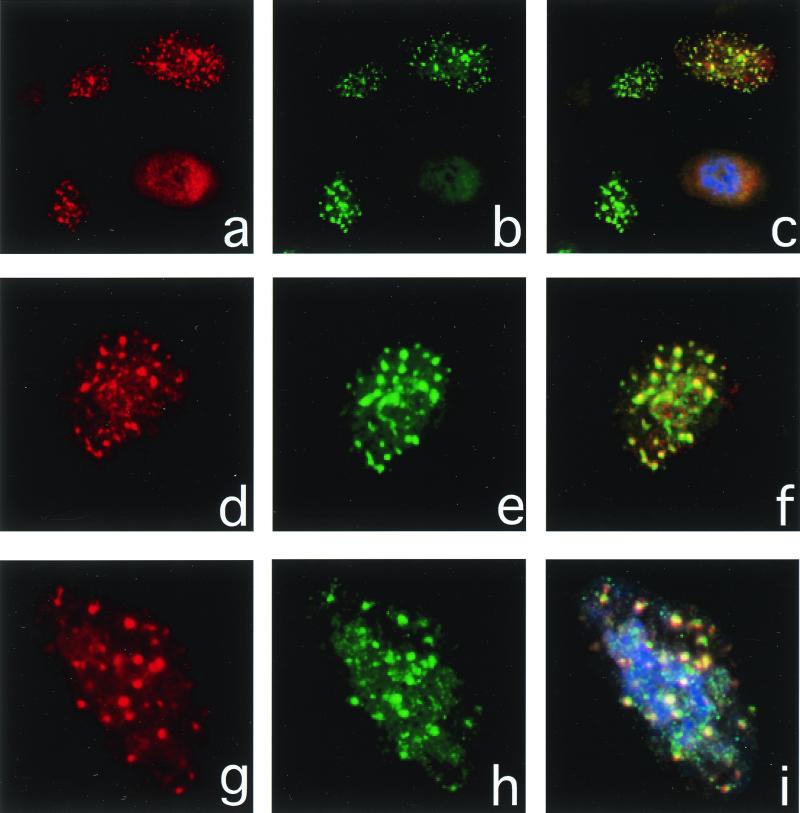
Figure 2.
Immunofluorescence of hMre11 and PCNA after UV irradiation. XP30R0(sv) cells were fixed 4 h after 13 J⋅m−2 of UV light and processed for the visualization of PCNA (red rhodamine signal, a, d, and g) and hMre11 (green fluorescein signal, b, e, and h). Merged images (e, f, and i) show the colocalization of each protein, and the one-to-one correspondence of nuclei showing hMre11 foci with those showing PCNA foci indicate these foci are formed exclusively in S phase cells.
Figure 3.
Induction of hMre11 foci in transformed normal (●, GM637), excision-defective XPA (▴, XP12R0), and XPV (■, XP30R0) cells by various doses of UV light. The frequencies of foci were determined at 4 and 8 h after UV irradiation, and the data were combined because the values were not significantly different at these doses.
Influence of Apoptosis on hMre11 Foci in XP30R0 Cells.
We observed that UV damage to SV40-transformed cells caused apoptosis and detachment from the substrate within a cell cycle (Table 3), and there was a possibility that the foci we observed could have been a consequence of apoptosis. We therefore exposed UV-damaged XPV cells to agents that we observed to stimulate (caffeine) or inhibit (zVAD) apoptosis, as determined by the percentage of detached and floating cells (Table 3). XP30R0 cells were examined for the formation hMre11 foci after UV exposure and showed no significant change in the yield of foci-positive cells (Table 3). Although the presence of caffeine increased the level of apoptosis by an average of 65% (over 1.3 to 13 J⋅m−2) and results in greatly increased levels of cell killing (28), it increased the formation of hMre11 foci by an average of only 11%.
Table 3.
