Abstract
The Helicobacter pylori ureE gene product was previously shown to be required for urease expression, but its characteristics and role have not been determined. The UreE protein has now been overexpressed in Escherichia coli, purified, and characterized, and three altered versions were expressed to address a nickel-sequestering role of UreE. Purified UreE formed a dimer in solution and was capable of binding one nickel ion per dimer. Introduction of an extra copy of ureE into the chromosome of mutants carrying mutations in the Ni maturation proteins HypA and HypB resulted in partial restoration of urease activity (up to 24% of the wild-type levels). Fusion proteins of UreE with increased ability to bind nickel were constructed by adding histidine-rich sequences (His-6 or His-10 to the C terminus and His-10 as a sandwich fusion) to the UreE protein. Each fusion protein was overexpressed in E. coli and purified, and its nickel-binding capacity and affinity were determined. Each construct was also expressed in wild-type H. pylori and in hypA and hypB mutant strains for determining in vivo urease activities. The urease activity was increased by introduction of all the engineered versions, with the greatest Ni-sequestering version (the His-6 version) also conferring the greatest urease activity on both the hypA and hypB mutants. The differences in urease activities were not due to differences in the amounts of urease peptides. Addition of His-6 to another expressed protein (triose phosphate isomerase) did not result in stimulation of urease, so urease activation is not related to the level of nonspecific protein-bound nickel. The results indicate a correlation between H. pylori urease activity and the nickel-sequestering ability of the UreE accessory protein.
Helicobacter pylori, a spiral, gram-negative, microaerophilic bacterium, has been shown to be the etiologic agent of chronic gastritis and peptic ulcers (3, 19) which can subsequently develop into gastric cancer (9). Since approximately half of the world's population is colonized by H. pylori, it is a major public health problem (9). One of the factors essential for gastric colonization is the urease enzyme (EC 3.5.1.5), the most abundant protein in H. pylori (up to 10% of the total protein content of the cell [2]), which raises the pH in the microenvironment surrounding the cell by producing ammonia (10, 34, 38).
Active urease is a multimeric enzyme that consists of six UreA and six UreB subunits coordinating twelve Ni2+ ions as a cofactor (8, 12, 13). It is known that nickel ions are transported into the cell by a high-affinity nickel transporter protein, NixA (21), and a battery of accessory proteins are thought to be required for synthesis of a catalytically active urease. Indeed, the ureI, ureE, ureF, ureG, and ureH genes need to be coexpressed with the structural genes ureA and ureB in Escherichia coli to get fully active urease (7), and it has recently been shown that H. pylori ureE, ureF, ureG, and ureH mutants are severely deficient in urease activity (35), but the specific roles of the corresponding proteins are still poorly understood. One exception is UreI, which has been shown to form a urea-specific pore (37).
In addition, two of the accessory genes (hypA and hypB) required for the activity of the membrane-bound Ni-containing hydrogen uptake hydrogenase enzyme have also been shown to be required for urease activity in H. pylori (26). Indeed, hypA and hypB mutants of H. pylori are severely affected in urease activity (26). Since the expression levels of the urease apoenzyme were not affected in these mutants but there was a four- and fivefold decrease in the nickel content of the urease in the hypA and hypB mutants, respectively, compared to the wild type, it was concluded that HypA and HypB were playing roles as Ni-specific metallochaperone proteins, involved in nickel sequestration and mobilization events leading to incorporation of the metal into the urease metallocenter. Some biochemical characteristics of H. pylori HypA and HypB have recently been reported (20).
In Klebsiella aerogenes, extensive biochemical studies revealed that three accessory proteins, UreD (homologous to H. pylori UreH), UreF, and UreG are required in vivo for the assembly of the nickel metallocenter within the urease (22, 23, 27, 28). A fourth accessory protein, UreE, was shown to be a metallochaperone that delivers nickel to urease (6, 16). It contains a histidine-rich carboxyl terminus (10 of the last 15 residues are histidine) and is able to bind five to six Ni2+ ions per dimer (6, 16). Since a truncated form of UreE lacking the histidine-rich region was still able to bind two Ni2+ ions per dimer and was as competent as wild-type UreE for activating urease, a His-rich region of K. aerogenes UreE was proposed to be mainly involved in nickel sequestering (4) unrelated to direct enzyme activation. Internal nickel-binding residues of UreE were shown to participate in urease activation (6), and recent structural analysis of the truncated UreE variant revealed that three internal histidine residues are likely to coordinate not two but three Ni2+ ions per dimer (30).
Whereas other UreE proteins (such as UreE from Proteus mirabilis [31]) have also been shown to have His-rich regions, the H. pylori UreE does not possess such a motif and is therefore thought to possess decreased nickel-binding properties compared to these proteins (Fig. 1). In this study, the nickel-binding ability of H. pylori UreE was determined, and it was increased by creating UreE proteins with additional histidine residues. This was done by adding a His tag motif or the C-terminal tail of K. aerogenes UreE (Fig. 1) to H. pylori UreE. The effect of the nickel-sequestering ability of UreE and its variants on urease activity was analyzed in vivo in H. pylori hypA and hypB mutants (mutants deficient in nickel sequestration) by introducing ureE or one of the newly generated variants into the chromosome. Interestingly, the presence of additional UreE alone was enough to partially restore urease activity in both mutants, and synthesis of His-rich UreE chimeric proteins resulted in higher urease activities. Analysis of the nickel-binding properties of the proteins overexpressed in Escherichia coli and purified revealed that H. pylori UreE was able to bind one Ni2+ ion per dimer and that the His-rich UreE proteins were able to sequester more nickel than the parent UreE. The results suggest a correlation between the nickel-sequestering ability of UreE and urease activity levels in H. pylori.
FIG. 1.
