Abstract
An Escherichia coli strain, ECOR28, was found to have insertions of an identical sequence (1,279 bp in length) at 10 loci in its genome. This insertion sequence (named IS621) has one large open reading frame encoding a putative protein that is 326 amino acids in length. A computer-aided homology search using the DNA sequence as the query revealed that IS621 was homologous to the piv genes, encoding pilin gene invertase (PIV). A homology search using the amino acid sequence of the putative protein encoded by IS621 as the query revealed that the protein also has partial homology to transposases encoded by the IS110/IS492 family elements, which were known to have partial homology to PIV. This indicates that IS621 belongs to the IS110/IS492 family but is most closely related to the piv genes. In fact, a phylogenetic tree constructed on the basis of amino acid sequences of PIV proteins and transposases revealed that IS621 belongs to the piv gene group, which is distinct from the IS110/IS492 family elements, which form several groups. PIV proteins and transposases encoded by the IS110/IS492 family elements, including IS621, have four acidic amino acid residues, which are conserved at positions in their N-terminal regions. These residues may constitute a tetrad D-E(or D)-D-D motif as the catalytic center. Interestingly, IS621 was inserted at specific sites within repetitive extragenic palindromic (REP) sequences at 10 loci in the ECOR28 genome. IS621 may not recognize the entire REP sequence in transposition, but it recognizes a 15-bp sequence conserved in the REP sequences around the target site. There are several elements belonging to the IS110/IS492 family that also transpose to specific sites in the repeated sequences, as does IS621. IS621 does not have terminal inverted repeats like most of the IS110/IS492 family elements. The terminal sequences of IS621 have homology with the 26-bp inverted repeat sequences of pilin gene inversion sites that are recognized and used for inversion of pilin genes by PIV. This suggests that IS621 initiates transposition through recognition of their terminal regions and cleavage at the ends by a mechanism similar to that used for PIV to promote inversion at the pilin gene inversion sites.
Insertion sequences (ISs) are small transposable elements, 0.7 to 2.5 kb in length, which are present in bacterial chromosomes and plasmids (for reviews, see references 27 and 32). These elements encode transposases that promote their transposition. More than 600 IS elements have been identified from 171 bacteria and classified into about 20 families based on homology among transposases (see reference 27). IS elements in most of these families have terminal inverted repeat sequences (IRs), 10 to 40 bp in length, which are recognized by transposases and generate duplication of a target site sequence (2 to 13 bp in length) upon transposition. Most of the IS elements belonging to the IS110/IS492 family are, however, atypical, because they have no terminal IRs and do not generate duplication of the target site sequence upon transposition. These elements encode transposases with significant homology to one another (19, 25). The most characteristic feature of these elements is that their transposases have partial homology with the pilin gene invertase (PIV) encoded by the piv gene that was first identified in Moraxella lacunata (25, 28). PIV recognizes the 26-bp IRs of pilin gene inversion sites (invL and invR) present in two pilin genes (tfpQ and tfpI) and promotes site-specific recombination at the two sequences, resulting in inversion of a segment between invL and invR (12, 18, 47). tfpQ and tfpI encode type IV pilin proteins; of these, the protein encoded by tfpQ is pathogenic because it is localized in the outer membrane. A promoter is present to express only one of two genes, thus determining pathogenicity to bacteria (18). PIV does not have the amino acid motifs conserved in the proteins of the λ-integrase family and in the recombinases of the Hin/Res family, some of which are involved in inversion of a DNA segment. It has been reported that PIV may have part of the catalytic motif in reverse transcriptases (19, 24, 26). PIV has, however, recently been shown to have a D-E-D triad motif corresponding to the catalytic D-D-E motif that is conserved in integrases encoded by retroviruses related to avian sarcoma virus (46).
The full genome sequences of Escherichia coli K-12 MG1655 and enterohemorrhagic E. coli (EHEC) O157:H7 have been determined (8, 16, 17, 36). We have been searching for mutations that occur from rearrangements in various E. coli strains, including E. coli C and six ECOR strains from an E. coli collection, focusing on the DNA segment (about 465 kb in length) corresponding to the 0- to 10-min region of the E. coli K-12 map by using PCR with primers that hybridize to the MG1655 sequence at positions spaced in 5-kb intervals. DNA sequencing of the polymorphic fragments generated from the E. coli strains revealed that the polymorphism is due to the presence of mutations, such as insertions, deletions, substitutions, and duplications of a DNA segment. Of these mutations, most insertions were identified by a computer-aided homology search to have homology with known IS elements.
In this study, we report that an E. coli strain, ECOR28, has a repeated sequence with homology to piv genes at three loci in the 0- to 10-min region of the E. coli K-12 map. We show that this sequence is a novel insertion element, named IS621, which does not have terminal inverted repeats and which encodes transposase with partial homology to those encoded by the IS110/IS492 family elements. The N-terminal regions of PIV proteins and transposases encoded by the IS110/IS492 family elements, including IS621, appear to have four acidic amino acid residues constituting a tetrad motif, D-E (or D)-D-D, rather than the triad motif as the catalytic center. IS621, which shows the highest homology to piv, has terminal sequences that have homology to the 26-bp IRs of pilin gene inversion sites, suggesting that IS621 initiates transposition through recognition of their terminal regions and cleavage at the ends by a common mechanism used by PIV to promote inversion at the pilin gene inversion sites. Interestingly, IS621 was found to be present in repetitive extragenic palindromic (REP) sequences located at three loci in the ECOR28 genome. REP sequences are bacterial short repeats, 35 to 40 bp in length, with imperfect palindromic sequences (for a review, see reference 5). In most cases, REP sequences at each locus occur in clusters called bacterial interspersed mosaic elements, which contain 2 to 12 REP sequences, with other short conserved sequences in positions spaced at intervals (5, 13). The number of copies and the arrangement of REP sequences vary among strains (45, 49). We show that IS621 is inserted into the same site in one of two copies of the REP sequences located at each of the three loci identified in the 0- to 10-min region as well as at seven loci identified in other regions of the ECOR28 genome. There are several elements belonging to the IS110/IS492 family which also transpose to specific sites in the repeated sequences, as does IS621. We discuss the possibility that IS621 and other IS110/IS492 family elements recognize a sequence of about 15 bp with the insertion site in the repeated sequences with full or partial homology.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used were E. coli K-12 MG1655, E. coli C, six ECOR strains (ECOR11, ECOR23, ECOR28, ECOR36, ECOR43, and ECOR46 [31]), and EHEC O157:H7 (NIID accession no. 960220; isolated from Sakai, Japan).