Dependence of UV-induced hMre11 foci on agents that modulate apoptosis in transformed XPV (XP30R0) cells
| Treatment | UV, J⋅m−2 | Apoptotic cells, %* | Time, h | Foci-positive cells/total cells | % Foci-positive cells |
|---|---|---|---|---|---|
| Sham UV | 0 | 2.6 | 7 | 1/1000 | 0.1 |
| Caffeine† | 0 | 2.5 | 5 | 0/1000 | 0 |
| zVAD‡ | 0 | 2.1 | 7 | 0/1000 | 0 |
| UV | 1.3 | 11 | 8 | 17/500 | 3.4 |
| UV + caffeine | 1.3 | 28 | 8 | 11/500 | 2.2 |
| UV | 5.2 | 39 | 5 | 97/1000 | 9.7 |
| UV + caffeine | 5.2 | 56 | 5 | 63/412 | 15 |
| UV | 10.4 | 45 | 5 | 74/600 | 12 |
| UV + caffeine | 10.4 | 54 | 5 | 145/900 | 16 |
| UV | 13 | 44 | 4 | 32/232 | 14 |
| UV + caffeine | 13 | 63 | 4 | 29/213 | 14 |
| UV | 13 | ND | 7 | 93/600 | 16 |
| UV + zVAD | 13 | 2.5 | 7 | 171/1000 | 17 |
ND, not determined.
Apoptosis measured 16 h after UV irradiation by the percentage of cells detached from the substrate. Floating cells were confirmed as apoptotic by annexin V fluorescence and poly(ADP-ribose) polymerase cleavage.
Caffeine (1 mM) added directly after UV irradiation to stimulate apoptosis.
zVAD (1 μg/ml) added directly after UV irradiation to inhibit caspase 3-mediated apoptosis.
The anti-apoptotic peptide zVAD blocked the action of caspase 3 and prevented cell detachment (Table 3). UV-irradiated XP30R0 cells that were treated with zVAD showed a large reduction in the frequency of floating apoptotic cells but there was no change in the number of foci-positive cells (Table 3).
hMre11 Foci Coincide with PCNA at Arrested Replication Forks After UV Irradiation.
XPV cells are defective in a polymerase-involved bypass replication of damaged DNA templates, and the defect is expressed as an enhancement in DNA replication arrest or the S phase checkpoint (29–32). Experiments therefore were undertaken to determine whether the hMre11 foci we observed after UV irradiation were formed in replicating cells. S phase cells were visualized through immunofluorescence of PCNA, and XP30R0 cells exposed to either x-rays or UV light were subjected to fixation conditions that solubilized unbound PCNA, leaving only the fraction bound in replication complexes (33). Cells were fixed at least 4 h after irradiation, so that excision repair was essentially over and PCNA would no longer be associated with excision repair sites (33).
XP30R0 cells were exposed to UV light and subjected to dual immunofluorescent staining to determine the number of cells staining positively for PCNA and hMre11. There was a one-to-one correspondence between cells staining positive for both hMre11 and PCNA foci (Fig. 2). The rhodamine (red) signal (Fig. 2 a, d, and g) corresponds to PCNA, the fluorescein (green) signal (Fig. 2 b, e, and h) corresponds to hMre11, and the merged signals (Fig. 2 c, f, and i) show the colocalization of each protein. In a scored population of 1,710 XP30R0 cells, 29.7% were PCNA-positive and 14.7% were hMre11-positive. But all of the hMre11 foci were in PCNA-positive cells, representing 49.4% of the PCNA-positive population. These results demonstrate that UV-induced hMre11 foci occur exclusively in replicating XP30R0 cells and colocalize with PCNA. However, not all S phase cells contain hMre11 foci. The colocalization observations suggest that hMre11 foci are formed at the sites of DNA replication arrest and are associated with PCNA. In contrast, in our preliminary experiments, hRAD51 and Brca1 did not colocalize with PCNA in XP30R0 cells after UV irradiation (R. Scully and J.E.C., unpublished observations).
hMre11 Foci Do Not Coincide with PCNA After X-Irradiation.
XP30R0 cells were exposed to 6-Gy x-rays, fixed 4 h later, and subjected to dual immunofluorescent staining. The majority of cells (>95%) showing x-ray-induced hMre11 foci did not show PCNA foci, whereas the majority of cells (>95%) showing PCNA foci did not show hMre11 foci (Fig. 1). For these experiments, the rhodamine (red) signal (Fig. 1 a, d, and g) corresponds to hMre11, the fluorescein (green) signal (Fig. 1 b, e, and h) corresponds to PCNA, and the merged signals (Fig. 1 c, f, and i) show the lack of colocalization of each protein. In sharp contrast to the images after UV irradiation (Fig. 1), most of the x-ray-damaged cells showed either hMre11 foci or PCNA foci (Fig. 1 a–f). Less than 5% of the PCNA-positive cells also showed hMre11 foci. When the same cell did contain both foci, the two protein complexes were not colocalized (Fig. 1 g–i). The lack of colocalization and paucity of cells showing both kinds of foci after x-ray damage is in marked contrast to the colocalization and the high frequency of cells exhibiting both foci after UV irradiation.