Protein sequences of H. pylori UreE, UreE variants constructed in this study, and other UreE proteins. The carboxyl-terminal region (residues 80 through 170) of H. pylori 43504 (Hp 43504), as deduced from the DNA sequence ([this study], and shown to be identical to H. pylori 85P [7]) is compared to the homologous UreE regions from H. pylori 26695 (Hp 26695 [32]), Bacillus pasteurii (Bp [5]), Proteus mirabilis (Pm [14]), and Klebsiella aerogenes (Ka [24]). Also shown are the sequences of the UreE variants engineered in this study. Histidine residues shown to be involved in nickel binding in K. aerogenes are indicated in boldface type (30). One of these residues also shown to be involved in nickel binding in B. pasteurii (29) and conserved in H. pylori is shaded. The histidine-rich carboxyl-terminal tail of K. aerogenes (used in this study) is underlined. The hexahistidine tag and additional amino acids added by the construction are shown in italics.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The H. pylori and E. coli strains used in this study are listed in Table 1. H. pylori strain ATCC 43504 was the parent strain. H. pylori was grown on Brucella agar (Difco) plates supplemented with 10% defibrinated sheep blood (BA plates) at 37°C under microaerophilic conditions (5% CO2, 4% O2, and 91% N2). DNA manipulations were performed in E. coli Top10 (Invitrogen, Carlsbad, Calif.). E. coli was grown aerobically in Luria-Bertani (LB) medium or plates at 37°C. Kanamycin (20 μg/ml), ampicillin (100 μg/ml), chloramphenicol (20 μg/ml), and isopropyl-β-d-thiogalactopyranoside (IPTG) (0.2 mM) were added as needed.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmidb | Relevant characteristicsa | Source or reference |
|---|---|---|
| H. pylori 43504 | Parent strain for all H. pylori strains | American Type Culture Collection |
| hypA mutant | hypA::aphA3 Kanr | 26 |
| hypB mutant | hypB::aphA3 Kanr | 26 |
| ureE mutant | ureE::aphA3 Kanr | 35 |
| 43504[Cm] | 43504 hp0405::cat Cmr | This study |
| 43504[HPE] | 43504 hp0405::Φ(ureAp-ureEHp-cat) Cmr | This study |
| 43504[HPE6] | 43504 hp0405::Φ(ureAp-ureEHp[His]6-cat) Cmr | This study |
| 43504[HPE10] | 43504 hp0405::Φ(ureAp-ureEHp-ureE′Ka-cat) Cmr | This study |
| 43504[HPE10SF] | 43504 hp0405::Φ(ureAp-ureE′Hp-ureE′Ka-ureE′Hp-cat) Cmr | This study |
| 43504[KAE] | 43504 hp0405::Φ(ureAp-ureEKa-cat) Cmr | This study |
| hypA[Cm] | hypA hp0405::cat Kanr Cmr | This study |
| hypA[HPE] | hypA hp0405::Φ(ureAp-ureEHp-cat) Kanr Cmr | This study |
| hypA[HPE6] | hypA hp0405::Φ(ureAp-ureEHp[His]6-cat) Kanr Cmr | This study |
| hypA[HPE10] | hypA hp0405::Φ(ureAp-ureEHp-ureE′Ka-cat) Kanr Cmr | This study |
| hypA[HPE10SF] | hypA hp0405::Φ(ureAp-ureE′Hp-ureE′Ka-ureE′Hp-cat) Kanr Cmr | This study |
| hypA[KAE] | hypA hp0405::Φ(ureAp-ureEKa-cat) Kanr Cmr | This study |
| hypB[Cm] | hypB hp0405::cat Kanr Cmr | This study |
| hypB[HPE] | hypB hp0405::Φ(ureAp-ureEHp-cat) Kanr Cmr | This study |
| hypB[HPE6] | hypB hp0405::Φ(ureAp-ureEHp[His]6-cat) Kanr Cmr | This study |
| hypB[HPE10] | hypB hp0405::Φ(ureAp-ureEHp-ureE′Ka-cat) Kanr Cmr | This study |
| hypB[HPE10SF] | hypB hp0405::Φ(ureAp-ureE′Hp-ureE′Ka-ureE′Hp-cat) Kanr Cmr | This study |
| hypB[KAE] | hypB hp0405::Φ(ureAp-ureEKa-cat) Kanr Cmr | This study |
| E. coli | ||
| TOP10 | Cloning strain | Novagen |
| Rosetta(DE3)(pLysS) | BL21 derivative, host for protein overproduction, Cmr | Novagen |
| Plasmids | ||
| pET21b | Cloning and expression vector, Ampr | Invitrogen |
| pETWTΔG | pET21b with K. aerogenes ureEFG′ | S. Mulrooney |
| pEU39cm | Vector used for homologous recombination into H. pylori, Cmr | 26 |
| pPA | ureAp cloned into BglII-NdeI sites of pET21b deleted for T7p | This study |
| pPA-HP | H. pylori ureE cloned into NdeI-SalI sites of pPA | This study |
| pPA-HP6 | H. pylori ureE (no stop codon) cloned into NdeI-SalI sites of pPA | This study |
| pPA-KA | K. aerogenes ureE cloned into NdeI-XhoI sites of pPA | This study |
| pPA-HP10 | HindIII-ScaI fragment of pPA-KA (encoding [His]10) into pPA-HP6 deleted for HindIII-ScaI fragment (encoding [His]6) | This study |
| pPA-HP10SF | H. pylori ureE′-K. aerogenes ureE′ (encoding [His]10)-H. pylori ureE′ sandwich fusion cloned into NdeI-BamHI sites of pPA | This study |
| pEU-HP | BglII-XhoI fragment of pPA-HP (encoding UreE) cloned into pEU39Cm | This study |
| pEU-HP6 | BglII-BlpI fragment of pPA-HP6 (encoding UreE-His6) into pEU39Cm | This study |
| pEU-HP10 | BglII-XhoI fragment of pPA-HP10 (encoding UreE-His10) into pEU39Cm | This study |
| pEU-HP10SF | BglII-XhoI fragment of pPA-HP10SF (encoding UreE-His10SF) into pEU39Cm | This study |
| pEU-KA | BglII-XhoI fragment of pPA-KA (encoding K. aerogenes UreE) into pEU39Cm | This study |
| pEU-Tpi | H. pylori tpi (no stop codon) cloned into SmaI-XhoI sites of pEU-HP6 deleted for ureAp-ureEHp | This study |
| pET-HP | NdeI-SalI fragment of pPA-HP (encoding UreE) cloned into pET21b | This study |
| pET-HP6 | NdeI-SalI fragment of pPA-HP6 (encoding UreE-His6) into pET21b | This study |
| pET-HP10 | NdeI-XhoI fragment of pPA-HP10 (encoding UreE-His10) into pET21b | This study |
| pET-HP10SF | NdeI-BamHI fragment of pPA-HP10SF (encoding UreE-His10SF) into pET21b | This study |
Hp, Helicobacter pylori; Ka, Klebsiella aerogenes.
HPE refers to H. pylori UreE, with E6 meaning 6 histidines (hexahistidine tag) and E10 meaning 10 histidines (encoded within ureE of K. aerogenes) were added to the protein.
DNA techniques.
All DNA manipulations were performed as described by Maniatis et al. (18). Chromosomal DNA was extracted from H. pylori with the Aquapure genomic DNA extraction kit (Bio-Rad, Hercules, Calif.). Plasmid DNA preparations were carried out with the QiaPrep Spin mini kit (Qiagen, Valencia, Calif.). DNA fragments or PCR products were purified from agarose gels with the Qiaquick gel extraction kit (Qiagen). PCR was performed in a Perkin-Elmer 2400 thermal cycler with Taq polymerase (Fischer, Fair Lawn, N.J.). All oligonucleotide primers were synthesized by Integrated DNA Technologies, Coralville, Iowa, and are listed in Table 2. All recombinant plasmids described in this study were sequenced at the Molecular Genetics Instrumentation Facility, University of Georgia, Athens, to ensure that no error had been introduced by PCR.