Media.
The culture media used were L broth and L-rich broth (51), SOC medium (40), and φ-medium (51). The L agar plates used contained 1.5% (wt/vol) agar (Wako) in L broth.
DNA preparation.
Genomic DNA was extracted from a 5-ml bacterial culture by the cetyltrimethylammonium bromide-NaCl method described previously (4). Plasmid DNA was extracted from cells cultured at 37°C for 16 h in 3 ml of L broth containing 100 μg of ampicillin/ml by using a Quantum prep kit (Bio-Rad).
PCR.
The chemically synthesized oligonucleotide primers used are listed in Table 1. PCR was performed according to the standard protocol in a 25-μl solution containing a 0.4 mM concentration of each deoxyribonucleoside triphosphate, a 0.24 μM concentration of each pair of primers, 1.5 U of LA-Taq DNA polymerase (Takara), and 0.2 μg of genomic DNA as the template. The PCR conditions were as follows: denaturation at 98°C for 40 s, annealing at 55°C for 30 s, and extension at 72°C for 2 min for a total of 30 cycles. PCR was done with a DNA thermal cycler model PJ2000 (Perkin-Elmer). PCR products were electrophoresed in a 1.0% agarose gel (Wako) in TAE buffer (40 mM Tris-acetate, 1.0 mM EDTA [pH 8.0]) at 100 V for 1 h.
TABLE 1.
Oligonucleotide primers used
| Primera | Sequence (5′ to 3′) | Positionb |
|---|---|---|
| P1F | CGGCTGGTGGAAACAACATT | 5395-5414 |
| P1R | TAAGAAAGCACGCGGTCTGA | 5817-5836 |
| P2F | GGTAATGTCAGCGAAGTGCT | 138511-138530 |
| P2R | TGACCTATGAAGTGAATGGTAA | 138932-138953 |
| P3F | TGGAGATGCTCGATCGTGAA | 215618-215637 |
| P3R | TGCGTTTAAGCCAGAAACGC | 216253-216272 |
| P4F | GAACGCGTGATCGACTTTAG | 856716-856735 |
| P4R | AACTGGGAAGGACGCTTTAC | 857090-857071 |
| P5F | AGGGATTGTGGAAGCTGACA | 1814038-1814057 |
| P5R | ACCTACGCATCTTGATAGCC | 1814787-1814806 |
| P6F | GGAAATGGCACAAAACGCTG | 1841540-1841559 |
| P6R | CGGCGGAATTATCATGGATG | 1842155-1842174 |
| P7F | ATTGCGGATGTGAGCGTTAC | 1952279-1952298 |
| P7R | GGCTTGTGTAAAAACGGCAG | 1952744-1952763 |
| P8F | CGCGAACATATCCGCGCAAT | 2840201-2840220 |
| P8R | GACCCAGCCGATTAAAGATG | 2840695-2840676 |
| P9F | CATAAATAGCCGTACTCACTC | 4371904-4371924 |
| P9R | ACAAACGCTGGCAAACCTGA | 4372425-4372444 |
| P10F | ATCTGCAATTACAGGCGCTG | 4467774-4467793 |
| P10R | ACGAAGCCTTCTTCACCGAA | 4468197-4468216 |
| P1stF | CTTTTGCGGGTCTGACACCACGG | 880*-902* |
| P1stR | ACTCAGCGACAGGTTCCATATAGGT | 381*-405* |
| P2ndF | AGCTGGCACAGGTGGCGTATGGA | 1093*-1115* |
| P2ndR | CCATAGTGAAACCAACCTTGCAAATAC | 169*-195* |
Primers P1st with F or R and P2nd with F or R were used for ADL-PCR (see Materials and Methods).
Positions not marked with an asterisk are indicated by coordinates given relative to the MG1655 sequence (8), and those marked with an asterisk are indicated by coordinates given relative to the IS621 sequence.
Adaptor-ligation-based (ADL) PCR (44) was done with LA-Taq DNA polymerase as follows. The total DNA of ECOR28 was digested with BamHI and BglII (New England Biolabs), neither of which cut the IS621 sequence. T4 DNA ligase (New England Biolabs) was used to ligate the digested DNA with the oligonucleotide adaptor. PCR was first done with a ligated sample as the template and with primers that hybridize to the adaptor and the IS621 sequence to obtain fragments with an end region of IS621 and its flanking sequence. PCR was then done with primers that hybridize to the adaptor and an end region of the IS621 sequence (see Table 1 for the primers used). Fragments that included the entire IS621 sequence in strain ECOR28 were obtained by PCR using primers (Table 1) that hybridize to the flanking sequence of each identified member.
Purification and cloning of DNA fragments.
The PCR-amplified fragments were cut out of an agarose gel, recovered by using a centrifuge tube with a filter (Suprec-01; Takara), and ethanol precipitated. The DNA fragments were cloned by dATP tailing, followed by ligation to a TA cloning vector as follows: dATP tailing was performed in 10 μl of solution containing 1× buffer, 2.5 mM MgCl2, 375 μM deoxyribonucleoside triphosphates, 5 U of LA-Taq DNA polymerase, and 5 μl of the purified DNA fragment at 72°C for 15 to 30 min; ligation was performed in 12 μl of solution containing 2 μl of dATP-tailed DNA solution, 50 ng of pGEM-T easy vector (Promega), and 400 U of T4 DNA ligase at 4°C for 16 to 20 h. The sample DNA was transformed into E. coli strain JM109 by using the method described previously (40). The white colonies were selected on L agar plates containing 100 μg of ampicillin/ml, 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and 100 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per ml.
DNA sequencing.
DNA sequencing was performed by the dideoxynucleotide chain termination method with oligonucleotide primers and an ABI BigDye Terminator DNA sequencing kit (Applied Biosystems). The PCR products were purified with a Centri-Sep spin column (Princeton) and analyzed with an ABI 377 DNA sequencer (Applied Biosystems).
Computer analysis.
Nucleotide sequences were analyzed using Genetyx-Mac version 10.1 and HarrPlot version 2.0 software. A homology search was performed by using the search engines FASTA (34), BLAST (1), and SSEARCH (33, 43) with the DDBJ homepage (http://www.ddbj.nig.ac.jp). Amino acid sequences of PIV proteins and transposases were aligned with YooEdit version 1.71, Clustal W version 1.7, and SeAl version 1.d1 software. A phylogenetic tree was constructed with Phylip version 3.572, njplot, and TreeviewPPC software. The secondary structures of proteins were analyzed with the software program PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred) (22, 29).