Discussion
The results we have described reveal an unexpected connection between the mechanisms involved in arrest of DNA replication by UV damage and those involved in double-strand break repair and recombination. The absence of polymerase η in transformed XPV cells, and to a lesser extent in the corresponding primary fibroblasts, resulted in accumulation of protein complexes involving the three components hMre11, hRad50, and Nbs1. These complexes were coincident with PCNA and therefore had accumulated at the replication forks as a result of the enhanced replication arrest seen in these cells. We need to place the results in the context of what is now known about the XPV complementation group, which includes: (i) the mechanism of DNA replication arrest, (ii) the roles of the damage-specific polymerases, (iii) the presence of multiple factors in the DNA replication fork complexes, (iv) the functions of the hMre11 complex, (v) the S phase checkpoint, and (vi) the role of p53.
(i) The Mechanism of DNA Replication Arrest by UV Damage.
The mechanism by which human cells replicate UV-damaged DNA initially was studied on the basis of the sizes of newly replicated DNA (34, 35). Early results indicated that normal human cells had a very efficient bypass mechanism that could replicate damaged templates. During bypass, gaps could be detected in newly replicated DNA (16) that were subsequently filled in by patches up to 1 kb (36). XPV cells showed a larger reduction in the sizes of newly replicated DNA, indicating that the replication apparatus stopped immediately on encountering most pyrimidine dimers (29, 30). XPV cells subsequently were shown to be unable to replicate DNA past photoproducts on the leading strand of the replication fork, but the lagging strand was much less vulnerable (32). Leading strand blocks resulted in the lagging strand extending further, leaving long, single-stranded regions of parental DNA. These regions of single-stranded DNA are likely to be coated with phosphorylated replication protein A (37) that could then be involved in subsequent signal transduction processes. Because replication protein A interacts directly with both termini of p53 (38), there could be a direct route for signal transduction from an arrested replication fork to a p53-dependent pathway.
In early experiments, cells showed less replication arrest as time passed after irradiation, despite the absence of dimer excision during this time period (39). This increased replication capacity could partly be explained by rapid excision of 6–4 photoproducts, but also required activation of increased capacity to replicate pyrimidine dimers, even in XPV cells. When DNA fragments that were synthesized soon after irradiation were allowed to extend during subsequent incubation, the DNA increased to parental sizes at similar rates as those in normal and XPV cells (35). There was, therefore, early evidence that although XPV cells were defective in a major replication factor, they also retained significant capacities for reconstruction of nascent DNA.
(ii) The Roles of the Damage-Specific Polymerases.
The XPV gene hRAD30 encodes a new DNA polymerase, pol η, that has the capacity to replicate pyrimidine dimers in parental DNA (12, 13, 40). With the identification of pol η came the recognition that there are multiple damage-specific polymerases in mammalian cells, many of which are actively involved in error generation. These include the polymerases encoded by the genes hREV1 (41) and hREV3/7 (42) that are required for mutagenesis after UV damage. Loss of function of pol η in XPV cells reduced UV survival slightly (20, 43) but increased mutagenesis (44) because of loss of its accurate, damage-specific replication ability and the greater dependence of cells on the remaining error-prone polymerases (40).
The arrest of replication forks in XPV cells occurs immediately upon irradiation, suggesting that pol η is a constitutive part of the replication apparatus (39). The ability of extracts from normal cells, but not from XPV cells, to replicate damaged DNA also suggests that pol η is constitutively present in the replication complex. However, its activity must be tightly regulated, otherwise its lack of precision might cause greater genomic instability, even in undamaged cells (14). The gradual acquisition of increased replication capacity and the reassembly of short DNA fragments to high molecular weight (35, 39) may then depend on activation of other polymerases or recombinases. They might be damage-inducible and synthesized de novo through increased transcription or alterations in the mRNA transcripts. The hRev3 gene, for example, has several ATG start sites from which alternative transcripts may be generated, only one being authentic (45). The polymerases might, alternatively, be activated at the protein level by phosphorylation through ATM, ATR, Chk1, DNA-protein kinase, or other kinases (46).