TABLE 2.
Oligonucleotide primers used in this study
| Designation | Sequencea (5′→3′) | Siteb |
|---|---|---|
| HpPureA F | ggccagatctCTAAAGGGGTATTAAACGC | BglII |
| HpPureA R | gggccccatATGATTCTCCTATTCTTAAAG | NdeI |
| HpUreE F | ccggcagccatATGATCATAGAGCGTTTAATAG | NdeI |
| HpUreE R1 | caggtgagtcgacCCTTTATCCATTTG | SalI |
| HpUreE R2 | gacggcggtcgacTTTCATGACCACTTTAAAATC | SalI |
| KaUreE F1 | ggccgagccatATGCTTTATTTAACTC | NdeI |
| KaUreE R | caaataatctcgagCAGGCGTTGTTCCGC | XhoI |
| KaUreE F2 | agagcaagcttCACGGTCATCATCATGC | HindIII |
| P21R | CTGGTGAGTACTCAACCAAG | |
| OE F | CCCCATAGTGAGCCTAATCACGGTCATCATCATG | |
| OE R1 | CATGATGATGACCGTGATTAGGCTCACTATGGGG | |
| OE R2 | cgggatccCTATTTCATGACCACTTTAAAATCGCTCGCCAGTGAGACCTTAAAGTGGCTGTGAGCGTG | BamHI |
| HpTpi F | accttagatctAAAATCGCTTGACGC | BglII |
| HpTpi R | cgggcctcgagTAAAAATGAAATGATTG | XhoI |
| Hp 405 F | GTAACGGGAATTCTTACGCC | |
| Cm R | AATGGGTTATCTCGGCGGTCACTC |
Capital letters indicate H. pylori-derived sequences, bold capital letters represent K. aerogenes-derived sequences, lowercase letters represent nucleotides added for cloning purposes, and underlining indicates newly generated restriction sites.
Restriction enzyme recognition sites.
Constructions of plasmids used for inserting an additional copy of ureE or its variants into the H. pylori chromosome.
A three-step strategy was undertaken to recombine H. pylori ureE and its variants or K. aerogenes ureE into the H. pylori chromosome. First, a 213-bp sequence immediately upstream of the start codon of ureA, including the transcriptional and translational signals required for UreA expression, was amplified by PCR with primers HpPureA F and HpPureA R and H. pylori genomic DNA as a template. The PCR product was digested with NdeI and BglII and ligated into the expression vector pET21b (Novagen, Madison, Wis.) previously cut with the same enzymes and gel purified to get rid of the T7 promoter, generating plasmid pPA. This plasmid was used to clone the coding sequence of ureE and its variants, placing each of them under the control of ureAp when the start codon was inserted at the NdeI restriction site.
To amplify the ureE gene from H. pylori, we used either primers HpUreE F and HpUreE R1 (designed to amplify ureE with its own stop codon) or HpUreE F and HpUreE R2 (designed to amplify ureE without its stop codon, in order to generate an hexahistidine-tagged UreE) and H. pylori genomic DNA as a template. Each 0.5-kb PCR product was digested with NdeI and SalI and ligated into similarly cut plasmid pPA to generate pPA-HP and pPA-HP6, respectively. Primers KaUreE F1 and KaUreE R were used to amplify K. aerogenes ureE from plasmid pETWTΔG, a gift from Scott Mulrooney (Departments of Microbiology and Biochemistry, Michigan State University, East Lansing). The resulting 0.5-kb PCR product was digested with NdeI and XhoI and ligated into the corresponding restriction sites of pPA, generating pPA-KA.
To generate pPA-HP10, we used pPA-KA as a template and primers KaUreE F2 and P21R to amplify the 3′ end of K. aerogenes ureE, which encodes the histidine-rich carboxy-terminal tail (His10), as well as a part of the cloning vector. The resulting 1.1-kb product was digested with HindIII and ScaI and ligated with the 5-kb DNA fragment purified from similarly digested pPA-HP6, yielding pPA-HP10. To construct pPA-HP10SF, a two-step overlapping PCR technique was used. First, part of H. pylori ureE was amplified from H. pylori genomic DNA in the presence of primers HpUreE F and OE R1 (whose 5′ end matches part of the K. aerogenes ureE sequence encoding His10), yielding a 0.5-kb product, and part of K. aerogenes ureE (encoding His10) was amplified from plasmid pETWTΔG in the presence of primers OE F1 (complementary to OE R1) and OE R2 (whose 5′ end matches the 3′ end of H. pylori ureE), yielding a 0.12-kb product; the two overlapping PCR products were combined and mixed with primers HpUreE F and OE R2 for a final round of PCR to amplify a 0.57-kb product, which was cut with NdeI and BamHI and ligated into similarly digested plasmid pPA, yielding pPA-HP10SF.
Finally, plasmids pPA-HP, pPA-KA, pPA-HP10, and pPA-HP10SF were digested with BglII and XhoI, while plasmid pPA-HP6 was cut with BglII and BlpI, each releasing a 0.7- to 0.8-kb DNA fragment (depending upon the constructs) with a distinct ureE variant under the control of ureAp. Each DNA fragment from pPA-HP, pPA-HP6, or pPA-KA was blunt ended with T4 polymerase and ligated into pEU39Cm previously digested with EcoRV to generate pEU-HP, pEU-HP6, and pEU-KA, respectively. Similarly, plasmids pPA-HP10 and pPA-HP10SF were sequentially cut with BglII, blunt ended, gel purified, and digested with XhoI, and each 0.7-kb DNA fragment released was ligated into pEU39Cm previously digested with both SmaI and EcoRV to yield pEU-HP10 and pEU-HP10SF, respectively. Plasmid pEU39Cm has been shown to be an efficient tool with which to insert DNA by homologous recombination (25, 26) in the region of the chromosome corresponding to strain ATCC 26695 hp0405 (32). Disruption of this region creates no obvious phenotype (data not shown).
Construction of plasmid pEU-Tpi.
To determine whether the presence of a random protein fused to a hexahistidine tag would have any effect on the urease activity of H. pylori hypA and hypB mutants, the gene hp0194, encoding triose phosphate isomerase (32), was PCR amplified with its own promoter with primers HpTpi F and HpTpi R. The resulting 900-bp product was sequentially digested with BglII, blunt ended with T4 polymerase, gel purified, and finally digested with XhoI. The purified product was then ligated into pEU-HP6 which had been digested with SmaI and XhoI and gel purified to get rid of the ureAp-ureE insert, generating pEU-Tpi.
Constructions of plasmids for overproduction of UreE, UreE-His6, UreE-His10, and UreE-His10SF proteins.