RESULTS
Identification of IS621.
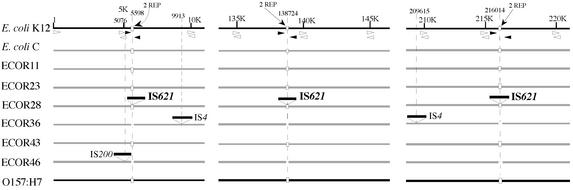
During the search for mutations that occurred from rearrangements in the 0- to 10-min regions (about 465 kb) of the genomes of nine E. coli strains, including E. coli K-12 MG1655 and EHEC O157:H7, we found that the PCR-amplified fragments from three regions showed polymorphisms with similar lengths upon gel electrophoresis (data not shown). Nucleotide sequencing of the polymorphic fragments revealed that an E. coli strain, ECOR28, had insertions of an identical sequence, 1,279 bp in length, in three loci at kb 5.6, 138.7, and 216.0 in the E. coli K-12 map (Fig. 1). Six E. coli strains (MG1655, C, ECOR11, ECOR23, ECOR43, and O157:H7) had the target sequence for the insertion, but the other two strains (ECOR36 and ECOR46) did not (Fig. 1), resulting in generation of fragments slightly shorter than those for the six strains (data not shown).
FIG. 1.
Physical maps of the DNA segments around the insertion sites of IS621 in the genomes of nine E. coli strains. The solid lines show the sequences of E. coli K-12 MG1655 and EHEC O157:H7. For the other E. coli strains, sequences identified by gel electrophoresis of PCR-amplified fragments and by sequencing are shown. Positions of IS element insertions (IS621, IS4, and IS200) and REP sequences (small rectangles) are shown. IS4 and IS200 have been identified in the regions shown. Numbers with or without K are coordinates given to the E. coli K-12 MG1655 sequence in kilobases or base pairs, respectively. The positions of the primers used for PCR are shown by small arrowheads; the solid arrowheads represent primers that were used to analyze polymorphism of the fragments with or without IS621 or REP. Note that IS621 is present in REP sequences in ECOR28 and that ECOR36 and ECOR46 have no REP sequences.
We have also identified many insertion mutations in the 0- to 10-min regions of the nine E. coli strains and characterized them by a computer-aided homology search, using nucleotide sequences as queries. They are almost identical to known IS elements, such as IS1, IS2, IS3, IS4, IS5, IS30, IS150, IS186, IS200, IS609, and IS911, except for the insertion sequence found at each of three loci in the ECOR28 genome, suggesting that this sequence is a novel IS element, here called IS621.
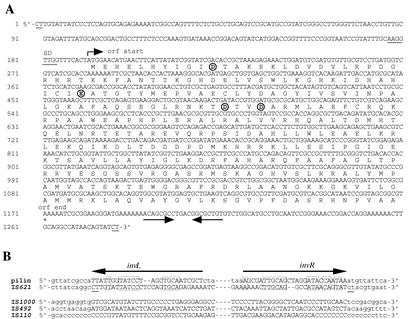
Comparison of the sequences flanking IS621 at each of three loci in ECOR28 with the MG1655 (or O157:H7) sequence having no IS621 showed that a 2-bp sequence CT appeared at the junction regions of IS621 with the target sequence (Fig. 2A). Note that IS621 has no IRs at its termini (Fig. 2A). IS621 has one large open reading frame, 981 bp in length, possibly encoding transposase (Fig. 3A).
FIG. 2.
Nucleotide sequences with IS621 at 10 loci in the ECOR28 chromosome. (A) Nucleotide sequences with IS621 at three loci in the region corresponding to the 0- to 10-min region of the E. coli K-12 map. The nucleotide sequence without IS621 at each position in the E. coli K-12 (MG1655) chromosome is shown for comparison. Two types of REP sequences that have the highest homology to the sequences at the 10 loci are shown in bold. Palindromic sequences in REP are indicated by short horizontal arrows. Note that each element is flanked by 2-bp sequences (in boxes). (B) Nucleotide sequences with IS621 at seven loci located outside of the 0- to 10-min region. IS621 is inserted into the 2-bp sequences shown in boxes. The nucleotide sequence without IS621 at each position in the E. coli K-12 (MG1655) chromosome is shown for comparison. Note that REP sequences at five positions (kb 1814.3, 1952.5, 2840.5, 4372.1, and 4468.0) are in a reverse orientation.
FIG. 3.
(A) The nucleotide sequence of IS621. The amino acid sequence of transposase encoded by one large open reading frame (orf) in IS621 is shown below the nucleotide sequence. A possible Shine-Dalgarno sequence preceding the initiation codon ATG is underlined. Palindromic sequences found in the region downstream of the coding region are shown by a pair of arrows. The CT sequences that appear at the junction regions of IS621 with the target sequence (Fig. 2) are underlined. Note that IS621 has no terminal inverted repeats. Four acidic residues constituting a catalytic motif are shown in circles. (B) Comparison of the pilin gene inversion site sequences and terminal sequences of IS621 and three other IS110/IS492 family elements that are closely related to IS621. Pilin gene inversion site sequences (horizontal arrows) and flanking sequences are indicated by uppercase and lowercase letters, respectively. Terminal sequences of IS621 at kb 5.6 (Fig. 1), IS492 (7), IS110 (9, 26), and IS1000 (3) and their flanking regions are shown by uppercase and lowercase letters, respectively. The CT sequences duplicated at the junction regions with IS621 are underlined. Homologous sequences between pilin gene inversion site sequences and terminal sequences of IS621 are shown in boxes.
Specific insertion of IS621 into REP sequences.
Interestingly, the sequences flanking IS621 at three loci were found to have homology with one another. A computer-aided homology search revealed that there were REP sequences present in two copies at each locus and that IS621 was present in one of the two copies (Fig. 1). Two E. coli strains (ECOR36 and ECOR46) had none of these REP sequences (Fig. 1). Six E. coli strains (MG1655, C, ECOR11, ECOR23, ECOR43, and O157:H7) had REP sequences without the IS621 insertion, in which the REP sequence at kb 216.0 exceptionally had a small deletion near the target site of insertion, but such a deletion was not present at the corresponding region with IS621 in ECOR28 (Fig. 2A). REP sequences have been classified into three types (6). IS621 at kb 5.6 was inserted in the Z2-type REP sequence, whereas IS621 members at kb 138.7 and 216.0 were inserted in the Z1-type REP sequence (Fig. 2A). Note that IS621 was inserted at a specific site in the sequences conserved in the two types of REP sequences (Fig. 2A).
