Abstract
Diatoms are a ubiquitous class of microalgae of extreme importance for global primary productivity and for the biogeochemical cycling of minerals such as silica. However, very little is known about diatom cell biology or about their genome structure. For diatom researchers to take advantage of genomics and post-genomics technologies, it is necessary to establish a model diatom species. Phaeodactylum tricornutum is an obvious candidate because of its ease of culture and because it can be genetically transformed. Therefore, we have examined its genome composition by the generation of approximately 1,000 expressed sequence tags. Although more than 60% of the sequences could not be unequivocally identified by similarity to sequences in the databases, approximately 20% had high similarity with a range of genes defined functionally at the protein level. It is interesting that many of these sequences are more similar to animal rather than plant counterparts. Base composition at each codon position and GC content of the genome were compared with Arabidopsis, maize (Zea mays), and Chlamydomonas reinhardtii. It was found that distribution of GC within the coding sequences is as homogeneous in P. tricornutum as in Arabidopsis, but with a slightly higher GC content. Furthermore, we present evidence that the P. tricornutum genome is likely to be small (less than 20 Mb). Therefore, this combined information supports the development of this species as a model system for molecular-based studies of diatom biology. The nucleotide sequence data reported has been deposited in GenBank Nucleotide Sequence Database (dbEST section) under accession nos. BI306757 through BI307753.
Diatoms are important components of marine phytoplankton, being particularly important for biogeochemical cycling of minerals such as silica, and for global carbon fixation (Werner, 1977; Tréguer et al., 1995). There are well over 250 genera of living diatoms, with perhaps as many as 100,000 species (Round et al., 1990). In toto, they may contribute as much as 25% of the total primary production on earth (Van Den Hoek et al., 1997). These figures illustrate the quantitative significance of diatoms for the functioning of “ecosystem Earth.”
The success of diatoms is not well understood, although it is known that they are remarkably flexible in adjusting their photosynthetic reactions to allow maximal growth rates over a wide range of light intensities (Falkowski and LaRoche, 1991), and that they may perform C4 photosynthesis (Reinfelder et al., 2000). In spite of their enormous ecological importance, only recently have they begun to attract the attention of molecular biologists (Scala and Bowler, 2001). As a consequence, knowledge of genome size and structure is extremely limited and only a few genes have been isolated. In November 2001, the sequences of less than 70 protein-encoding nuclear genes from diatoms had been deposited in GenBank (GenBank release 126, October 15, 2001).
Diatoms are brown algae belonging to the division Heterokonta and are thought to have arisen from a secondary endosymbiosis between a red alga (Rhodophyta) and a heterotrophic flagellate (related to the Oomycetes) around 300 million years ago (Gibbs, 1981; Delwiche and Palmer, 1997; Medlin et al., 2000). This can explain why diatom plastids are surrounded by four membranes rather than two. Therefore, any common evolution of the photosynthetic apparatus with higher plants and other chlorophytes is limited to the primary endosymbiotic event that gave rise to the chloroplast, probably more than 650 million years ago, thus, studies of diatom biology are likely to reveal many novel aspects.
To make diatoms accessible to powerful genomics and post-genomics technology platforms, it is important that the diatom research community focuses research efforts on a single species. Phaeodactylum tricornutum is an attractive model because of its apparently small genome (Darley, 1968; Veldhuis et al., 1997), short generation time, and ease of genetic transformation (Apt et al., 1997; Falciatore et al., 1999). However, little information is available about its genome and, before this study, only 21 sequences could be retrieved from GenBank (GenBank release 126, October 15, 2001). Of these, 10 correspond to the multigene family encoding the fucoxanthin, chlorophyll a/c-binding proteins (FCP A-F; Bhaya and Grossman, 1993).
A rapid method for identifying genes is by partial sequencing of random cDNAs to produce expressed sequence tags (ESTs). The popularity of this approach is clear from the EST database (http://www.ncbi.nlm.nih.gov/dbEST/), which currently contains more than five million sequences. From photosynthetic eukaryotes, the Viridiplantae (green plants) are the best represented, whereas only two EST projects have been reported from non-green macroalgae. One of these is from the red alga Porphyra yezoensis (Nikaido et al., 2000), and the other is from the brown alga Laminaria digitata (Crépineau et al., 2000). Before the current study, only one EST had been reported from a diatom (GenBank release 126 [15 October 2001]).
The generation of a cDNA catalog from P. tricornutum is useful for the sake of functional and phylogenetic comparisons of expressed gene populations with those of other organisms and because it will lead to a significant increase in the very limited amount of sequence information currently available from diatoms. Moreover, new metabolic routes may be discovered by means of EST cataloging. For example, diatoms have the unique characteristic of a silica-based rigid cell wall (Mann and Ozin, 1996; Zurzolo and Bowler, 2001), so the generation of large numbers of ESTs may eventually lead to the identification of genes encoding essential components involved in its formation. In this paper, we summarize an analysis of a population of approximately 1,000 ESTs from P. tricornutum and describe the compositional properties of its genome.
RESULTS
Sequence Analysis
The cDNA library used in this study consisted of 1.8 × 106 clones. A total of 997 single-pass nucleotide sequences were generated from the 5′ ends of randomly chosen cDNA clones. After deletion of vector sequences and ambiguous bases, an average length of 303 bp was used for the database searches. Table I shows a general summary of the BLASTX results obtained when using different filters of identity/similarity, sequence length, and E value. It can be seen that although a 40% filter on similarity does not change the number of BLASTX hits below specified E values, the selection of identity and a filter on the amino acid length does affect the number of sequences identified. As a dataset for the remainder of this study, we used the sequences obtained with the most stringent criteria, i.e. 40% identity, a length of more than 50 amino acids, and E values of less than 0.0001 (see “Materials and Methods”). A comprehensive list of the results of the BLASTX analysis can be found at http://193.205.231.39/Phaeodactylum/DW1.htm.
Table I.