Plasmids pPA-HP and pPA-HP6 were digested with NdeI and SalI, plasmid pPA-HP10 was digested with NdeI and XhoI, and plasmid pPA-HP10SF was digested with NdeI and BamHI, and each DNA fragment containing ureE or a variant (ranging from 0.5 to 0.7 kb depending upon the construct) was inserted into expression vector pET21b to generate pET-HP, pET-HP6, pET-HP10, and pET-HP10SF, respectively. These recombinant pET plasmids were then transformed into E. coli strain Rosetta(DE3)(pLysS) (Novagen, Madison, Wis.).
Overproduction and purification of UreE and UreE variants.
Rosetta(DE3)(pLys) cells harboring a pET recombinant plasmid were grown at 37°C to an optical density at 600 nm of 0.5 in 500 ml of LB medium with 100 μg of ampicillin and 30 μg of chloramphenicol per ml. Expression of each recombinant protein was induced by adding 0.2 mM IPTG in the medium, and cells were grown for an additional 3 h at 37°C and harvested by centrifugation (5,000 × g, 15 min, 4°C). All subsequent steps were performed at 4°C. For purification of UreE-His6, UreE-His10, and UreE-His10SF proteins, the following protocol was used. Cells were washed with 200 ml of 20 mM Na2HPO4 (pH 7.4)-500 mM NaCl-5 mM imidazole (buffer A) and resuspended in 5 ml of the same buffer. Phenylmethylsulfonyl fluoride was added to a final concentration of 0.5 mM. Bacteria were lysed by three passages through a cold French pressure cell at 18,000 lb/in2, cell debris was removed by centrifugation at 20,000 × g, and the supernatant was subjected to ultracentrifugation at 100,000 × g for 2 h. The membrane-free supernatant, used as a source for purification of these three proteins, was applied to a nickel-nitrilotriacetic acid (Ni-NTA) affinity column (Qiagen), and buffer A was used to wash the resin until the A280 reached the baseline. Proteins were washed with buffer B (buffer A with 30 mM imidazole) until the A280 reached the baseline and finally eluted with buffer C (buffer A with 250 mM imidazole). Fractions were analyzed by gel electrophoresis (see below), and samples of interest were pooled. Protein concentration was determined with the BCA protein kit and the Coomassie dye binding kit (Pierce, Rockford, Ill.).
For the UreE protein, an alternative purification protocol was required because this protein did not bind to the Ni-NTA column. Cells were washed with 50 mM HEPES (pH 7.2) containing 50 mM NaCl (buffer D) and resuspended in 5 ml of the same buffer, and a membrane-free supernatant was isolated as described above. Solid ammonium sulfate was added to the extract to a final concentration of 2 M, and the pellet obtained after centrifugation (14,000 × g, 15 min) was resuspended in 5 ml of buffer D and dialyzed against the same buffer. The sample was applied to a 5-ml SP Sepharose cation exchange column, and the proteins were eluted with a linear gradient from 0 to 1 M NaCl in buffer D. UreE was eluted from the resin at approximately 0.4 M salt, giving nearly homogeneous protein. Samples of interest were pooled, and the protein concentration was determined with the BCA protein kit and the Coomassie dye binding kit. Purified UreE was used to raise polyclonal antiserum in a New Zealand White rabbit at Cocalico Biologicals, Reamstown, Pa.
Molecular mass determination of UreE by size exclusion chromatography and SDS-PAGE.
The apparent molecular mass of purified UreE was estimated by gel filtration with a Superose 6 HR 10/30 column chromatography (Amersham Pharmacia Biotech, Piscataway, N.J.) column at a flow rate of 0.4 ml/min. The column was equilibrated with 50 mM Tris-HCl (pH 7.5)-100 mM KCl and calibrated with the following reference proteins (Sigma GF 200 kit markers): β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa). The column void volume was determined with blue dextran.
The apparent molecular mass of denatured purified UreE and UreE variants as well as the proteins' purity were determined on sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis (SDS-PAGE) with a Mini-Protean II apparatus (Bio-Rad, Hercules, Calif.), according to the method of Laemmli (15). Proteins were visualized with Coomassie brilliant blue R-250. Molecular size markers were purchased from Bio-Rad Laboratories.
Immunoblotting.
Proteins were transferred to nitrocellulose (0.45-μm-pore-size membrane; Osmonics, Westborough, Mass.) as described by Towbin et al. with a Bio-Rad Trans Blot Cell (33). After transfer, proteins were visualized with Ponceau red (0.1% Ponceau red dye in 1.0% acetic acid). Immunoblotting was performed by a chemiluminescence method according to the manufacturer's instructions (Amersham Pharmacia Biotech, Piscataway, N.J.). Antisera were used at the following dilutions: anti-UreE rabbit antiserum, 1:750; anti-UreA or anti-UreB rabbit antiserum (a gift from Harry Mobley, Department of Medicine, Maryland School of Medicine, Baltimore), 1:2,000; and goat anti-rabbit immunoglobulin G-peroxidase (ICN-Cappel, Aurora, Ohio), 1:5,000.
Equilibrium dialysis and nickel determination.
The ability of the purified UreE, UreE-His6, UreE-His10, and Ure-His10SF recombinant proteins to bind nickel was determined by graphite furnace atomic absorption spectrophotometry (Shimadzu AA-6701F) following equilibrium dialysis. Briefly, 6 to 7 μM protein was dialyzed against 1 liter of 50 mM NaCl (pH 8.25) containing increasing concentrations of NiCl2 for 48 h at 4°C. After dialysis, the protein concentration was determined again in each dialysis bag with the BCA protein assay kit (Pierce), and the nickel concentration of the protein solution (bound plus free Ni2+) and the dialysis buffer (free Ni2+) was measured. The concentration of bound nickel was estimated by subtracting the two values.
Introduction of ureE variants into H. pylori chromosome.
Preparation of H. pylori competent cells and transformation by electroporation were done as previously described (26). Plasmids pEU39Cm, pEU-HP, pEU-HP6, pEU-HP10, pEU-HP10SF, pEU-KA, and pEU-Tpi were used to transform the parental strain 43504 hypA and hypB mutants. Cells were plated onto a nonselective BA plate, incubated for 48 h, harvested, and plated onto BA supplemented with chloramphenicol. Resistant colonies appeared in 72 to 96 h. This procedure yielded strains that had the original ureE and a construct (ureE or one of its variants) at an unrelated site. The presence of each construct in the chromosome was confirmed by PCR amplification with primers Hp 405 F (annealing upstream of the disrupted region) and Cm R (specific for the cat gene) and genomic DNA from each of the isolated colonies as a template.
Urease assays.