The results described above suggest that IS621 recognizes REP sequences and is inserted into specific sites in their sequences. To confirm this, we carried out ADL PCR (see Materials and Methods) to identify and characterize more IS621 members that are supposed to be present in the ECOR28 chromosome. We found nine IS621 members at different loci, two of which were the same as those identified at kb 138.7 and 216.0 in the E. coli K-12 map. Seven new IS621 members had sequences identical to the three members initially identified or had substitutions of 3 bp at most. All the new members were found to be present at specific sites in the Z1-type REP sequences (Fig. 2B), confirming the above suggestion.
IS621, an IS110/IS492 family element most closely related to piv.
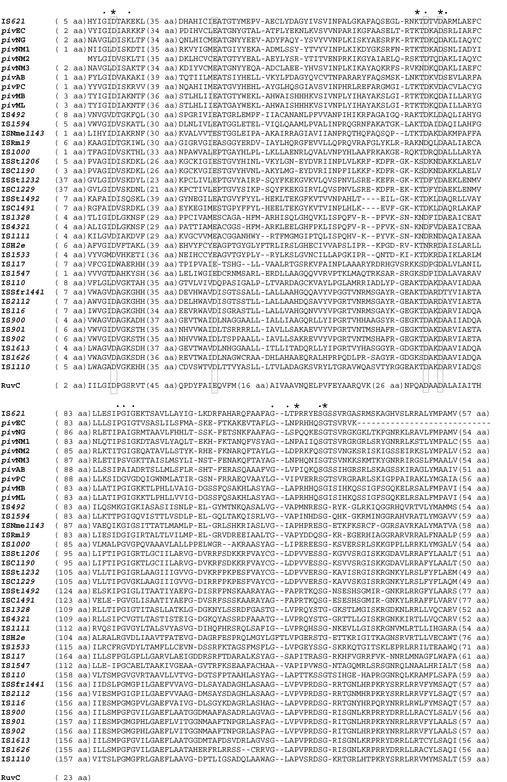
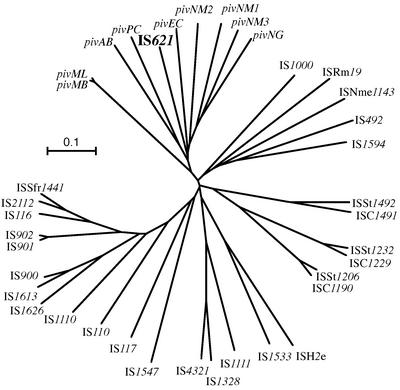
A computer-aided homology search based on the nucleotide sequence of IS621 as the query revealed that IS621 is homologous to the piv genes encoding PIV from various bacteria (Table 2). A homology search based on the amino acid sequence of the putative protein encoded by IS621 as the query, however, revealed that IS621 has partial homology not only to PIV proteins but also to transposases encoded by the IS110/IS492 family elements in various bacteria, including even archaebacteria (Table 2 and Fig. 4). This finding is consistent with the fact that transposases encoded by the IS110/IS492 family elements have partial homology to PIV (25) and indicates that IS621 is a new member of the IS110/IS492 family. A phylogenetic tree based on the amino acid sequences of PIV proteins and transposases revealed that piv genes form a group, whereas IS elements form several groups distinct from the piv gene group (Fig. 5). Note that IS621 belongs to the piv gene group but not to the IS groups (Fig. 5).
TABLE 2.
piv genes and IS110/IS492 family elements
| Elementa | Accession no. | Origin | Length (bp)b |
|---|---|---|---|
| piv genes | |||
| pivML | M34367 | Moraxella lacunata ATCC17956 | 969 |
| pivMB | M32345 | Moraxella bovis EPP63 | 969 |
| pivNG | U65994 | Neisseria gonorrhoeae | 963 |
| pivNM-1 | AE002505 | Neisseria meningitidis MC58 | 957 |
| pivNM-2 | AE002525 | Neisseria meningitidis MC58 | 951 |
| pivNM-3 | AL162754 | Neisseria meningitidis Z2491 | 966 |
| pivEC | AB024946 | Escherichia coli plasmid pB171 | (828) |
| pivAB | AF282240 | Acinetobacter sp. strain SE19 | 975 |
| pivPC | AF011334 | Pectobacterium chrysanthemi | 990 |
| IS elements | |||
| IS621 | This paper | Escherichia coli ECOR28 | 1,279 |
| IS110 | Y00434 | Streptomyces coelicolor | 1,558 |
| IS116 | M31716 | Streptomyces clavuligerus | 1,421 |
| IS117 | X15942 | Streptomyces coelicolor | 2,527 |
| IS492 | M24471 | Pseudomonas atlantica | 1,202 |
| IS900 | X16293 | Mycobacterium paratuberculosis | 1,451 |
| IS901 | X59272 | Mycobacterium avium | 1,472 |
| IS902 | X58030 | Mycobacterium avium | 1,470 |
| IS1000 | M33159 | Thermus thermophilus HB8 | 1,196 |
| IS1110 | Z23003 | Mycobacterium avium | 1,457 |
| IS1111 | M80806 | Coxiella burnetii | 1,450 |
| IS1328 | Z48244 | Yersinia enterocolitica | 1,353 |
| IS1533 | M82880 | Leptospira borgpetersenii | 1,464 |
| IS1547 | Y16254 | Mycobacterium tuberculosis 9504 | 1,346 |
| IS1594 | AF047044 | Anabaena sp. strain PCC7120 | 1,471 |
| IS1613 | AJ011837 | Mycobacterium avium | 1,453 |
| IS1626 | AF071067 | Mycobacterium avium | 1,418 |
| IS2112 | AF060871 | Rhodococcus rhodochrous | 1,415 |
| IS4321 | U60777 | Enterobacter aerogenes plasmid pR751 | 1,347 |
| ISSfr1441 | AF164961 | Streptomyces fradiae T#2717 | (1,441) |
| ISNme1143 | AL162755 | Neisseria meningitidis Z2491 | 1,143 |
| ISH2e | AE000092 | Rhizobium sp. strain NGR234 | (1,201) |
| ISRm19 | AL603647 | Sinorhizobium meliloti | (1,224) |
| ISC1190 | AE006641 | Sulfolobus solfataricus P2 | 1,187 |
| ISC1229 | AE006641 | Sulfolobus solfataricus P2 | 1,229 |
| ISC1491 | AE006641 | Sulfolobus solfataricus P2 | 1,488 |
| ISSt1206 | AP000985 | Sulfolobus tokodaii 7 | 1,206 |
| ISSt1232 | AP000985 | Sulfolobus tokodaii 7 | 1,232 |
| ISSt1492 | AP000985 | Sulfolobus tokodaii 7 | 1,492 |
The piv genes and IS elements underlined are named in this study. The IS elements listed do not have terminal IRs, except for IS1000, IS1111, IS1328 and IS1533, which are reported to have imperfect or perfect IRs of 6, 7, 12, and 31 bp, respectively (3, 20, 38, 52). IS elements do not generate duplication of the target sequence upon insertion, except for IS492, IS1000, and IS1533, which are reported to generate duplication of 5-, 5-, and 2-bp sequences, respectively (3, 35, 52).