Overview of BLASTX search results with different filters
| Identity/Similarity | Length of Filter on Amino Acids | No. of BLASTX Matches
|
||
|---|---|---|---|---|
| E ≤ 0.0001 | E ≤ 0.001 | E ≤ 0.01 | ||
| No filter | None | 314 | 339 | 352 |
| 40% Similarity | None | 314 | 339 | 352 |
| 40% Identity | None | 264 | 274 | 280 |
| 40% Similarity | >50 | 278 | 295 | 304 |
| 40% Identity | >50 | 230 | 232 | 234 |
Out of the 997 ESTs analyzed in this report, 819 represented unique or nonredundant sequences. A redundancy of 17.8% was found in this set of sequences (Table II); these redundant sequences could be transcripts of the same gene or cognate genes. We found 194 nonredundant sequences (23.7%) that had significant amino acid sequence similarities to sequences registered in protein databases (Table II). The remainder showed similarities to other protein sequences below our threshold criteria (69.1%) or showed no matches at all with any sequences currently present in the databases (7.2%; Table II).
Table II.
Overview of results from P. tricornutum EST program
| No. | % | |
|---|---|---|
| Total ESTs sequenced | 997 | – |
| No. of unique sequences | 819 | 82.2 |
| Redundant sequences | 178 | 17.8 |
| Average length (bp) | 303 | – |
| Putatively identified genes | 194 | 23.7a |
| Unidentified genes | 625 | 76.3a |
| No significant matchesb | 566 | 69.1a |
| No matches at all in databases | 59 | 7.2a |
Percentages of identified and unidentified genes in the unique sequences.
No significant matches according to our filtering method (see “Materials and Methods”).
Several ESTs matched genes previously identified in P. tricornutum or in other diatoms (see Table III in Supplementary Information at http://www.plantphysiol.org). For example, a total of 74 ESTs encoded fucoxanthin, chlorophyll a,c-binding proteins (FCP), the major protein components of the light-harvesting antenna complexes of photosystems I and II within diatom plastids (Grossman et al., 1990), or showed similarities to the functionally homologous light-harvesting chlorophyll a,b-binding proteins (LHC) of green algae and higher plants (Jansson, 1999). Twenty-two of these sequences were nonredundant (see Table III and Fig. 1 in Supplementary Information at http://www.plantphysiol.org). After the FCP/LHC-encoding cDNAs, the next most abundant ESTS encoded glyceraldehyde-3-P dehydrogenase (13 sequences).
Another well-represented family of proteins encoded by our ESTs are the frustulins, important calcium-binding glycoprotein components of the diatom siliceous cell wall (Kröger et al., 1994). We found a total of 12 ESTs encoding α- or ε-frustulins, of which seven were nonredundant. Frustulins had not been previously identified in P. tricornutum, nor had any silicon transporters (Hildebrand et al., 1997), which we also found (PTSS0913; see Table III in Supplementary Information at http://www.plantphysiol.org). The identification of such sequences reinforces the hypothesis that P. tricornutum could be a good model species for studying the unique aspects of diatom cell biology such as silica metabolism and frustule cell wall formation (see “Conclusions”).
EST programs from photosynthetic organisms normally result in the identification of numerous sequences encoding the small subunit of Rubisco, the principal enzyme in carbon fixation (Höfte et al., 1993). The reason why we have not identified this gene in P. tricornutum is simply because it is known to be plastid encoded in diatoms (Kowallik et al., 1995).
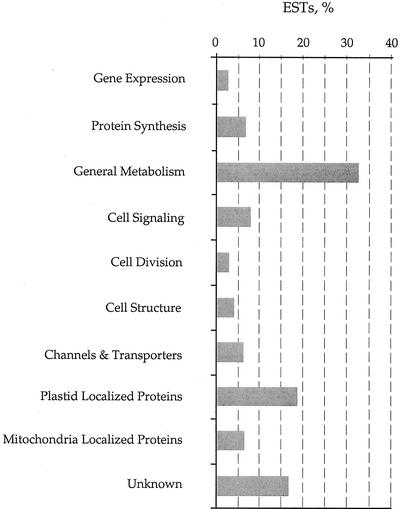
All of the ESTs encoding proteins similar to known proteins in the public databases above our predefined threshold levels are listed in Table III in Supplementary Information at http://www.plantphysiol.org. They have been divided into 10 functional groups based on their biochemical function. Further details can be found at http://www.szn.it/plant/PhaeodactylumEST. A comprehensive list of the results of the BLASTX analysis can also be found at this site. The percentage of sequences falling into different functional groups has been summarized in Figure 1. As expected, the majority of ESTs can be classified as encoding proteins involved in general cellular metabolism (32%) or as being localized to the plastid (18%). However, from this arbitrary classification it is apparent that a high number of ESTs encode proteins involved in cell signaling (8%) or channels and transporters (6%). This latter finding presumably reflects the importance of maintaining cellular ion homeostasis in a marine environment.
Figure 1.
Functional classification of derived coding sequences from P. tricornutum ESTs. The nonredundant BLASTX hits shown in Table III (see Supplementary Information at http://www.plantphysiol.org) were classified manually into the different functional groups shown.
The large number of sequences encoding cell signaling components points to the importance of stimulus perception and signal transduction mechanisms for regulating diatom cell physiology. Furthermore, from our small sample population it would appear that diatoms contain many of the components of signaling pathways found in other organisms such as small GTP-binding proteins (e.g. PTSS0963 and PTSS0715), protein kinases (e.g. PTSS0457), protein phosphatases (e.g. PTSS0311 and PTSS0103), and calcium-regulated enzymes (e.g. PTSS0773). The importance of calcium as a second messenger has already been inferred from studies of signal perception in transgenic diatoms expressing the calcium-sensitive photoprotein aequorin (Falciatore et al., 2000).
However, a major surprise is that we have found ESTs encoding the catalytic and regulatory subunits of cAMP-dependent protein kinase (PTSS0234 and PTSS0368). In addition, other ESTs within our collection show “Twilight Zone” similarity to adenylyl cyclases and to cAMP response element-binding protein transcription factors (data not shown). This implicates the importance of cAMP signaling in diatoms, a major divergence from higher plants, which so far have not been found to contain any of these enzymes.
Many other interesting similarities can be found within the Twilight Zone. For example, similarities could be found between some of the ESTs and nitric oxide synthase, salt- and submergence-induced proteins from plants, and aquaporins. Furthermore, it was surprising that diatom ESTs could be found with similarity to all the major components of the extracellular matrix of mammalian cells: collagen, laminin, elastin, fibronectin, and tenascin (data not shown).