(iii) The Presence of Multiple Fidelity Factors at the DNA Replication Fork.
Waga and Stillman (47) showed that in vitro replication of eukaryotic viral DNA required only a minimal set of proteins consisting of a helicase (T antigen), replication protein A, PCNA, replication factor C, and DNA polymerases α and δ. These factors, however, contained no additional provision for monitoring fidelity of replication other than the editing functions of the DNA polymerases. PCNA, however, is regulated by p21 activation in irradiated cells, providing one pathway for an S phase checkpoint that is not targeted at the sites of damage, but through a phosphorylation and activation cascade (46, 48).
DNA replication in the nucleus occurs at large macromolecular complexes, “factories,” that appear to involve multiple replication sites and can be identified by BrdUrd and PCNA immunofluorescence (49). Our results, and others, indicate that these factories may involve many fidelity factors. Other factors reported include 5-MeC transferase (50), poly(ADP-ribose) polymerase (51), uracil DNA glycosylase (52), and Rad51 and Brca1 (53). Our results indicate that the replication apparatus also requires pol η and can recruit the hMre11 recombination complex under certain circumstances as an additional component to the checkpoint pathways. A major function of replication factories therefore may involve localization of a large number of factors that are required for monitoring the accuracy of DNA replication subsequent to replication fork progression. A similar concept recently has been advocated for “rebooting” DNA replication in damaged Escherichia coli cells (54).
(iv) Functions of the Mre11/Rad50/Nbs Complex in Repair.
The hMre11 complex is involved in signaling the presence of x-ray-induced DNA double-strand breaks and some other kinds of DNA damage, but not pyrimidine dimers (refs. 21 and 22; and C.L.L., unpublished observations). Mutations in components of the complex are associated with the human diseases ataxia telangiectasia and Nijmegen breakage syndrome (55). The functions of the complex have been defined in human cells and in S. cerevisiae, and it is unclear whether the complexes have identical functions in both systems. Clearly, the greater dependence of human cells for nonhomologous endjoining (NHEJ) suggests that the functions of the hMre11 complex may be very cell-type dependent (22).
The hMre11 complex is involved in both DNA repair and telomere maintenance (56). It localizes at telomeres in yeast (56, 57) and seems to mediate an alternative, recombinational mechanism of telomere maintenance involving Rad50 and Rad51 (57). Although the hMre11 complex seems involved in NHEJ (22), it may have a dual role. Ku70 was required to direct hMre11 into foci involved in NHEJ, but in its absence, hMre11 was involved in homologous break repair (58). NHEJ involving hMre11 is error-prone in Saccharomyces pombe, possibly because of mispairing in the short regions of overlap (56).
The hMre11 complex has several properties that may be important for resolving blocked DNA replication forks. The complex has ATP-dependent DNA strand unwinding activity, endonuclease activity at DNA hairpin structures, and a 3′–5′ exonuclease capacity (59, 60). The hMre11 complex that we have identified at arrested replication forks, in combination with the remaining damage-specific polymerases, may therefore be responsible for processing the DNA structures into high-molecular weight DNA, recombination products, and SCEs. hMre11 complexes also could be involved in a signal transduction pathway resembling that seen in cells containing DNA breaks, in which there are both upstream (e.g., ATM, ATR, Chk1, and DNA-protein kinase) and downstream effectors to hMre11 (46, 61). One question that remains to be resolved is whether the immunofluorescence we have identified at replication forks represents the complete hMre11/hRad50/Nbs heterotrimer, because this will influence the enzymatic and signal transduction properties displayed by the complex. Phosphorylation of the hMre11 complex seems to require the presence of the complete heterotrimer, especially Nbs1 (62), and is carried out by the ATM protein. If this occurs in XPV cells, then it suggests that there is a role for ATM in the UV-induced S phase checkpoint, which was previously attributed to ATR, unless ATR also can phosphorylate the hMre11 complex (63, 64).