The sizes of IS1547 and IS1594 have been previously reported to be 1,351 and 2,265 bp in length, respectively, but are redefined here. The numbers in parentheses indicate only approximate sizes, due to uncertainty of the defined sequence, except for pivEC, which is truncated in its distal region by deletion mediated by an IS element.
FIG. 4.
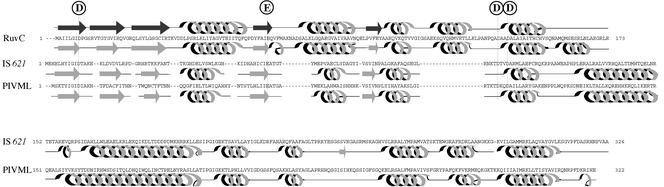
An alignment of PIV proteins and transposases. Only three regions that are well conserved are shown. A RuvC Holliday junction resolvase (accession no. P24239) is aligned to show that four acidic amino acid residues [D, E (or D), D, and D, shown in boxes] in the PIV and transposase proteins are present in positions corresponding to those constituting the catalytic center in the RuvC protein. Amino acid residues present in all the proteins are indicated by asterisks. Other conserved amino acid residues are indicated by dots.
FIG. 5.
A phylogenetic tree of piv genes and IS110/IS492 family elements. The tree was constructed by the neighbor-joining method based on amino acid sequences of PIV proteins and transposases (Fig. 4). The scale bar equals a distance of 0.1.
PIV recognizes the 26-bp IRs of pilin gene inversion sites that are present in two genes (tfpQ and tfpI). PIV promotes recombination at the inverted repeat sequences, invL and invR, resulting in the inversion of a DNA segment between them (12, 18). Terminal sequences of IS621 were found to have significant homology with the 26-bp sequences, whereas those of the other IS110/IS492 family elements were not (Fig. 3B).
The presence of four acidic amino acids conserved in PIV proteins and transposases encoded by IS110/IS492 family elements.
Retroviral integrases and transposases encoded by many IS elements with terminal IRs have three amino acid residues constituting the catalytic D-D-E motif, which is responsible for the strand transfer reaction (see references 11 and 15). Recently, PIV has been reported to have a triad motif, D-E-D, which corresponds to the D-D-E motif conserved in integrases encoded by retroviruses related to avian sarcoma virus (46). The N-terminal regions of transposases encoded by the IS110/IS492 family elements, including IS621, appeared to have the D-E-D (or D-D-D) motif at corresponding positions in transposases, as occurs in PIV proteins (see the first three acidic amino acid residues shown in the boxes in Fig. 4). Interestingly, PIV and transposase proteins had another D residue conserved at the position, three amino acids downstream of the third D residue in the D-E (or D)-D motif (Fig. 4). This leads us to assume that these proteins may have a tetrad motif, D-E (or D)-D-D, like the D-E-D-D motif identified as the catalytic center in the RuvC Holliday junction resolvases (2, 21, 39). In fact, four acidic amino acid residues conserved in the PIV and transposase proteins were present in positions corresponding to those constituting the catalytic center in a RuvC protein (Fig. 4; see Fig. 3A for the positions of four acidic amino acids constituting the tetrad motif in the IS621 transposase).
The tertiary structure of a RuvC Holliday junction resolvase from E. coli has been determined by X-ray crystallography (2). The secondary structure of the RuvC protein, based on the tertiary structure, is shown schematically in Fig. 6. Note that the secondary structure of the RuvC protein was generally similar to that deduced by using the software program PSIPRED (Fig. 6). Therefore, the secondary structures of the IS621 transposase and PIV proteins were analyzed with PSIPRED and compared with those of RuvC. The secondary structures deduced for IS621 transposase and PIV proteins were found to be similar to each other and to RuvC in the regions with four acidic amino acid residues constituting the D-E-D-D motif (Fig. 6). This supports the above assumption that IS621 transposase and PIV proteins are closely related to each other and to RuvC with the D-E-D-D motif.
FIG. 6.
Comparison of secondary structures of RuvC, IS621 transposase, and PIV proteins. The secondary structure of RuvC, based on the tertiary structure (PDB code 1HJR), is shown above the polypeptide sequence. α helices are indicated by ribbons, and β sheets are indicated by arrows. Other features are indicated by straight solid lines. Positions of acidic amino acids constituting the D-E-D-D motif are indicated by thin vertical lines. The secondary structures of RuvC, IS621 transposase, and PIV proteins were predicted by PSIPRED and are shown under the polypeptide sequence of each protein.
Identification of other IS110/IS492 family elements that transpose to a specific site in repeated sequences.
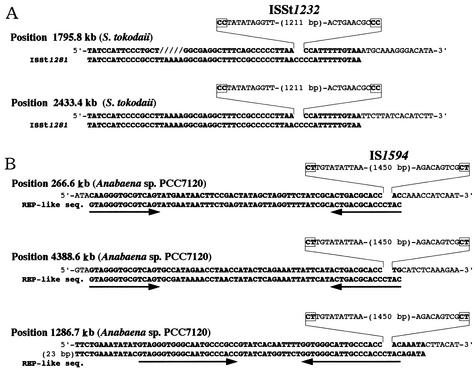
As described above, IS621 is inserted into specific sites within REP sequences. This leads us to assume that some other elements belonging to the IS110/IS492 family may also transpose to specific sites in repeated sequences, like IS621. In fact, two members of ISSt1232, which was identified in this study as an IS110/IS492 family element in the genome of archaebacterium Sulfolobus tokodaii (Table 2), were found to be inserted into a specific site within another IS element (named ISSt1281) repeated in the S. tokodaii genome (Fig. 7A). Comparison of the sequences flanking ISSt1232 to the ISSt1281 sequence having no ISSt1232 showed that a 2-bp sequence, CC, was present at the junction regions (Fig. 7A). ISSt1232 appears to have no IRs at its termini (Fig. 7A). There are several truncated members of ISSt1232, which do not, however, seem to be inserted into ISSt1281 (data not shown).