Codon Usage
A table showing codon usage in P. tricornutum derived from the identified diatom ESTs is shown in Table IV (see Supplementary Information at http://www.plantphysiol.org). Codon usage in P. tricornutum is not strongly biased as it is in P. yezoensis (Nikaido et al., 2000) and L. digitata (Crépineau et al., 2000), which indicates that GC content of coding sequences in P. tricornutum is not high as it is in these other two algae (see below).
Compositional Distribution of Genes
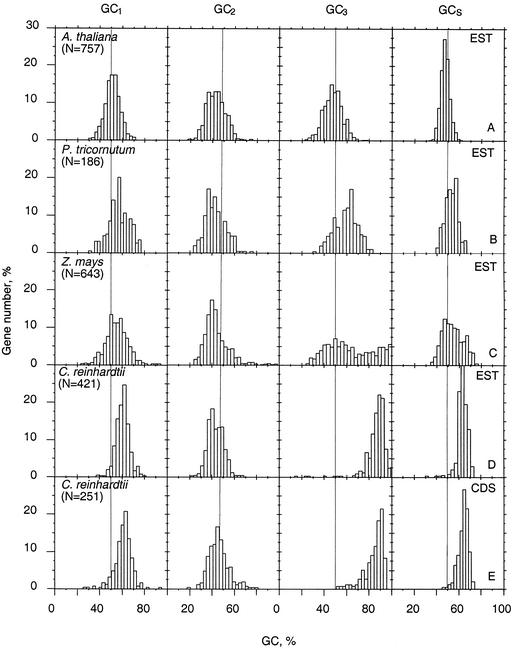
To study compositional distribution within diatom open reading frames derived from the ESTs shown in Table III (see Supplementary Information at http://www.plantphysiol.org), we compared GC composition at the three codon positions in the nonredundant P. tricornutum sequences with randomly selected ESTs from Arabidopsis, maize (Zea mays), and C. reinhardtii (see “Materials and Methods”). To examine whether ESTs can be used to assess the base composition of coding sequences (CDS) at a genome level, we also compared GC distribution within CDS and ESTs from the same organism (C. reinhardtii). The data in Figure 2 show that estimations of GC contents within the codons are qualitatively very similar, regardless of whether ESTs or CDS are used (Fig. 2, D and E). This can also be seen when comparing the data for maize and Arabidopsis ESTs with previously published data using a limited number of CDS (Carels et al., 1998), as well as with a larger data set of maize and Arabidopsis CDS (data not shown). Therefore, from these observations we conclude that EST data sets can provide useful information for genome characterization and that full-length coding sequences are not necessary to generate qualitative reliable information in P. tricornutum with respect to these other organisms.
Figure 2.
Distribution of coding sequences of Arabidopsis (A), P. tricornutum (B), maize (C), and C. reinhardtii (D and E) according to the GC1, GC2, GC3, and GCs levels. GC1, GC2, GC3, and GCs are the GC levels of first, second, and third codon positions and of the whole coding sequences, respectively. For C. reinhardtii, coding sequences derived from ESTs (D) and CDS (E) are shown. n is the size of the sequence sample.
Table V shows the average GC values for each codon position. The GC profile at the third codon position within the diatom ESTs is within the same compositional interval as that of Arabidopsis (Fig. 2, A and B). However, the whole distribution tends to be shifted by around 5% toward higher GC levels, with a slight asymmetry to the right in such a way that the peak of the EST distribution is about 10% higher at GC3 compared with Arabidopsis.
Table V.
Average GC levels and cesium chloride buoyant densities in maize, Arabidopsis, C. reinhardtii, and P. tricornutum
| Species | CDS, GCs | CDS, GC1 | CDS, GC2 | CDS, GC3 | Buoyant Density of DNA | Total DNA, GC |
|---|---|---|---|---|---|---|
| % | g cm−3 | % | ||||
| C. reinhardtii | 64.3 | 60 | 40 | 90 | 1.723a, 1.7218b | 62.1c, 63.1d |
| Maize | 60.3 | 60 | 40 | 70e | 1.7021 | 47.0 |
| P. tricornuum | 53.7 | 55 | 40 | 65 | 1.7075 | 48.5 |
| Arabidopsis | 46.4 | 50 | 40 | 45 | 1.695 | 35.7 |
GC levels of total DNA were obtained by calculation of GC level at the peak of the cesium chloride profiles. In the case of maize, we used the value reported by Carels et al. (1995).
Buoyant densities from C. reinhardtii were taken from Chiang and Sueoka (1967).
Buoyant densities from C. reinhardtii were taken from our results.
Corresponding GC levels were taken from Sager and Ishida (1963).
Corresponding GC levels were calculated from our measurements of the DNA buoyant density using Schildkraut's formula (Schildkraut et al., 1962).
The average GC3 level is inappropriate for comparison in maize because two classes of genes must be taken into consideration (Carels and Bernardi, 2000). The values previously reported are 68.6% for GC-poor genes and 89% for GC-rich genes. However, the value for GC-poor genes is biased by the small size of the sample and is more likely to be around 50% to 55% GC3 (see Fig. 3).
The overall percentage of GC of the diatom coding sequences (GCs) is 53.7%, which is 7.3% higher than was found in the random population of Arabidopsis sequences, and 10.6% and 6.6% lower than in the C. reinhardtii and maize sequences, respectively (Table V).
Average Base Composition at the Whole Genome Level
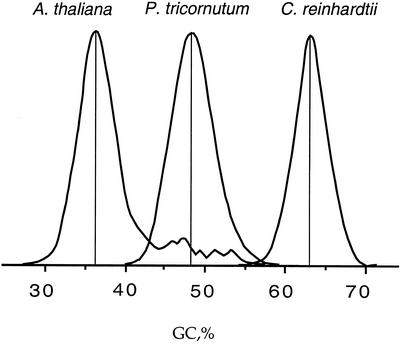
In addition to examining GC content of coding sequences, it was of interest to determine the GC content of the whole genome of P. tricornutum. The average GC content of a genome can be calculated from its buoyant density in cesium chloride (Thiery et al., 1976; Cortadas et al., 1977) using the formula of Schildkraut (Schildkraut et al., 1962). Therefore, we performed cesium chloride gradient analyses of genomic DNA from P. tricornutum and found a buoyant density of 1.7072 g cm−3. Because the genome of this diatom is hardly methylated (below 3%; Jarvis et al., 1992), we can conclude that the average GC content derived from this analysis is 48.5% (Table V; Fig. 3). The cesium chloride profile of the diatom DNA falls between the Arabidopsis and C. reinhardtii profiles (Fig. 3).