Our observations that hMre11 foci are seen in approximately half of S phase, PCNA-positive nuclei raise the question of whether the foci are formed at specific stages in the S phase. These foci could represent a late attempt of cells to resolve arrested replication forks toward the latter part of S phase. The foci would then occur at a stage known to be involved in the repair of x-ray damage by homologous recombination, involving proteins such as Rad51, XRCC2, and XRCC3 (65). The very low proportion of cells with foci seen in normal SV40-transformed cells, and in primary XPV fibroblasts, would indicate that these cells preferentially resolve arrested replication forks by bypass mechanisms using pol ζ or hREV1 (40).
(v) S Phase Checkpoint Controls.
The functions of cell-cycle checkpoints in G1 and G2 and at phase transitions have received major attention, but the functions of checkpoints within the S phase have been less clear. This lack of clarity may be because there are multiple pathways for regulating the S phase at all stages including replicon initiation, chain growth, and macromolecular assembly (46). Initiation of replication at the G1/S boundary and during S at individual replicons may be under different regulation than that required for continuation of DNA chain growth (48, 66).
Our observations suggest that arrested replication forks in XPV cells recruit components of the homologous repair system. The arrested replication forks represent targeted events, occurring at the sites of DNA damage. The arrested replication fork, and the proteins that accumulate there, also may act as the starting points of signal transduction pathways that exert effects elsewhere in the cell in a nontargeted manner. Arrested replication, for example, results in changes in the phosphorylation status of replication protein A (37) and in p21 binding to PCNA (48), which could be a starting point for signal transduction.
Early observations also showed that UV damage in human cells inhibited replicon initiation during S and the G1/S checkpoint, both of which represent nuclear-wide events that are not targeted at the sites of damage (66–68). These S phase and G1/S checkpoints appear to involve the ATM/ATR and other kinases (46). Cells from patients with ataxia telangiectasia or Nijmegen breakage syndrome, who have defects in the ATM kinase or the hMre11 complex, are unable to down-regulate DNA replication after x-ray or UV damage (67, 68). This inability is because of a failure of the ATM kinase to phosphorylate a number of downstream targets, including p53, Mre11, Nbs1, Brca1, etc. (61). But there are major differences in the x-ray and UV signal transduction pathways, because p53 is phosphorylated on different residues after these two kinds of DNA damage, and x-rays induce less targeted replication arrest (63). In addition, caffeine increases S phase arrest after UV damage (69) but reverses the G1/S and G2/M checkpoint delays in x-ray-damaged cells by inhibiting the ATM/ATR kinases (70).
(vi) SCEs, p53, and Recombination.
Our previous observation of high levels of SCEs in UV-irradiated p53-null XPV cells (20) suggested to us that SCEs represented a recombinational pathway during postreplication repair (25, 71). We initially considered that this might involve Rad51 and Brca1, which are inhibited by p53 (27). This pathway appears to be involved in SCE formation and postreplication repair in chick cells, which have a much more prominent homologous recombination system (71). However, we did not find any evidence for a role of these proteins in UV-damaged XPV cells that differed from normal cells (R. Scully and J.E.C., unpublished observations). Instead, we now have discovered that postreplication repair defects and high levels of SCEs in XPV cells are correlated with the formation of hMre11 complexes. These complexes may mediate the formation of SCEs through dual scission and exchange of duplex DNA (25). Our results with XPA cells (Table 2 and Fig. 3) suggest that there may be multiple pathways for SCE formation, because these cells show high SCE frequencies after UV irradiation (72) but only intermediate levels of hMre11 foci (Fig. 3). The hMre11 foci correlate more closely with the functional deficit in postreplication repair in XPA and XPV cells (73).
The previous observation that high levels of SCEs were formed by UV damage in p53-negative XPV cells (20) suggests that some of the functions of DNA replication arrest and hMre11 complex formation are under p53 control. The inactivation of p53 consequently uncovers an alternative pathway for resolving an arrested replication fork involving DNA double-strand breaks and recombination. The suppression of this pathway by the presence of wild-type p53 accounts for why previous attempts to demonstrate recombination during postreplication repair in human cells were inconclusive. Further investigations of the role of polymerase η, p53, ATM/ATR, and hMre11 therefore are warranted to understand the mechanisms of the S phase checkpoint after UV damage.