The urease activity of fresh lysates was determined by measuring ammonia production from urea hydrolysis with the phenol-hypochlorite assay as described by Weatherburn (36). Briefly, cells were grown for 2 days on BA plates, harvested with a swab, resuspended in 50 mM HEPES-NaOH (pH 7.5), washed once, and resuspended in the same buffer. Lysates of freshly sonicated cells (0.5 to 10.0 μg of protein) were incubated for 20 min at room temperature in 1 ml of the same buffer plus 25 mM urea, and the amount of ammonia released was then assayed with the phenol-hypochlorite assay (36). A standard ammonium chloride concentration curve was used to convert the absorbance at 625 nm to nanomoles of ammonia. Data are presented as urease specific activity, defined as micromoles of ammonia (NH3) produced per minute per milligram of protein. Stated values are the mean ± standard variation for three to four independent cultures, with each assay made in duplicate.
Hydrogenase assays.
Hydrogen uptake activity was determined amperometrically for whole cells as described previously (17).
RESULTS AND DISCUSSION
Introduction of UreE variants into H. pylori.
In order to study the effect of UreE and its genetically modified histidine-rich variants on urease activity, we introduced a copy of the ureE gene or one of the variants into the chromosome of wild-type H. pylori and its hypA and hypB mutants. Because ureE is part of the ureIEFGH operon and does not possess its own promoter (1), our first step was to ensure proper expression of this gene as well as all the other constructs (including the K. aerogenes ureE gene) by placing each of them under the control of the H. pylori ureA promoter, as described in the Materials and Methods section. By cloning H. pylori ureE without its stop codon into the vector pPA (Table 1), we generated a hexahistidine-tagged UreE, UreE-His6 (Fig. 1). UreE-His10 was generated by cloning the sequence encoding the histidine-rich carboxyl-terminal tail of K. aerogenes UreE (last 15 residues) in frame with the H. pylori ureE gene. To investigate the effects of an alternative location of this K. aerogenes histidine-rich region on UreE properties, this sequence was also inserted within H. pylori UreE, generating a sandwich fusion protein, UreE-His10SF (Fig. 1). Finally, to determine whether a hexahistidine-tagged random protein would have any effect on urease expression, a triose phosphate isomerase (Tpi)-His6 fusion protein was generated.
Each construct was inserted into pEU39 cm, and the resulting plasmid was introduced by electroporation into H. pylori wild-type, hypA, and hypB mutant strains. Use of plasmid pEU39 cm allowed insertion of each construct along with the chloramphenicol resistance gene (cat) into the same (but unrelated to Ni enzyme maturation) region of the chromosome (corresponding to gene hp0405) (32) by homologous recombination. Each gene insertion was confirmed by PCR amplification of the fragment of the expected size with genomic DNA purified from each of the chloramphenicol-resistant strains and primers specific for the hp0405 and cat genes. Insertion of the cat gene only into hp0405 yielded a 670-bp PCR product, whereas insertion of any of the ureE variants (under the control of ureA promoter) in the hp0405 region gave a unique 1,450- to 1,550-bp PCR product, depending upon the construct (data not shown).
The expression levels of the introduced UreE proteins were compared by immunoblotting with antiserum raised against purified UreE. The different constructs, Ure-His6, Ure-His10, and Ure-His10SF, could be separated as larger species from native UreE on SDS-PAGE and were expressed at about the same levels whether present in hypB (Fig. 2) or hypA (data not shown) mutants. Although the level of expression of additional UreE alone is hard to estimate because the strains also possessed the original copy of the ureE gene, it is reasonable to predict that the expression level of additional UreE was the same as that of Ure-His6, Ure-His10, and Ure-His10SF, since all constructs were under the control of the same promoter. Although the modified versions of UreE appeared to be expressed at a lower level than the original, the antibodies used in this study were raised against native UreE, and their specificity for the engineered versions was shown to be lower than for native UreE (data not shown).
FIG. 2.
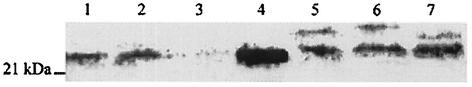
Western blot analysis of UreE expression in H. pylori. Whole-cell lysate (25 μg) of H. pylori and purified UreE (500 ng) were subjected to SDS-PAGE, transferred to nitrocellulose, and blotted with antibodies raised against purified UreE. Lane 1, hypB[Cm]; lane 2, hypB[HPE]; lane 3, ureE mutant; lane 4, purified UreE; lane 5, hypB[HPE6]; lane 6, hypB[HPE10]; lane 7, hypB[HPE10SF]. The position of a molecular mass standard is indicated on the left.
Effect of introduced UreE and histidine-rich versions of UreE on urease activities of wild-type H. pylori and hypA and hypB mutants.
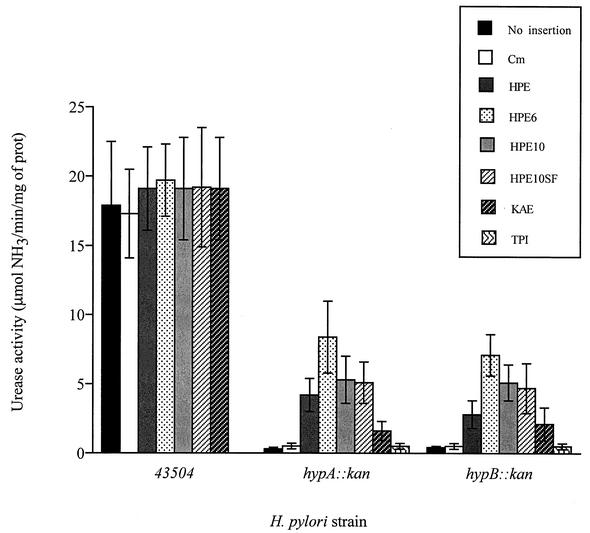
Wild-type strain 43504 and hypA and hypB mutants expressing each of the UreE variant proteins were grown on BA plates for 2 days, and cell extracts were assayed for urease activity (Fig. 3). As expected, disruption of hp0405 by the cat gene did not cause a change in urease activity levels in the wild-type or the mutant strains, and therefore it was considered a control for all the insertion experiments. Insertion of an additional copy of ureE or one of its variants into the chromosome of the wild-type strain had no or a slight effect on the urease activity (up to 10% increase), presumably because the urease activity was already near or at its optimal level under these conditions (with all the accessory proteins synthesized). However, expression of additional UreE showed an eightfold and sixfold increase in urease expression compared to the controls in the hypA and hypB mutant, respectively. This suggested that in these mutants affected for Ni-sequestering activity, the presence of an additional UreE itself was enough to partially restore urease activity (24% and 16% of the wild-type activity for hypA[HPE] and hypB[HPE], respectively).
FIG. 3.
Urease activities of H. pylori 43504 and hypA and hypB mutants harboring an additional copy of ureE or one of its variants in the chromosome. Constructs introduced by homologous recombination within the hp0405 gene region of the chromosome of either strain are indicated. Results shown are the averages of three to four independent growth experiments; error bars denote standard deviations. TPI, triose phosphate isomerase.