Figure 3.
Comparison of cesium chloride profiles of genomic DNA from Arabidopsis, P. tricornutum, and C. reinhardtii. Buoyant density data are expressed in GC level to facilitate the comparison. GC distributions are equally homogeneous in all three organisms, except for the tail in the Arabidopsis profile, which is derived from plastid DNA (Barakat et al., 1998).
The C. reinhardtii genome is also not significantly methylated in vegetative cells (Blamire et al., 1974; Sano et al., 1980) and, in addition, is very GC rich (62.1%; Table V; Fig. 3; Sueoka, 1960; Sager and Ishida, 1963). In spite of this difference, the P. tricornutum and C. reinhardtii CsCl profiles are similarly homogeneous and symmetrical (like Arabidopsis, but not maize [Carels et al., 1995]; Fig. 3). In the case of P. tricornutum, this was expected because of the high homogeneity in GC distribution observed in the codons of the EST-derived sequences. On average, P. tricornutum ESTs are 53.7% GC, i.e. around 5% higher than the average of the whole genome (GC = 48.5%).
Determination of Genome Size
P. tricornutum genome size was calculated by analytical ultracentrifugation of cesium chloride density gradients (Macaya et al., 1976). The area calculated under the peak of triplicate samples corresponding to three million cells was 0.015798 OD cm−1 with an sd ± 12.0%. The maxima was close to 0.3 OD. In a second trial on six samples of three million cells, we obtained a value of 0.011959 OD cm−1 with an sd ± 17.8%. When combined, these experiments give an average of 0.013462 OD cm−1 with an sd ± 20%. In two separate experiments, we verified that the surface covered by the peak corresponding to six million cells was double this value. In this case, the OD peaked at around 0.6, the expected value. The peak covered by the phage DNA reference was 0.005045 OD cm−1 with a maximal OD value of 0.2. From these results, we can calculate that three million cells of P. tricornutum correspond to 40 ng of DNA and, therefore, that one cell corresponds to an average of 0.0133 pg. Because 1 bp corresponds to approximately 1.05 × 10−9 pg, one can calculate that the genome size of P. tricornutum is about 13 Mb.
DISCUSSION
EST Analysis
Although this work represents only a pilot-scale study of diatom ESTs, it has revealed a lot of new information about diatom biology. For example, the number of nonredundant sequences that could be functionally defined (194 out of 819; 23.7%) is much lower than was observed in P. yezoensis (33.1%; Nikaido et al., 2000) and L. digitata (39% of gametophyte ESTs and 48% of sporophyte ESTs; Crépineau et al., 2000). By contrast, 62.3% of deduced amino acid sequences from the legume Lotus japonicus could be assigned a function (Asamizu et al., 2000). Although this reflects the more stringent criteria used to assign protein function (see “Materials and Methods”) compared with these previous studies, it also suggests that diatoms may contain a large number of divergent sequences.
Because of the high stringency used to identify similarities, it is highly probable that the vast majority of matches that have been assigned will be confirmed by subsequent biochemical experiments. Although this is clearly advantageous, it does also mean that other ESTs encoding proteins with similarities slightly below our threshold levels are not described. Notable examples from this study included ESTs encoding proteins with similarity to P450 monooxygenases, DNA helicases, ankyrins, the D1-processing protease, and several receptor kinases, which were only excluded from Table III (see Supplementary Information at http://www.plantphysiol.org) because the regions of similarity were slightly lower than 50 amino acids (see http://www.szn.it/plant/PhaeodactylumEST for a comprehensive list of BLASTX results).
We found similarities with all the major components of the extracellular matrix of mammalian cells: collagen, laminin, elastin, fibronectin, and tenascin. Such extracellular components do not appear to be present in higher plants. As a consequence, in addition to revealing important clues about the proteinaceous components of diatom cell walls, the identification of such sequences suggests that diatoms may have more similarities with animal cells than has been previously appreciated. This is further supported by the fact that similar numbers of the EST-encoded proteins reported in Table III (see Supplementary Information at http://www.plantphysiol.org) share similarities with metazoan and plant counterparts (see http://www.szn.it/plant/PhaeodactylumEST) and by the fact that cAMP signaling appears to have a clear role in diatoms, as it does in metazoans and fungi, but not in plants. These results are likely a reflection of the different phylogenetic histories of diatoms and higher plants, and they suggest that much of the repertoire of diatom genes derive from the ancestral heterotrophic flagellate that was the host for the secondary endosymbiosis (Van Den Hoek et al., 1997; Medlin et al., 2000). This information suggests that the placing of diatoms in eukaryotic phylogenetic trees may require some revision (Baldauf et al., 2000).
Compositional Distribution of Genes
Comparison of the genome properties of Arabidopsis, maize, and C. reinhardtii provides an interesting reference for the characterization of other photosynthetic eukaryotes (Figs. 2 and 3; Table V). These three species summarize the three basic levels of compositional transition in GC content that can be observed in chlorophytes with respect to the average GC level (36%–40%) at the root of their evolution, i.e. no compositional transition (Arabidopsis), partial transition (maize), and complete transition (C. reinhardtii).
It is interesting to note that because of the determining role of the second position in protein synthesis, GC2 is always centered on 40%, whatever the level of compositional transition. This observation, which is true for CDS- and for EST-derived sequences (compare the current study with Carels et al., 1998), demonstrates that the diatom ESTs are in the correct reading frame. GC1 is less restricted by protein-coding constraints and can follow to some extent the trend promoted by compositional bias toward higher GC values. The third codon position, being the most degenerate, absorbs the majority of compositional variation.