Acknowledgments
We are grateful for the assistance of patients and their families in the early development of this study. We also thank James C. Corcoran, James P. Carney, John H. J. Petrini, and Ralph Scully for antibodies, discussions, and preliminary investigations. J.E.C. also is very grateful to his mentor, Dr. Robert B. Painter, whose discovery of unscheduled and radioresistant DNA synthesis provided the basis for his subsequent discovery of the repair deficiency in XP, and who allowed him the freedom to explore a then uncharted field. This work was supported by American Cancer Society Grant CN-156 and National Institutes of Environmental Health Sciences Grant 1 RO1 ES 8061 (to J.E.C.), and by a grant from the Academic Senate Committee on Research of the University of California San Francisco, CA (to J.E.C.). We also are grateful to the XP Society, Poughkeepsie, NY, for their continued support and encouragement to J.E.C.
Abbreviations
- XP
xeroderma pigmentosum
- XPV
XP variant
- XPA
XP group A
- SCE
sister chromatid exchange
- NHEJ
nonhomologous endjoining
- SV40
simian virus 40
- PCNA
proliferating cell nuclear antigen
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 27, 1999.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130182897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130182897
References
- 1.Cleaver J E, Kraemer K H. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. III. New York: McGraw–Hill; 1995. pp. 4393–4419. [Google Scholar]
- 2.Kraemer K H, Lee M M, Scotto J. Carcinogenesis. 1984;5:511–514. doi: 10.1093/carcin/5.4.511. [DOI] [PubMed] [Google Scholar]
- 3.Kraemer K H, Lee M M, Andrews A D, Lambert W C. Arch Dermatol. 1994;130:1018–1021. [PubMed] [Google Scholar]
- 4.Hessel A, Siegel R J, Mitchell D L, Cleaver J E. Arch Dermatol. 1992;128:1233–1237. [PubMed] [Google Scholar]
- 5.Cleaver J E. J Invest Dermatol. 1972;58:124–128. doi: 10.1111/1523-1747.ep12538913. [DOI] [PubMed] [Google Scholar]
- 6.Cleaver J E, Greene A E, Coriell L L, Mulivor R A. Cytogenet Cell Genet. 1981;31:188–192. doi: 10.1159/000131646. [DOI] [PubMed] [Google Scholar]
- 7.Maher V M, Oulette L M, Curren R D, McCormick J J. Nature (London) 1976;261:593–595. doi: 10.1038/261593a0. [DOI] [PubMed] [Google Scholar]
- 8.Maher V M, Oulette L M, Curren R D, McCormick J J. Biochem Biophys Res Commun. 1976;71:228–234. doi: 10.1016/0006-291x(76)90272-2. [DOI] [PubMed] [Google Scholar]
- 9.Waters H L, Seetharam S, Seidman M M, Kraemer K H. J Invest Dermatol. 1993;101:744–748. doi: 10.1111/1523-1747.ep12371686. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y C, Maher V M, McCormick J J. Proc Natl Acad Sci USA. 1991;88:7810–7814. doi: 10.1073/pnas.88.17.7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y C, Maher V M, Mitchell D L, McCormick J J. Mol Cell Biol. 1993;13:4276–4283. doi: 10.1128/mcb.13.7.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson R E, Kondratick C M, Prakash S, Prakash L. Science. 1999;264:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 13.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokol M, Yuasa M, Araki M, Iwa S, Takio K, Hanoaka F. Nature (London) 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 14.Johnson R E, Washington M T, Prakash S, Prakash L. J Biol Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 15.Friedberg E C, Walker G, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 16.Menighini R. Biochim Biophys Acta. 1976;425:419–427. doi: 10.1016/0005-2787(76)90006-x. [DOI] [PubMed] [Google Scholar]
- 17.Cleaver J E. Biochim Biophys Acta. 1978;516:489–516. doi: 10.1016/0304-419x(78)90020-3. [DOI] [PubMed] [Google Scholar]