Expression of hexahistidine-tagged UreE (HPE6) from the chromosome in the hypA and hypB backgrounds resulted in the highest urease activities measured, 17-fold and 14-fold more than the urease activity of the controls, respectively. This is 49% and 41% of the wild-type activity for the hypA mutant expressing HPE6 (hypA[HPE6]) and hypB[HPE6], respectively. The result for the His6 version was significantly greater than all the other introduced versions (for both hypA and hypB mutants). Still, expression of the UreE-His10 (HPE10) and UreE-His10SF (HPE10SF) chimeric proteins produced a significant increase in urease activity in both mutants compared with expression of additional UreE only (Fig. 3).
Since the urease activities for both constructs were similar, the location of the K. aerogenes UreE nickel-binding sequence within H. pylori UreE did not seem to have an effect on the ability of this protein to assist in urease assembly. Insertion of the K. aerogenes ureE gene (under the control of the H. pylori ureA promoter) into the H. pylori chromosome resulted in a moderate increase in urease activity in both hypA and hypB mutants compared to the other histidine-rich proteins, suggesting that this heterologous UreE protein could not play an efficient role in H. pylori. Expression of a hexahistidine-tagged triose phosphate isomerase (TPI-His6) chimera in the H. pylori hypA and hypB mutants (as verified by purification on an Ni-NTA column followed by SDS-PAGE analysis; data not shown) did not increase the urease activity (Fig. 3), indicating that the presence of a nickel-binding component was not enough to partially restore the urease activity. This is in good agreement with previous results from Gilbert and coworkers, who showed that the presence and absence of Hpn, a protein with 47% His residues and a very high Ni-binding capacity, had no effect on urease activity in H. pylori (11). Therefore, the Ni-binding specificity to the UreE component of the fusion protein is the important factor for urease activation.
From these experiments, it was clear that addition of any nickel-coordinating motif (hexahistidine tag and histidine-rich sequence from K. aerogenes UreE) to the H. pylori UreE protein resulted in increased urease activity in the hypA and hypB backgrounds, suggesting a correlation between the nickel-binding capacity of each UreE variant and its ability to activate the urease.
The addition of small amounts of nickel to the medium (0.5 μM to 2.5 μM) resulted in an increase in all urease activities with the same differences observed between the various strains (for instance, hypA, hypA/HPE, and hypA/HPE6 urease activities remained at 2%, 25%, and 50% of that of the wild-type strain, respectively). When more nickel was added to the medium (5 to 100 μM), urease activity was fully restored (data not shown), as reported previously (26).
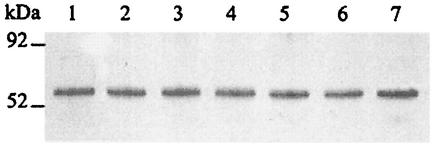
To determine whether differences in urease activity were linked to differences in urease synthesis, an immunoblot was performed with antibodies raised against the urease structural subunit UreB. As shown in Fig. 4, all hypA mutants had the same amount of UreB as wild-type strain 43504 regardless of the presence of additional UreE or one of its variants. The same results were seen with the hypB mutant and its derivatives (data not shown). In addition, another immunoblot experiment with antibodies against the other urease structural subunit, UreA, also showed that urease was expressed at the same level in all strains (data not shown). Therefore, the difference in urease activity described above between the strains is not related to a difference in urease expression.
FIG. 4.
Western blot analysis of UreB expression in H. pylori. Whole-cell lysate (6 μg) of H. pylori was subjected to SDS-PAGE, transferred to nitrocellulose, and blotted with antibodies directed to the urease large subunit UreB. Lane 1, 43504[Cm]; lane 2, hypA[HPE10SF]; lane 3, hypA[HPE10]; lane 4, hypB[HPE6]; lane 5, hypB[HPE]; lane 6, hypB[Cm]; lane 7, hypB. The positions of molecular mass standards are indicated on the left.
Effect of expression of UreE and histidine-rich versions of UreE on hydrogenase activities of hypA and hypB mutants.
Since mutations within hypA and hypB, encoding proteins known to be hydrogenase accessory proteins, showed a pleiotropic effect on both hydrogenase and urease activities (26), it was of particular interest to determine whether the presence of additional UreE and its variants would have any effect on hydrogenase activity. All the variants were introduced into the hypA and hypB backgrounds and tested for hydrogenase activity, and they all showed the same, almost not detectable hydrogenase level (less than 0.01 nmol of H2 oxidized/min/108 cells), suggesting that the presence of additional UreE or one of its variants did not have any effect on hydrogenase activity. In addition, hydrogenase activity assays were also performed on a ureE mutant (this strain kindly provided by David Scott, Department of Physiology, University of California at Los Angeles). These assays revealed that this mutant had the same hydrogenase level as wild-type strain 43504 (0.60 ± 0.15 nmol of H2 oxidized/min/108 cells). Therefore, a mutation in ureE does not have any effect on hydrogenase activity in H. pylori.
Purification of UreE and histidine-rich UreE variants.
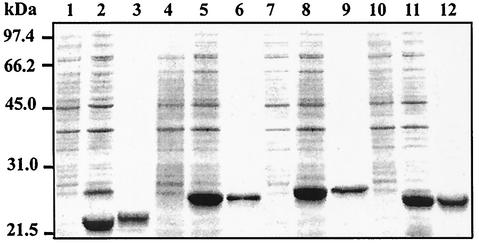
Genetically engineered UreE-His6, UreE-His10, and UreE-His10SF were expected to bind nickel because of the presence of all the histidines in these sequences, but the precise nickel-sequestering ability of each protein was unknown. Furthermore, as the nickel-binding properties of H. pylori UreE have not been reported to date, we decided to overexpress all four proteins in E. coli, purify them, and investigate their ability to bind nickel. Plasmids pPA-HP, pPA-HP6, pPA-HP10, and pPA-HP10SF were digested to release each distinct ureE variant. Each gene was inserted into the expression vector pET21b, and the newly generated pET-HP, pET-HP6, pET-HP10, and pET-HP10SF plasmids were introduced into E. coli strain Rosetta(pLys). Following overexpression, the UreE-His6, UreE-His10, and UreE-His10SF fusion proteins localized exclusively to the soluble fraction of the cell and were purified in a single step by Ni-NTA affinity chromatography, as revealed by SDS-PAGE (Fig. 5, lanes 6, 9, and 12, respectively).
FIG. 5.