Genome Size and Average Base Composition at the Whole-Genome Level
To our knowledge, the genome size of P. tricornutum had been previously examined by two different methods. Darley (1968) used a spectrophotometric method and estimated a DNA content of approximately 0.12 pg of DNA per cell, which corresponds to a genome size of 120 Mb. Veldhuis et al. (1997) subsequently combined flow cytometry with DNA-binding fluorochromes and obtained a value approximately twice this amount. However, this method is likely to overestimate DNA contents due to nonspecific binding of the dyes throughout the cell. The method that we use, based upon analytical ultracentrifugation of DNA derived from a fixed number of cells (Macaya et al., 1976), generated a value around 13 Mb, which is even smaller than the previous calculations.
The discrepancies evidenced above reflect the fact that genome size determinations are difficult and prone to error. Regarding our method, the various error sources are as follows: The percentage of mortality in our cell cultures was 2%, meaning that genome size determination could be underestimated by 0.2 Mb; the cell counting process may introduce an error of up to 5%; and the measurement itself generated a value with an sd of ±20%. The chloroplast genome, which is 120 kb in the diatom Odontella sinensis (Kowallik et al., 1995), is not a complicating factor in P. tricornutum because each cell contains only one plastid. Putting all sources of error together, we can conclude that our method estimates a genome size between 7 and 19 Mb. The haploid set of yeast (Saccharomyces cerevisiae) chromosomes is 12 Mb and contains 6,500 genes, the exons of which are 1 kb on average (Rubin et al., 2000). Yeast is among the simplest of eukaryotic organisms, therefore it is unlikely that the genome of P. tricornutum is smaller because it is an obligate phototroph. However, a value between 12 and 20 Mb for P. tricornutum has been confirmed by Mark Hildebrand (personal communication) using a method based on flow cytometry, thus, this value is likely to be correct. Therefore, the higher values reported previously may reflect the limited accuracy of the analytical techniques used with respect to more modern methods.
The fact that the cesium chloride profile of P. tricornutum DNA is homogeneous (Fig. 3) reflects the high homogeneity in GC distribution observed in the codons of the EST-derived sequences and indicates that very little satellite DNA is present within the genome. This latter finding was expected because of the small genome size. Moreover, because of the similar range of GC levels found within the cesium chloride profiles of Arabidopsis, P. tricornutum, and C. reinhardtii (Fig. 3), it can be concluded that the heterogeneity of base composition is similar in each organism. However, the average GC contents of these genomes are so different that their cesium chloride profiles do not overlap. Compared with Arabidopsis, which is typical of genomes that have not changed significantly in average base composition over the last 100 million years (see Bernardi and Bernardi, 1990, for other examples), the cesium chloride profiles of P. tricornutum and C. reinhardtii indicate that their genomes are examples of “horizontal shift.” However, a complete compositional transition is only observed in C. reinhardtii.
CONCLUSIONS
The small scale study of ESTs described here has yielded the following new information about the genome of the diatom P. tricornutum: Codon usage is not strongly biased in comparison with Arabidopsis; many genes are more similar to animal rather than plant counterparts; and genome size is similar to the yeast S. cerevisiae.
These characteristics, together with its ease of culture, short generation time, and ease of genetic transformation (Apt et al., 1997; Falciatore et al., 1999) suggest that P. tricornutum would be an appropriate model species for genomic research in diatoms. The availability of a protocol for genetic transformation is particularly important because reverse genetics approaches will be essential to elucidate new protein functions in diatoms.
However, P. tricornutum is a rather atypical diatom in that it is polymorphic. It exists as three different morphotypes: oval, fusiform, and triradiate, which are only partially silicified (Lewin, 1958; Lewin et al., 1958; Borowitzka et al., 1977; Borowitzka and Volcani, 1978). Therefore, submicrometer-scale silica structures are limited, thus its use for studies of bioinorganic pattern formation may not be ideal. However, our study has revealed that the usual diatom-specific cell wall components such as frustulins are present. Furthermore, phylogenetic analysis using 18S rRNA sequences place P. tricornutum in the middle of the pennate diatom lineage (D. Vaulot, personal communication). Therefore, the differences in genome sizes between different diatoms (Veldhuis et al., 1997) are more likely to be due to different contents of noncoding “junk” DNA, as is the case in other organisms (e.g. the genomes of some cereal plants are more than 200 times bigger than that of Arabidopsis, even though the number of genes is likely to be very similar in all plant species [The Arabidopsis Genome Initiative, 2000]). In spite of its atypical properties, in our opinion, P. tricornutum appears to be the most appropriate choice for a diatom genome sequencing project, and its potential for unraveling the secrets of diatom biology can be similar to the role played by Arabidopsis in studies of plant biology, physiology, and ecology.
MATERIALS AND METHODS
Strains and Media
Phaeodactylum tricornutum Bohlin clone CCMP 632 was obtained from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton (West Boothbay Harbor, ME). Cells were grown axenically in filtered seawater enriched with nutrients as in f/2 medium (Guillard, 1975) at 20°C in a 12-h light/12-h dark photoperiod (150 μmol m−2 s−1). Light sources were cool-white fluorescent tubes (TLD 58W/84; Philips, Eindhoven, The Netherlands). Cultures were periodically tested for bacterial contamination by culturing in the dark in medium (100% [w/v] seawater plus f/2 nutrients) supplemented with 1 g L−1 peptone (Oxoid S.p.A.).
cDNA Library Preparation
The cDNA library used in this study was generated from an exponentially growing culture. Diatom cells were collected by centrifugation and were frozen in liquid nitrogen. Total RNA was prepared from the cell pellet essentially according to Verwoerd et al. (1989). Total RNA was then treated with proteinase K and RNase-free DNase according to Monstein et al. (1995).
Poly(A) mRNA was isolated from total RNA with Dynabeads (Dynal Biotech, Lake Success, NY) according to the manufacturer's recommendations. cDNA was prepared using a cDNA synthesis kit (Stratagene, La Jolla, CA) and was directionally cloned into the Uni-ZAP vector (Stratagene). The resulting library contained 1.8 × 106 independent phage and was amplified to 4.9 × 109 pfu mL−1 according to the manufacturer's recommendations.
The Uni-ZAP XR vector allows in vivo excision of the pBluescript phagemid, allowing the insert to be characterized in a plasmid-based system. The λ phage library was converted into a plasmid library by performing a mass in vivo excision by superinfecting with ExAssist helper phage. In the resulting library, the cDNAs were cloned in the pBluescript SK plasmid. Conversion into a plasmid library simplified subsequent manipulation and handling of the library.