- 18.Clarkson J M, Hewitt R R. Biophys J. 1976;16:1155–1164. doi: 10.1016/S0006-3495(76)85764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menighini R, Hanawalt P C. Biochim Biophys Acta. 1976;425:428–437. doi: 10.1016/0005-2787(76)90007-1. [DOI] [PubMed] [Google Scholar]
- 20.Cleaver J E, Afzal V, Feeney L, McDowell M, Sadinski W, Volpe J P G, Busch D, Yu Y, Nagasawa H, Little J B. Cancer Res. 1999;59:1102–1108. [PubMed] [Google Scholar]
- 21.Petrini J H J. Am J Hum Genet. 1999;64:1264–1269. doi: 10.1086/302391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haber J E. Cell. 1998;95:585–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 23.Volpe J P G, Cleaver J E. Mutat Res. 1995;337:111–117. doi: 10.1016/0921-8777(95)00015-c. [DOI] [PubMed] [Google Scholar]
- 24.Szollosi J, Lockett S J, Balazs M, Waldman F M. Cytometry. 1995;20:356–361. doi: 10.1002/cyto.990200412. [DOI] [PubMed] [Google Scholar]
- 25.Painter R B. Mutat Res. 1980;70:337–341. doi: 10.1016/0027-5107(80)90023-8. [DOI] [PubMed] [Google Scholar]
- 26.Fujiwara Y, Tatsumi M. Mutat Res. 1976;37:91–110. doi: 10.1016/0027-5107(76)90058-0. [DOI] [PubMed] [Google Scholar]
- 27.Buchhop S, Gibson M K, Wang X W, Wagner P, Sturzbechter H-W, Harris C W. Nucleic Acids Res. 1997;25:3868–3874. doi: 10.1093/nar/25.19.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cleaver J E. Teratogen Carcinogen Mutagen. 1989;9:147–155. doi: 10.1002/tcm.1770090303. [DOI] [PubMed] [Google Scholar]
- 29.Cleaver J E, Thomas G H, Park S D. Biochim Biophys Acta. 1979;564:122–131. doi: 10.1016/0005-2787(79)90193-x. [DOI] [PubMed] [Google Scholar]
- 30.Lehmann A R. Nucleic Acids Res. 1979;7:1901–1912. doi: 10.1093/nar/7.7.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cordonnier A M, Lehmann A R, Fuchs R P. Mol Cell Biol. 1999;19:2206–2211. doi: 10.1128/mcb.19.3.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cordiero-Stone M, Zaritskaya L S, Price L K, Kaufmann W K. J Biol Chem. 1997;272:13945–13954. doi: 10.1074/jbc.272.21.13945. [DOI] [PubMed] [Google Scholar]
- 33.Balajee A S, May A, Dianova I, Bohr V A. Mutat Res. 1988;409:135–146. doi: 10.1016/s0921-8777(98)00051-2. [DOI] [PubMed] [Google Scholar]
- 34.Lehman A R, Kirk-Bell S, Arlett C F, Paterson M C, Lohman P H M, de Weerd-Kastelein E A, Bootsma D. Proc Natl Acad Sci USA. 1975;72:219–235. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park S D, Cleaver J E. Proc Natl Acad Sci USA. 1979;76:3927–3931. doi: 10.1073/pnas.76.8.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehmann A R. J Mol Biol. 1972;66:319–337. doi: 10.1016/0022-2836(72)90418-4. [DOI] [PubMed] [Google Scholar]
- 37.Zernik-Kobak M, Vasunia K, Connelly M, Anderson C W, Dixon K. J Biol Chem. 1997;272:23896–23904. doi: 10.1074/jbc.272.38.23896. [DOI] [PubMed] [Google Scholar]
- 38.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 39.Park S D, Cleaver J E. Nucleic Acids Res. 1979;6:1151–1159. doi: 10.1093/nar/6.3.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cleaver J E. Science. 1999;285:212–213. doi: 10.1126/science.285.5425.212. [DOI] [PubMed] [Google Scholar]
- 41.Lin W, Xin H, Zhang Y, Wu X, Yuan F, Wang Z. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibbs P E, McGregor W G, Maher V M, Nisson P, Lawrence C W. Proc Natl Acad Sci USA. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arlett C F, Harcourt S A, Broughton B C. Mutat Res. 1975;33:341–346. doi: 10.1016/0027-5107(75)90209-2. [DOI] [PubMed] [Google Scholar]
- 44.McGregor W G, Wei D, Maher V M, McCormick J J. Mol Cell Biol. 1999;19:147–154. doi: 10.1128/mcb.19.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murakumo Y, Roth T, Ishii H, Rasio D, Numata S, Croce C M, Fishel R. J Biol Chem. 2000;275:4391–4397. doi: 10.1074/jbc.275.6.4391. [DOI] [PubMed] [Google Scholar]