FIG. 7.
Nucleotide sequences with or without another IS110/IS492 family element, ISSt1232 or IS1594. (A) Nucleotide sequences of ISSt1281 with or without ISSt1232 at two loci. ISSt1281 sequences are shown in bold. (B) Nucleotide sequences with or without IS1594 at three loci. REP-like sequences are shown in bold for comparison. The two types of REP-like sequences that have the highest homology to the sequences at the three loci are shown. Palindromic sequences in the REP-like sequences are indicated by horizontal arrows. Note that REP-like sequences at kb 266.6 and 4388.6 are identical but are in a reverse orientation. Note also that each element is flanked by 2-bp sequences shown in boxes.
IS1594 is an Anabaena IS element, which was identified in this study as belonging to the IS110/IS492 family (Table 2). Analysis of 12 members of IS1594 present in the Anabaena genome revealed that this element is actually 1,473 bp in length, not 2,265 bp as was previously reported (accession no. AF047044). Interestingly, 10 members of IS1594 were found to be present in two types of REP-like sequences in the Anabaena genome (Fig. 7B). IS1594 was inserted into a particular site in the IRs with homology in one REP-like sequence to the other. Note that a 2-bp sequence, CT, was present at the junction regions of IS1594 with REP-like sequences and that IS1594 has no IRs at its termini (Fig. 7B).
DISCUSSION
In this study, we have shown that ISs found at 10 loci in the ECOR28 genome are a novel IS element, IS621, which belongs to the IS110/IS492 family. We have also shown that a 2-bp sequence, CT, was present at the junction regions of IS621 with the target sequence (Fig. 2). It is possible that the 2-bp sequence is used as the target and duplicated upon insertion of 1,277-bp-long IS621 (Fig. 2). This possibility may be supported by the observation that each member of the other two elements belonging to the IS110/IS492 family, ISSt1232 and IS1594, was flanked by a 2-bp target site sequence, CC or CT (Fig. 7). However, based on previous reports that many IS110/IS492 family elements do not generate duplication of the target site sequence upon insertion (14, 23, 30), we cannot exclude the possibility that 1,279-bp-long IS621 is inserted into the target site without duplication. We have been trying to develop a system for transposing IS621 into the target plasmid with or without a REP sequence by using methods that have been used previously to transpose IS elements, such as IS1, IS3, and ISY100 (41, 42, 48), but we have failed to do so. IS492, an IS110/IS492 family element, is known to generate circular IS492 molecules with an extra 5-bp sequence immediately adjacent to the element (35). Circular molecules of IS621 were not, however, detected in ECOR28 cells (our unpublished results), and therefore, the circle formation could not be used as the assay system for IS621 transposition.
The IS110/IS492 family includes elements with divergent nucleotide sequences, and transposases encoded by them show only partial homology to one another and to PIV (Fig. 4). A phylogenetic tree constructed on the basis of amino acid sequences of transposases and PIV proteins shows that the IS110/IS492 family elements are classified into several groups, which are distinct from the group consisting of piv genes and IS621 (Fig. 5). This distinction is the reason why piv genes could be exclusively identified by the homology search by using the nucleotide sequence of IS621 as the query.
PIV has been reported to have a triad motif, D-E-D, which corresponds to the catalytic D-D-E motif that is conserved in retroviral integrases and transposases encoded by IS elements with IRs. We have shown in this study that transposases encoded by the IS110/IS492 family elements, including IS621, appear to have the D-E-D (or D-D-D) motif at positions in their N-terminal regions that correspond to those in PIV proteins (Fig. 4). We have also shown that PIV and transposase proteins have another D residue conserved at a position downstream of the triad motif (Fig. 4) and that the four acidic amino acid residues in these proteins are present in positions corresponding to those which constitute the D-E-D-D motif identified as the catalytic center in the RuvC Holliday junction resolvase (Fig. 4 and 6). These findings strongly suggest that these proteins have a tetrad motif, D-E (or D)-D-D, as does RuvC (2, 21, 39). We have also shown in this study that PIV and transposase proteins have several amino acid residues conserved at corresponding positions in their C-terminal half regions (Fig. 4). This finding suggests that the C-terminal half of these proteins may have a domain(s) that is perhaps responsible for DNA binding to the pilin gene inversion sites or the end regions of IS elements, whereas their N-terminal half has the catalytic domain with the tetrad motif.
We have shown in this study that IS621 and two other elements, ISSt1232 and IS1594, do not have terminal IRs, which is consistent with the fact that most IS110/IS492 family elements are atypical and do not have terminal IRs. This finding and the finding that transposases encoded by the IS110/IS492 family elements have a catalytic motif that is similar to that in transposases encoded by the IR-carrying IS elements suggest that the transposases encoded by the IS110/IS492 family elements with no IRs catalyze the strand transfer reaction, as do those encoded by the IR-carrying IS elements.
We have shown that transposase encoded by IS621 has the highest homology with PIV and that terminal sequences of IS621 show significant homology with the 26-bp sequences of the pilin gene inversion sites. These findings suggest that IS621 initiates transposition through recognition of their terminal regions and cleavage at its ends by a similar mechanism to that used for PIV to promote site-specific recombination at the pilin gene inversion sites.
Interestingly, in this study we have shown that IS621 is present at a specific site in each of the REP sequences at 10 loci in the ECOR28 genome. We have also shown that ISSt1232 is inserted into a specific site within an IS element repeated in the genome of the archaebacterium S. tokodaii, whereas IS1594 is inserted into REP-like sequences repeated in the Anabaena genome. Note that it is usually difficult to determine the sequences of IS elements, particularly those which do not have terminal IRs and transpose to specific sites in repeated sequences; therefore, not only the sequences of several IS copies but also those around the target sites have to be carefully examined to define the elements.