SDS-PAGE of purified UreE variants. Lanes 1, 4, 7, and 10 contain noninduced cell extracts of strain Rosetta harboring pET-HP (for expression of UreE), pET-HP6 (UreE-His6), pET-HP10 (UreE-His10), and pET-HP10SF (UreE-His10SF), respectively. Lanes 2, 5, 8, and 11 contain cell extracts after 3 h of IPTG induction of UreE, UreE-His6, UreE-His10, and UreE-His10SF, respectively. UreE was purified by ammonium sulfate precipitation and cation exchange chromatography (lane 3). UreE-His6, UreE-His10, and UreE-His10SF were purified through an Ni-NTA column (lanes 6, 9, and 12, respectively). The positions of molecular mass standards are indicated on the left.
Since UreE did not bind to the Ni-NTA column (data not shown), this protein was purified from the soluble fraction by ammonium sulfate precipitation followed by cation exchange chromatography (Fig. 5, lane 3). As reported previously for other UreE proteins (16, 31), H. pylori UreE migrated as a heavy peptide (about 23,000 Da) compared with the size (Mr = 19,419) predicted by DNA sequence analysis. This was also observed for each of the variants generated in this study, UreE-His6 (Mr = 21,171), UreE-His10 (Mr = 22,055), and UreE-His10SF (Mr = 21,599). Nevertheless, the migration of each UreE variant compared to the others was in good agreement with their amino acid sequences, as deduced from the DNA sequence of strain 43504 (Fig. 1).
In addition, the molecular mass of UreE was determined by gel filtration chromatography. It was shown to be 40.9 kDa, which is consistent with a protein existing as a dimer in solution. Whereas addition of Ni2+ has been shown to promote tetramerization of the Bacillus pasteurii UreE protein (5), addition of nickel ions did not change the gel filtration elution pattern of H. pylori UreE (data not shown), suggesting that this protein formed only dimeric species in the presence of nickel.
Nickel-binding ability of UreE and genetically engineered histidine-rich UreE proteins.
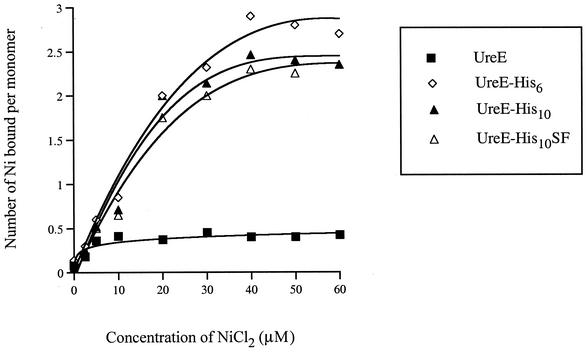
The number of Ni2+ ions bound to UreE and each of the variants engineered for nickel binding was determined over a range of nickel ion concentrations, as shown in Fig. 6. Equilibrium dialysis of UreE showed that this protein was capable of binding about 0.5 nickel ion per monomer (1 nickel ion per dimer). This is lower than that for the previously characterized K. aerogenes UreE protein, but this is not surprising, given the lack of homology between the two protein sequences. Indeed, H. pylori UreE shares only one of the residues shown to be involved in nickel binding in K. aerogenes UreE, His102 (Fig. 1), corresponding to His96 of K. aerogenes UreE (6, 30). In addition, study of the Bacillus pasteurii UreE crystal structure also showed the central role of the same conserved His residue in Ni binding (29). It needs to be pointed out that two other His residues (His110 and His112 of K. aerogenes UreE) shown to be involved in metal coordination in K. aerogenes are not conserved in H. pylori UreE.
FIG. 6.
Nickel ion binding by UreE, UreE-His6, UreE-His10, and UreE-His10SF proteins based on equilibrium dialysis. Each purified protein (at 6 to 7 μM monomer concentration) in 50 mM NaCl (pH 8.25) was equilibrated with the indicated concentrations of NiCl2, and the number of nickel ions bound per monomer was determined by graphite furnace atomic absorption spectrophotometry. Standard deviations were ≤10% for each set of measurements.
As expected, genetically modified, histidine-rich versions of H. pylori UreE had increased nickel-binding abilities compared with wild-type UreE, with about 3 nickel ions bound per monomer of UreE-His6 and about 2.5 nickel ions bound per monomer of both Ure-His10 and UreE-His10SF. These results can be compared to those of previous studies showing that K. aerogenes UreE (containing 10 His residues at the C terminus) was able to bind 5 to 6 nickel ions per dimer (6, 16), while the same protein deleted for its carboxyl-terminal tail could still coordinate 2 nickel ions per dimer (4) and more likely 3, as suggested by crystal structure analysis of this truncated variant (30). Kd values, as determined from curves, were estimated to be 1 μM for UreE, 15 μM for UreE-His6, 13 μM for UreE-His10, and 15 μM for UreE-His10SF.
Interestingly, we could see a correlation between the in vitro nickel-binding ability of each UreE variant and its capacity to activate the urease in vivo in the mutants. Indeed, the maximal urease activity seen in both hypA and hypB mutants as well as in the wild type was when UreE-His6, the protein shown to have the highest nickel-binding ability, was expressed in the cell. Likewise, Ure-His10 and UreE-His10SF were less efficient than UreE-His6 for both nickel binding and urease activation, while expression of additional wild-type UreE had the lowest effect of all H. pylori constructs on the urease activity, in agreement with its lower nickel-sequestering capacity.
Finally, even though the H. pylori UreE protein does not have high nickel-binding ability (unlike the case in other microorganisms, such as K. aerogenes and Proteus mirabilis), the stomach-colonizing bacterium has a high nickel demand and has found a complementary way of sequestering nickel, by using both the hydrogenase (HypA and HypB) and urease accessory proteins to achieve full nickel-urease maturation.
Acknowledgments
We thank Scott Mulrooney and Robert Hausinger (Michigan State University, East Lansing) for the gift of plasmid pETWTΔG, David Scott (Department of Physiology, UCLA, Los Angeles) for the gift of the H. pylori ureE mutant, and Harry Mobley (Department of Medicine, Maryland School of Medicine, Baltimore) for the gift of anti-UreA and anti-UreB antisera. We also thank all members of the Maier laboratory for helpful discussions and constant support.
This work was supported by the Georgia Research Foundation and by National Institutes of Health grant RO1DKK60061.