- 46.Carr A M. Science. 2000;287:1765–1766. doi: 10.1126/science.287.5459.1765. [DOI] [PubMed] [Google Scholar]
- 47.Waga S, Stillman B. Nature (London) 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 48.Waga S, Hannon G J, Beach D, Stillman B. Nature (London) 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 49.Zink D, Bornfleth H, Visser A, Cremer C, Cremer T. Exp Cell Res. 1999;247:176–188. doi: 10.1006/excr.1998.4311. [DOI] [PubMed] [Google Scholar]
- 50.Adams R L. BioEssays. 1995;17:139–145. doi: 10.1002/bies.950170209. [DOI] [PubMed] [Google Scholar]
- 51.Simbulan-Rosenthal C M, Rosenthal D S, Hilz H, Hickey R, Malkas L, Applegren N, Wu Y, Bers G, Smulson M E. Biochemistry. 1996;35:11622–11633. doi: 10.1021/bi953010z. [DOI] [PubMed] [Google Scholar]
- 52.Nilsen, H., Rosewell, I., Robins, P., Slupphaug, G., Krokan, H. E., Lindahl, T. & Barnes, D. E. (2000) Mol. Cell, in press. [DOI] [PubMed]
- 53.Scully R, Chen J, Ochs R L, Keegan K, Hoekstra M, Fuenten J, Livingston D M. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 54.Cox M M, Goodman M F, Kreuzer K N, Sherrat D J, Sandler S J, Marians K J. Nature (London) 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 55.Stewart G S, Maser R S, Stanovic T, Bressan D A, Kaplan M I, Jaspers N G J, Raams A, Byrd P J, Petrini J H J, Taylor A M R. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 56.Wilson S, Warr N, Taylor D L, Watts F Z. Nucleic Acids Res. 1999;27:2655–2661. doi: 10.1093/nar/27.13.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le S, Moore J K, Haber J E, Greider C W. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goedecke W, Eijpe M, Offenberg H H, van Aalderen M, Heyting C. Nat Genet. 1999;23:194–198. doi: 10.1038/13821. [DOI] [PubMed] [Google Scholar]
- 59.Paull T T, Gellert M. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 60.Paull T T, Gellert M. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim S T, Lim D S, Canman C E, Kastan M B. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 62.Dong Z, Zhong Q, Chen P-L. J Biol Chem. 1999;274:19513–19516. doi: 10.1074/jbc.274.28.19513. [DOI] [PubMed] [Google Scholar]
- 63.Hirao A, Kong Y-Y, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge S J, Mak T W. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 64.Chehab N H, Malikzay A, Appel M, Halazonetis T D. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson L H. Mutat Res. 1996;363:77–88. doi: 10.1016/0921-8777(96)00008-0. [DOI] [PubMed] [Google Scholar]
- 66.Kaufmann W K, Wilson S J. Mutat Res. 1994;314:67–76. doi: 10.1016/0921-8777(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 67.Painter R B. Mutat Res. 1981;84:183–190. doi: 10.1016/0027-5107(81)90061-0. [DOI] [PubMed] [Google Scholar]
- 68.Kaufmann W K, Cleaver J E. J Mol Biol. 1981;149:171–187. doi: 10.1016/0022-2836(81)90297-7. [DOI] [PubMed] [Google Scholar]
- 69.Domon M, Rauth A M. Radiat Res. 1969;39:207–221. [PubMed] [Google Scholar]
- 70.Sarkaria J N, Busby E C, Tibbetts R S, Roos P, Taya Y, Karnitz L M, Abraham R T. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 71.Sonoda E, Sasaki M S, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Mol Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Weerd-Kastelein E A, Keijzer E A, Rainaldi G, Bootsma D. Mutat Res. 1977;45:253–261. doi: 10.1016/0027-5107(77)90025-2. [DOI] [PubMed] [Google Scholar]
- 73.Lehmann A R, Kirk-Bell S, Arlett C F, Harcourt S A, de Weerd-Kastelein E A, Keijzer W, Hall-Smith P. Cancer Res. 1977;37:904–910. [PubMed] [Google Scholar]