We assume that IS621 spontaneously transposed to the same site in the REP sequences at the 10 loci in the ECOR28 genome by the action of transposase encoded by itself. It is, however, possible that IS621, once inserted in a REP sequence at one locus, transposed to the REP sequence at another locus by a gene conversion mechanism through recombination between the homologous REP sequences. This possibility is, however, unlikely, because all the IS621 members present at 10 loci are almost identical in their nucleotide sequences, whereas the REP sequences nested by IS621 are of two kinds, Z1 and Z2, which are only partially homologous to each other (Fig. 2). This leads us to assume that IS621 does not recognize the entire REP sequence in transposition but recognizes a short homologous sequence, 15 bp in length, with the target site of insertion in the REP sequences (Table 3). Similarly, in the case of IS1594, it may not recognize the entire REP-like sequence, but it recognizes a homologous 15-bp sequence with the target site in the REP-like sequences (Table 3), which are partially homologous to one another (Fig. 7B). ISSt1232 may also not recognize the entire sequence of an IS element (named ISSt1281) repeated in the S. tokodaii genome, but it recognizes a homologous sequence of 15 bp in length (Table 3), which can be identified in ISSt1281 by comparison with the target site sequences flanking each of the truncated members of ISSt1232 present in the S. tokodaii genome.
TABLE 3.
Target sequences possibly recognized by IS110/IS492 family elements
| IS element | Target sequencea |
|---|---|
| IS621 | CTTATCAGGCCTACG |
| IS1594 | ACTGACGCACCCTAC |
| ISSt1232 | GCCCCCTTAACCCCAT |
| IS900 | CATGNNNNTCYCCTT |
| IS901 | CATNNGYNGNTTCCNTTC |
| IS902 | CATNNNCARNTTCCTTTC |
| IS1626 | CATGCGTTTCTCCTATGTC |
| IS2112 | CATGGTGTCCTCGCTT |
| IS492 | GTTTNAANNNCTTGTTAG |
Consensus sequences were derived from published nucleotide sequences (IS900 [14], IS901 [24], IS902 [30], IS1626 [37], IS2112 [23], and IS492 [35]). The sequences that appear at the two junction regions of each element with the target sequence are shown in bold. Note that these sequences are not necessarily meant to be duplicated upon insertion of each element. The sequences complementary to start codons and the ribosome-binding sequences in the target sequences for IS900-related elements are underlined.
It should be noted that in the other IS110/IS492 family elements, IS900 and its related elements, such as IS901 and IS902, were shown to be present in the 13- to 16-bp-long sequences with partial homology to one another (24, 30). The homologous sequences are not, however, located within the repeated sequences, such as the REP- or IS-like sequences, but are supposed to be in the proximal region of a gene, in which the IS elements are inserted into a site between the ribosome-binding sequence and the start codon (Table 3). These data lead us to assume that the IS110/IS492 family elements generally recognize a short DNA sequence and are inserted into a specific site within it. In fact, members of each of several IS110/IS492 family elements, including IS492, that are not closely related to IS900 seem to be present in the sequences of about 16 bp in length with homology to one another (Table 3). Note that the homologous sequences around the insertion sites of IS492 are not located in the proximal region of a gene with the start codon and the ribosome-binding sequence, like those around the insertion sites of IS621, IS1594, and ISSt1232 (Table 3).
It has been reported that an IS element, IS1397, which belongs to the IS3 family, is inserted into REP sequences (10, 50), like IS621. IS1397 appears to recognize a different region in REP from that recognized by IS621, because IS1397 is present in the center of the region flanked by palindromic sequences in REP, whereas IS621 is inserted into a site in the region outside of a palindromic sequence in REP (Fig. 2). In spite of this difference, IS1397 may recognize a short sequence and transpose into a particular target site within it, as does IS621.
Acknowledgments
We thank J. Amemura-Maekawa for providing us the total DNA of EHEC O157:H7.
This research was supported by a Grant-in-Aid of Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ariyoshi, M., D. G. Vassylyev, H. Iwasaki, H. Nakamura, H. Shinagawa, and K. Morikawa. 1994. Atomic structure of the RuvC resolvase: a Holliday junction-specific endonuclease from E. coli. Cell 78:1063-1072. [DOI] [PubMed] [Google Scholar]
- 3.Ashby, M. K., and P. L. Bergquist. 1990. Cloning and sequence of IS1000, a putative insertion sequence from Thermus thermophilus HB8. Plasmid 24:1-11. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology, p. 2.4.1-2.4.5. Green Publishing Associates and Wiley-Interscience, New York, N.Y.
- 5.Bachellier, S., E. Gilson, M. Hofnung, and C. W. Hill. 1996. Repeated sequences, p. 2012-2040. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 6.Bachellier, S., W. Saurin, D. Perrin, M. Hofnung, and E. Gilson. 1994. Structural and functional diversity among bacterial interspersed mosaic elements (BIMEs). Mol. Microbiol. 12:61-70. [DOI] [PubMed] [Google Scholar]
- 7.Bartlett, D. H., and M. Silverman. 1989. Nucleotide sequence of IS492, a novel insertion sequence causing variation in extracellular polysaccharide production in the marine bacterium Pseudomonas atlantica. J. Bacteriol. 171:1763-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 9.Bruton, C. J., and K. F. Chater. 1987. Nucleotide sequence of IS110, an insertion sequence of Streptomyces coelicolor A3(2). Nucleic Acids Res. 15:7053-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clément, J.-M., C. Wilde, S. Bachellier, P. Lambert, and M. Hofnung. 1999. IS1397 is active for transposition into the chromosome of Escherichia coli K-12 and inserts specifically into palindromic units of bacterial interspersed mosaic elements. J. Bacteriol. 181:6929-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig, N. L. 1996. Transposition, p. 2339-2362. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 12.Fulks, K. A., C. F. Marrs, S. P. Stevens, and M. R. Green. 1990. Sequence analysis of the inversion region containing the pilin genes of Moraxella bovis. J. Bacteriol. 172:310-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilson, E., W. Saurin, D. Perrin, S. Bachellier, and M. Hofnung. 1991. The BIME family of bacterial highly repetitive sequences. Res. Microbiol. 142:217-222. [DOI] [PubMed] [Google Scholar]