Clones were randomly picked for template preparations with the idea of generating an overall picture of the genes expressed in the diatom cells. Clones from the oriented library were sequenced from the 5′ end to target preferentially the coding region of cDNAs and to avoid premature sequence termination resulting from long poly(A) sequences at the 3′ end of the cDNAs.
Plasmid DNA from the resulting pBluescript SK-based clones was prepared using a spin column miniprep kit. Automated cycle sequencing was performed on plasmid DNA, at the 5′ end of the cDNA insert, using the T3 inverse sequencing primer (5′-TTTAATTGGGAGTGATTTCCC-3′). ESTs were produced by single passes on an automated sequencer (ABI377; Applied Biosystems, Foster City, CA) with base caller ABI200. The quality score was between 95% and 99% accuracy per sequence, and the average percentage of error for all the sequences was 2%. Sequences were further edited manually to eliminate unwanted regions of vector, poly(A) tails, and lower quality data from the end of the sequencing run. The average length of readable sequence was 303 bp after the deletion of vector-derived sequences. The reasons for such a short sequence length are because the sequencing was done in 1999, when sequencing lengths were not as long as they are today, and because of financial constraints.
Similarity Searches
The sequences were first checked for redundancy using the program BLASTN (Altschul et al., 1990). We considered sequences to be redundant when more than 90% identity was observed over sequences more than 250 nucleotides long. The nonredundant sequences were translated into three reading frames and were then compared with the protein sequences contained within SWISS PROT, SPTREMBL+REM GENPEPT, and PIR databases within NAL3D and PDB databases using the BLASTX algorithm (Altschul et al., 1990) at the Infobiogen server (see http://www.infobiogen.fr).
We considered significant similarities to known proteins as having probability values of less than 10−4, a percentage of identity of more than 40% and a length of the region of similarity of more than 50 amino acids. These values were based on the following considerations.
Probability
It is difficult to come up with a measure of “biological significance.” In general, one tries to infer biological significance from statistical significance, and most often, an E or P value of less than 0.05 is considered to be statistically significant. However, because the current version of BLAST sometimes underestimates E values (S.F. Altschul, personal communication), we decided to impose smaller E values before claiming statistical significance. As suggested by Azam et al. (1996) and P. Dessen (personal communication), we decided to consider E values less than 0.0001 as being of biological significance.
Percentage of Identity
The percentage of identity is the number of amino acids that are identical within the region of similarity with respect to the length of this region. To reduce ambiguous results, we defined a protein to be similar to another when a percentage of identity higher than 40% was found between them. This is just above the Twilight Zone defined by Doolittle (1987).
Region of Similarity
A length of 50 amino acids was chosen arbitrarily as the cut-off length for a region of similarity. If more than one EST showed similarity to the same gene, we analyzed the corresponding sequences by clustalW alignment (Thompson et al., 1994). When the number of nucleotides within this region exceeded 100 bases but the differences in nucleotide sequences were greater than 10%, they were considered to be different isoforms of the same protein, e.g. clones PTSS0141 and PTSS0701. When the sequences overlapped perfectly, only one EST was included in Table III (see Supplementary Information at http://www.plantphysiol.org) and the overlapping sequences were used where possible to reconstruct a longer sequence.
Automated analyses were performed using software implemented at the Stazione Zoologica based on Perl programming language (Wall et al., 2000). GC level was calculated in the three codon positions using ANALSEQ (Gautier and Jacobzone, 1989). For comparison, we analyzed coding sequences derived from random samples of ESTs from Arabidopsis (n = 1,000), maize (Zea mays; n = 2,000), and Chlamydomonas reinhardtii (n = 5,000) retrieved from GenBank using the ACNUC retrieval protocol (GenBank release 117 [April 2000]). The maize sample was double that of Arabidopsis because of the two classes of genes in this species (Carels and Bernardi, 2000). For C. reinhardtii, 5,000 random ESTs were necessary because of the high number of non-unique sequences in the ESTs from this alga.
Cesium Chloride Gradient Analysis of Genomic DNA
Genomic DNA from P. tricornutum and C. reinhardtii were analyzed by sedimentation in cesium chloride as previously described (Thiery et al., 1976; Cortadas et al., 1977). The formula of Schildkraut et al. (1962), ρ = (GC × 0.098)/100 + 1.66, was used for conversion of buoyant densities (g cm−3) into GC contents (percentage). Original cesium chloride profiles from maize (Carels et al., 1995) and Arabidopsis (Barakat et al., 1998) were used without modification. For maize, the relationship between buoyant density and GC level [GC = (1102 × ρ) − 1829.5] was taken from Carels et al. (1995).
Determination of Genome Size
Analysis of genome size was performed on log-phase cultures that had passed through at least five successive rounds of growth in f/2 medium (Guillard, 1975) at 20°C and in a 12-h light/12-h dark photoperiod (150 μmol m−2 s−1). Three and six million cells were collected by centrifugation. The cell pellets were then resuspended in 400 μL of distilled water and frozen in liquid nitrogen. DNA extraction and cesium chloride density gradient analytical ultracentrifugation were performed as described in Macaya et al. (1976).
In two separate experiments, we measured the surface area under the DNA profile (above the base line) after analytical centrifugation of samples corresponding to three and six million actively growing cells. The cesium chloride profiles produced by the optical system of the XL-A analytical ultracentrifuge (Beckman Coulter, Fullerton, CA) were measured by integrating the optical densities (OD) obtained for each 10-μm distance, which are listed in the ASCII file produced by the instrument. The unit of surface is expressed in OD cm−1. The quantity of DNA was determined by comparison with a phage 2c DNA buoyant density reference. The phage was loaded onto the gradient to a final quantity of 15 ng. Following the determination of DNA quantity per cell, genome size was calculated on the basis that 1 bp is equivalent to 660 D.