REFERENCES
- 1.Akada, J. K., M. Shirai, H. Takeuchi, M. Tsuda, and T. Nakazawa. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071-1084. [DOI] [PubMed] [Google Scholar]
- 2.Bauerfeind, P., R. Garner, B. E. Dunn, and H. L. Mobley. 1997. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut 40:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaser, M. J. 1990. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J. Infect. Dis. 161:626-633. [DOI] [PubMed] [Google Scholar]
- 4.Brayman, T. G., and R. P. Hausinger. 1996. Purification, characterization, and functional analysis of a truncated Klebsiella aerogenes UreE urease accessory protein lacking the histidine-rich carboxyl terminus. J. Bacteriol. 178:5410-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciurli, S., N. Safarov, S. Miletti, A. Dikiy, S. K. Christensen, K. Kornetzky, D. A. Bryant, I. Vandenberghe, B. Devreese, B. Samyn, H. Remaut, and J. van Beeumen. 2002. Molecular characterization of Bacillus pasteurii UreE, a metal-binding chaperone for the assembly of the urease active site. J. Biol. Inorg. Chem. 7:623-631. [DOI] [PubMed] [Google Scholar]
- 6.Colpas, G. J., T. G. Brayman, L. J. Ming, and R. P. Hausinger. 1999. Identification of metal-binding residues in the Klebsiella aerogenes urease nickel metallochaperone UreE. Biochemistry 38:4078-4088. [DOI] [PubMed] [Google Scholar]
- 7.Cussac, V., R. L. Ferrero, and A. Labigne. 1992. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol. 174:2466-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn, B. E., G. P. Campbell, G. I. Perez-Perez, and M. J. Blaser. 1990. Purification and characterization of urease from Helicobacter pylori. J. Biol. Chem. 265:9464-9469. [PubMed] [Google Scholar]
- 9.Dunn, B. E., N. B. Vakil, B. G. Schneider, M. M. Miller, J. B. Zitzer, T. Peutz, and S. H. Phadnis. 1997. Localization of Helicobacter pylori urease and heat shock protein in human gastric biopsies. Infect. Immun. 65:1181-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton, K. A., C. L. Brooks, D. R. Morgan, and S. Krakowka. 1991. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect. Immun. 59:2470-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert, J. V., J. Ramakrishna, F. W. Sunderman, Jr., A. Wright, and A. G. Plaut. 1995. Protein Hpn: cloning and characterization of a histidine-rich metal-binding polypeptide in Helicobacter pylori and Helicobacter mustelae. Infect. Immun. 63:2682-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawtin, P. R., H. T. Delves, and D. G. Newell. 1991. The demonstration of nickel in the urease of Helicobacter pylori by atomic absorption spectroscopy. FEMS Microbiol. Lett. 61:51-54. [DOI] [PubMed] [Google Scholar]
- 13.Hu, L. T., and H. L. Mobley. 1990. Purification and N-terminal analysis of urease from Helicobacter pylori. Infect. Immun. 58:992-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, B. D., and H. L. Mobley. 1989. Proteus mirabilis urease: nucleotide sequence determination and comparison with jack bean urease. J. Bacteriol. 171:6414-6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Lee, M. H., H. S. Pankratz, S. Wang, R. A. Scott, M. G. Finnegan, M. K. Johnson, J. A. Ippolito, D. W. Christianson, and R. P. Hausinger. 1993. Purification and characterization of Klebsiella aerogenes UreE protein: a nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 2:1042-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maier, R. J., C. Fu, J. Gilbert, F. Moshiri, J. Olson, and A. G. Plaut. 1996. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol. Lett. 141:71-76. [DOI] [PubMed] [Google Scholar]
- 18.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i: 1311-1315. [DOI] [PubMed] [Google Scholar]
- 20.Mehta, N., J. W. Olson, and R. J. Maier. 2003. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J. Bacteriol. 185:726-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mobley, H. L., R. M. Garner, and P. Bauerfeind. 1995. Helicobacter pylori nickel-transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol. Microbiol. 16:97-109. [DOI] [PubMed] [Google Scholar]
- 22.Moncrief, M. B., and R. P. Hausinger. 1997. Characterization of UreG, identification of a UreD-UreF-UreG complex, and evidence suggesting that a nucleotide-binding site in UreG is required for in vivo metallocenter assembly of Klebsiella aerogenes urease. J. Bacteriol. 179:4081-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moncrief, M. B., and R. P. Hausinger. 1996. Purification and activation properties of UreD-UreF-urease apoprotein complexes. J. Bacteriol. 178:5417-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulrooney, S. B., and R. P. Hausinger. 1990. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J. Bacteriol. 172:5837-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson, J. W., J. N. Agar, M. K. Johnson, and R. J. Maier. 2000. Characterization of the NifU and NifS Fe-S cluster formation proteins essential for viability in Helicobacter pylori. Biochemistry 39:16213-16219. [DOI] [PubMed] [Google Scholar]
- 26.Olson, J. W., N. S. Mehta, and R. J. Maier. 2001. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 39:176-182. [DOI] [PubMed] [Google Scholar]
- 27.Park, I. S., M. B. Carr, and R. P. Hausinger. 1994. In vitro activation of urease apoprotein and role of UreD as a chaperone required for nickel metallocenter assembly. Proc. Natl. Acad. Sci. USA 91:3233-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, I. S., and R. P. Hausinger. 1995. Evidence for the presence of urease apoprotein complexes containing UreD, UreF, and UreG in cells that are competent for in vivo enzyme activation. J. Bacteriol. 177:1947-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Remaut, H., N. Safarov, S. Ciurli, and J. Van Beeumen. 2001. Structural basis for Ni2+ transport and assembly of the urease active site by the metallochaperone UreE from Bacillus pasteurii. J. Biol. Chem. 276:49365-49370. [DOI] [PubMed] [Google Scholar]
- 30.Song, H. K., S. B. Mulrooney, R. Huber, and R. P. Hausinger. 2001. Crystal structure of Klebsiella aerogenes UreE, a nickel-binding metallochaperone for urease activation. J. Biol. Chem. 276:49359-49364. [DOI] [PubMed] [Google Scholar]
- 31.Sriwanthana, B., M. D. Island, D. Maneval, and H. L. Mobley. 1994. Single-step purification of Proteus mirabilis urease accessory protein UreE, a protein with a naturally occurring histidine tail, by nickel chelate affinity chromatography. J. Bacteriol. 176:6836-6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 33.Towbin, H., T. Staehelin, and J. Gordon. 1992. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Bio/Technology 24:145-149. [PubMed]
- 34.Tsuda, M., M. Karita, M. G. Morshed, K. Okita, and T. Nakazawa. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 62:3586-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voland, P., D. L. Weeks, E. A. Marcus, C. Prinz, G. Sachs, and D. Scott. 2003. Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. Am. J. Physiol. Gastrointest. Liver Physiol. 284:G96-G106. [DOI] [PubMed] [Google Scholar]
- 36.Weatherburn, M. W. 1968. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 39:971-974. [Google Scholar]
- 37.Weeks, D. L., S. Eskandari, D. R. Scott, and G. Sachs. 2000. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 287:482-485. [DOI] [PubMed] [Google Scholar]
- 38.Wirth, H. P., M. H. Beins, M. Yang, K. T. Tham, and M. J. Blaser. 1998. Experimental infection of Mongolian gerbils with wild-type and mutant Helicobacter pylori strains. Infect. Immun. 66:4856-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]