- 14.Green, E. P., M. L. Tizard, M. T. Moss, J. Thompson, D. J. Winterbourne, J. J. McFadden, and J. Hermon-Taylor. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haren, L., B. Ton-Hoang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245-281. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8(Suppl.):47-52. [DOI] [PubMed] [Google Scholar]
- 18.Heinrich, D. W., and A. C. Glasgow. 1997. Transcriptional regulation of type 4 pilin genes and the site-specific recombinase gene, piv, in Moraxella lacunata and Moraxella bovis. J. Bacteriol. 179:7298-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez-Perez, M., N. G. Fomukong, T. Hellyer, I. N. Brown, and J. W. Dale. 1994. Characterization of IS1110, a highly mobile genetic element from Mycobacterium avium. Mol. Microbiol. 12:717-724. [DOI] [PubMed] [Google Scholar]
- 20.Hoover, T. A., M. H. Vodkin, and J. C. Williams. 1992. A Coxiella burnetti repeated DNA element resembling a bacterial insertion sequence. J. Bacteriol. 174:5540-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichiyanagi, K., H. Iwasaki, T. Hishida, and H. Shinagawa. 1998. Mutational analysis on structure-function relationship of a Holliday junction specific endonuclease RuvC. Genes Cells 3:575-586. [DOI] [PubMed] [Google Scholar]
- 22.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 23.Kulakov, L. A., G. J. Poelarends, D. B. Janssen, and M. J. Larkin. 1999. Characterization of IS2112, a new insertion sequence from Rhodococcus, and its relationship with mobile elements belonging to the IS110 family. Microbiology 145:561-568. [DOI] [PubMed] [Google Scholar]
- 24.Kunze, Z. M., S. Wall, R. Appelberg, M. T. Silva, F. Portaels, and J. J. McFadden. 1991. IS901, a new member of a widespread class of atypical insertion sequences, is associated with pathogenicity in Mycobacterium avium. Mol. Microbiol. 5:2265-2272. [DOI] [PubMed] [Google Scholar]
- 25.Lenich, A. G., and A. C. Glasgow. 1994. Amino acid sequence homology between Piv, an essential protein in site-specific DNA inversion in Moraxella lacunata, and transposases of an unusual family of insertion elements. J. Bacteriol. 176:4160-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leskiw, B. K., M. Mevarech, L. S. Barritt, S. E. Jensen, D. J. Henderson, D. A. Hopwood, C. J. Bruton, and K. F. Chater. 1990. Discovery of an insertion sequence, IS116, from Streptomyces clavuligerus and its relatedness to other transposable elements from actinomycetes. J. Gen. Microbiol. 136:1251-1258. [DOI] [PubMed] [Google Scholar]
- 27.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marrs, C. F., F. W. Rozsa, M. Hackel, S. P. Stevens, and A. C. Glasgow. 1990. Identification, cloning, and sequencing of piv, a new gene involved in inverting the pilin genes of Moraxella lacunata. J. Bacteriol. 172:4370-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 30.Moss, M. T., Z. P. Malik, M. L. Tizard, E. P. Green, J. D. Sanderson, and J. Hermon-Taylor. 1992. IS902, an insertion element of the chronic-enteritis-causing Mycobacterium avium subsp. silvaticum. J. Gen. Microbiol. 138:139-145. [DOI] [PubMed] [Google Scholar]
- 31.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohtsubo, E., and Y. Sekine. 1996. Bacterial insertion sequences. Curr. Top. Microbiol. Immunol. 204:1-26. [DOI] [PubMed] [Google Scholar]
- 33.Pearson, W. R. 1991. Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics 11:635-650. [DOI] [PubMed] [Google Scholar]
- 34.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perkins-Balding, D., G. Duval-Valentin, and A. C. Glasgow. 1999. Excision of IS492 requires flanking target sequences and results in circle formation in Pseudoalteromonas atlantica. J. Bacteriol. 181:4937-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 37.Puyang, X., K. Lee, C. Pawlichuk, and D. Y. Kunimoto. 1999. IS1626, a new IS900-related Mycobacterium avium insertion sequence. Microbiology 145:3163-3168. [DOI] [PubMed] [Google Scholar]
- 38.Rakin, A., and J. Heesemann. 1995. Virulence-associated fyuA/irp2 gene cluster of Yersinia enterocolitica biotype 1B carries a novel insertion sequence IS1328. FEMS Microbiol. Lett. 129:287-292. [DOI] [PubMed] [Google Scholar]
- 39.Saito, A., H. Iwasaki, M. Ariyoshi, K. Morikawa, and H. Shinagawa. 1995. Identification of four acidic amino acids that constitute the catalytic center of the RuvC Holliday junction resolvase. Proc. Natl. Acad. Sci. USA 92:7470-7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Sekine, Y., K. Aihara, and E. Ohtsubo. 1999. Linearization and transposition of IS3 circles. J. Mol. Biol. 294:21-34. [DOI] [PubMed] [Google Scholar]
- 42.Shiga, Y., Y. Sekine, and E. Ohtsubo. 1999. Transposition of IS1 circles. Genes Cells 4:551-559. [DOI] [PubMed] [Google Scholar]
- 43.Smith, T. F., and M. S. Waterman. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195-197. [DOI] [PubMed] [Google Scholar]
- 44.Spertini, D., C. Beliveau, and G. Bellemare. 1999. Screening of transgenic plants by amplification of unknown genomic DNA flanking T-DNA. BioTechniques 27:308-314. [DOI] [PubMed] [Google Scholar]
- 45.Teanpaisan, R., and C. W. Douglas. 1999. Molecular fingerprinting of Porphyromonas gingivalis by PCR of repetitive extragenic palindromic (REP) sequences and comparison with other fingerprinting methods. J. Med. Microbiol. 48:741-749. [DOI] [PubMed] [Google Scholar]
- 46.Tobiason, D. M., J. M. Buchner, W. H. Thiel, K. M. Gernert, and A. C. Karls. 2001. Conserved amino acid motifs from the novel Piv/MooV family of transposases and site-specific recombinases are required for catalysis of DNA inversion by Piv. Mol. Microbiol. 39:641-651. [DOI] [PubMed] [Google Scholar]
- 47.Tobiason, D. M., A. G. Lenich, and A. C. Glasgow. 1999. Multiple DNA binding activities of the novel site-specific recombinase, Piv, from Moraxella lacunata. J. Biol. Chem. 274:9698-9706. [DOI] [PubMed] [Google Scholar]
- 48.Urasaki, A., Y. Sekine, and E. Ohtsubo. 2002. Transposition of cyanobacterium insertion element ISY100 in Escherichia coli. J. Bacteriol. 184:5104-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilde, C., S. Bachellier, M. Hofnung, and J.-M. Clément. 2001. Transposition of IS1397 in the family Enterobacteriaceae and first characterization of ISKpn1, a new insertion sequence associated with Klebsiella pneumoniae palindromic units. J. Bacteriol. 183:4395-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshioka, Y., H. Ohtsubo, and E. Ohtsubo. 1987. Repressor gene finO in plasmids R100 and F: constitutive transfer of plasmid F is caused by insertion of IS3 into F finO. J. Bacteriol. 169:619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuerner, R. L. 1994. Nucleotide sequence analysis of IS1533 from Leptospira borgpetersenii: identification and expression of two IS-encoded proteins. Plasmid 31:1-11. [DOI] [PubMed] [Google Scholar]