On the same cell cultures used for DNA extraction, the percentage of mortality was quantified to reduce errors in genome size determination. Viability assays were performed in triplicate on different culture dilutions (1 × 105 and 5 × 105 cells mL−1) labeled with 20 μm Sytox green (Molecular Probes, Eugene, OR). Percentage of mortality was measured using a flow cytometer (FACSCalibur; BD Biosciences, Franklin Lakes, NJ; R. Casotti, unpublished data).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Mark Hildebrand and Daniel Vaulot for sharing unpublished information. We thank Arthur Grossman for his generous gift of C. reinhardtii genomic DNA, Margherita Branno for help with library preparation, Raffaella Casotti for help with cell viability assays, Alessandro Manfredonia for technical assistance, Alfredo Profeta for help with URL preparation, and Toplab (Martinsried, Germany) for performing the DNA sequencing under contract.
Footnotes
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010713.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Apt KE, Kroth-Pancic PG, Grossman AR. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol Gen Genet. 1997;252:572–579. doi: 10.1007/BF02172403. [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S. Generation of 7,37 non-redundant expressed sequence tags from a legume, Lotus japonicus. DNA Res. 2000;7:127–130. doi: 10.1093/dnares/7.2.127. [DOI] [PubMed] [Google Scholar]
- Azam A, Paul J, Sehgal D, Prasad J, Bhattacharya S, Bhattacharya A. Identification of novel genes from Entamoeba histolytica by expressed sequence tag analysis. Gene. 1996;181:111–116. doi: 10.1016/s0378-1119(96)00484-2. [DOI] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- Barakat A, Matassi G, Bernardi G. Distribution of genes in the genome of Arabidopsis thaliana and its implications for the genome organization of plants. Proc Natl Acad Sci USA. 1998;95:10044–10049. doi: 10.1073/pnas.95.17.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G, Bernardi G. Compositional transitions in the nuclear genomes of cold-blooded vertebrates. J Mol Evol. 1990;31:282–293. doi: 10.1007/BF02101123. [DOI] [PubMed] [Google Scholar]
- Bhaya D, Grossman AR. Characterization of gene clusters encoding the fucoxanthin chlorophyll proteins of the diatom Phaeodactylum tricornutum. Nucleic Acids Res. 1993;21:4458–4466. doi: 10.1093/nar/21.19.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blamire J, Flechtner VR, Sager R. Regulation of nuclear DNA replication by the chloroplast in Chlamydomonas. Proc Natl Acad Sci USA. 1974;71:2867–2871. doi: 10.1073/pnas.71.7.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowitzka MA, Chiappino ML, Volcani BE. Ultrastructure of a chain-forming diatom Phaeodactylum tricornutum. J Phycol. 1977;13:162–170. [Google Scholar]
- Borowitzka MA, Volcani BE. The polymorphic diatom Phaeodactylum tricornutum: ultrastructure of its morphotypes. J Phycol. 1978;14:10–21. [Google Scholar]
- Carels N, Barakat A, Bernardi G. The gene distribution of the maize genome. Proc Natl Acad Sci USA. 1995;92:11057–11060. doi: 10.1073/pnas.92.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carels N, Bernardi G. Two classes of genes in plants. Genetics. 2000;154:1819–1825. doi: 10.1093/genetics/154.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carels N, Hatey P, Jabbari K, Bernardi G. Compositional distribution of homologous coding sequences from plants. J Mol Evol. 1998;46:45–53. doi: 10.1007/pl00006282. [DOI] [PubMed] [Google Scholar]
- Chiang K-S, Sueoka N. Replication of chloroplast DNA in Chlamydomonas reinhardtii during vegetative life cycle: its mode and regulation. Proc Natl Acad Sci USA. 1967;57:1506–1513. doi: 10.1073/pnas.57.5.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortadas J, Macaya G, Bernardi G. An analysis of the bovine genome by density gradient centrifugation: fractionation in Cs2SO4/3,6 bis (acetato-mercurimethyl) dioxane density gradient. Eur J Biochem. 1977;76:13–19. doi: 10.1111/j.1432-1033.1977.tb11565.x. [DOI] [PubMed] [Google Scholar]
- Crépineau F, Roscoe T, Kaas R, Kloareg B, Boyen C. Characterization of complementary DNAs from the expressed sequence tag analysis of life cycle stages of Laminaria digitata (Phaeophyceae) Plant Mol Biol. 2000;43:503–513. doi: 10.1023/a:1006489920808. [DOI] [PubMed] [Google Scholar]
- Darley WM. Deoxyribonucleic acid content of the three cell types of Phaeodactylum tricornutum Bohlin. J Phycol. 1968;4:219–220. doi: 10.1111/j.1529-8817.1968.tb04718.x. [DOI] [PubMed] [Google Scholar]
- Delwiche CF, Palmer JD. The origin of plastids and their spread via secondary symbiosis. In: Bhattacharya D, editor. Origins of Algae and Their Plastids. Springer-Verlag, Vienna. 1997. pp. 53–86. [Google Scholar]
- Doolittle RF. Of Urfs and Orfs: A Primer on How to Analyze Derived Amino Acid Sequences. Mill Valley, NY: University Science Books; 1987. [Google Scholar]
- Falciatore A, Casotti R, Leblanc C, Abrescia C, Bowler C. Transformation of nonselectable reporter genes in marine diatoms. Mar Biotechnol. 1999;1:239–251. doi: 10.1007/pl00011773. [DOI] [PubMed] [Google Scholar]
- Falciatore A, Ribera D'Alcalà M, Croot P, Bowler C. Perception of environmental signals by a marine diatom. Science. 2000;288:2363–2366. doi: 10.1126/science.288.5475.2363. [DOI] [PubMed] [Google Scholar]
- Falkowski PG, LaRoche J. Acclimation to spectral irradiance in algae. J Phycol. 1991;27:8–14. [Google Scholar]
- Gautier C, Jacobzone M. Publication interne, UMR CNRS 5558 Biometrie, Genetique et Biologie des Populations. Lyon, France: Universite Claude Bernard; 1989. [Google Scholar]
- Gibbs SP. The chloroplasts of some algal groups may have evolved from endosymbiotic eukaryotic algae. Ann NY Acad Sci. 1981;361:193–208. doi: 10.1111/j.1749-6632.1981.tb46519.x. [DOI] [PubMed] [Google Scholar]
- Grossman AR, Manodori A, Snyder D. Light-harvesting proteins of diatoms: their relationship to the chlorophyll a/b binding proteins of higher plants and their mode of transport into plastids. Mol Gen Genet. 1990;224:91–100. doi: 10.1007/BF00259455. [DOI] [PubMed] [Google Scholar]
- Guillard RRL. Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chaney MH, editors. Culture of Marine Invertebrate Animals. New York: Plenum Press; 1975. pp. 29–60. [Google Scholar]
- Hildebrand M, Volcani BE, Gassmann W, Schroeder JL. A gene family of silicon transporters. Nature. 1997;385:688–689. doi: 10.1038/385688b0. [DOI] [PubMed] [Google Scholar]
- Höfte H, Desprez T, Amselm J, Chiapello H, Caboche M, Moisan A, Jourion MF, Charpenteau JL, Berthomieu P, Guerrier D et al. An inventory of 1152 expressed sequence tags obtained by partial sequencing of cDNA clones from Arabidopsis thaliana. Plant J. 1993;4:1051–1061. doi: 10.1046/j.1365-313x.1993.04061051.x. [DOI] [PubMed] [Google Scholar]
- Jansson J. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999;4:236–240. doi: 10.1016/s1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- Jarvis EE, Dunahay TG, Brown LM. DNA nucleoside composition and methylation in several species of microalgae. J Phycol. 1992;28:356–362. [Google Scholar]
- Kowallik KV, Stoebe B, Schaffran I, Kroth-Pancic P, Freier U. The chloroplast genome of a chlorophyll a+c-containing alga, Odontella sinensis. Plant Mol Biol Rep. 1995;13:336–342. [Google Scholar]
- Kröger N, Bergsdorf C, Sumper M. A new calcium binding glycoprotein family constitutes a major diatom cell wall component. EMBO J. 1994;13:4676–4683. doi: 10.1002/j.1460-2075.1994.tb06791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin JC. The taxonomic position of Phaeodactylum tricornutum. J Gen Microbiol. 1958;18:427–432. doi: 10.1099/00221287-18-2-427. [DOI] [PubMed] [Google Scholar]
- Lewin JC, Lewin RA, Philpott DE. Observations on Phaeodactylum tricornutum. J Gen Microbiol. 1958;18:418–426. doi: 10.1099/00221287-18-2-418. [DOI] [PubMed] [Google Scholar]
- Macaya G, Thiery J-P, Bernardi G. An approach to the organization of eukaryotic genomes at a macromolecular level. J Mol Biol. 1976;108:237–254. doi: 10.1016/s0022-2836(76)80105-2. [DOI] [PubMed] [Google Scholar]
- Mann S, Ozin GA. Synthesis of inorganic materials with complex form. Nature. 1996;382:313–318. [Google Scholar]
- Medlin LK, Kooistra WC, Schmid A-MM. A review of the evolution of the diatoms: a total approach using molecules, morphology and geology. In: Witkowski A, Sieminska J, editors. The Origin and Early Evolution of the Diatoms: Fossil, Molecular and Biogeographical Approaches. W. Szafer Institute of Botany, Polish Academy of Sciences, Cracow, Poland. 2000. pp. 13–35. [Google Scholar]
- Monstein HJ, Nylander AG, Chen D. RNA extraction from gastrointestinal tract and pancreas by a modified Chomczynski and Sacchi method. BioTechniques. 1995;19:340–343. [PubMed] [Google Scholar]
- Nikaido I, Asamizu E, Nakajima M, Nakamura Y, Saga N, Tabata S. Generation of 10,154 expressed sequence tags from a leafy gametophyte of a marine red alga, Porphyra yezoensis. DNA Res. 2000;7:223–227. doi: 10.1093/dnares/7.3.223. [DOI] [PubMed] [Google Scholar]
- Reinfelder JR, Kraepiel AML, Morel FMM. Unicellular C4 photosynthesis in a marine diatom. Nature. 2000;407:996–999. doi: 10.1038/35039612. [DOI] [PubMed] [Google Scholar]
- Round FE, Crawford RM, Mann DG. The Diatoms: Biology and Morphology of the Genera. London: Cambridge University Press; 1990. [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R, Ishida MR. Chloroplast DNA in Chlamydomonas. Proc Natl Acad Sci USA. 1963;50:725–730. doi: 10.1073/pnas.50.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Royer H-D, Sager R. Identification of 5-methylcytosine in DNA fragments immobilized on nitrocellulose paper. Proc Natl Acad Sci USA. 1980;77:3581–3585. doi: 10.1073/pnas.77.6.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala S, Bowler C. Molecular insights into the novel aspects of diatom biology. Cell Mol Life Sci. 2001;58:1666–1673. doi: 10.1007/PL00000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildkraut CL, Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J Mol Biol. 1962;4:430–443. doi: 10.1016/s0022-2836(62)80100-4. [DOI] [PubMed] [Google Scholar]
- Sueoka N. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1960;46:83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Thiery J-P, Macaya G, Bernardi G. An analysis of eukaryotic genomes by density gradient centrifugation. J Mol Biol. 1976;108:219–235. doi: 10.1016/s0022-2836(76)80104-0. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson JS. Improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific Gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tréguer P, Nelson DM, Van Bennekom AJ, DeMaster DJ, Leynaert A, Quéguiner B. The silica balance in the world ocean: a reestimate. Science. 1995;268:375–379. doi: 10.1126/science.268.5209.375. [DOI] [PubMed] [Google Scholar]
- Van Den Hoek C, Mann DG, Johns HM. Algae: An Introduction to Phycology. London: Cambridge University Press; 1997. [Google Scholar]
- Veldhuis MJW, Cucci TL, Sieracki ME. Cellular DNA content of marine phytoplankton using two new fluorochromes: taxonomic and ecological implications. J Phycol. 1997;33:527–541. [Google Scholar]
- Verwoerd TC, Decker BM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall L, Christiansen T, Orwant J. Programming Perl. Cambridge, UK: O'Reilly and Associates; 2000. [Google Scholar]
- Werner D. Silicate Metabolism. Los Angeles: Blackwell Scientific Publications; 1977. [Google Scholar]
- Zurzolo C, Bowler C. Exploring bioinorganic pattern formation in diatoms. Plant Physiol. 2001;127:1339–1345. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.