Abstract
A mannose (Man)-binding lectin has been isolated and characterized from the thallus of the liverwort Marchantia polymorpha. N-terminal sequencing indicated that the M. polymorpha agglutinin (Marpola) shares sequence similarity with the superfamily of monocot Man-binding lectins. Searches in the databases yielded expressed sequence tags encoding Marpola. Sequence analysis, molecular modeling, and docking experiments revealed striking structural similarities between Marpola and the monocot Man-binding lectins. Activity and specificity studies further indicated that Marpola is a much stronger agglutinin than the Galanthus nivalis agglutinin and exhibits a preference for methylated Man and glucose, which is unprecedented within the family of monocot Man-binding lectins. The discovery of Marpola allows us, for the first time, to corroborate the evolutionary relationship between a lectin from a lower plant and a well-established lectin family from flowering plants. In addition, the identification of Marpola sheds a new light on the molecular evolution of the superfamily of monocot Man-binding lectins. Beside evolutionary considerations, the occurrence of a G. nivalis agglutinin homolog in a lower plant necessitates the rethinking of the physiological role of the whole family of monocot Man-binding lectins.
Carbohydrate-binding proteins, also called lectins or agglutinins, are widespread among flowering plants. Biochemical, molecular, and structural studies revealed that virtually all known plant lectins can be classified in seven families of structurally and evolutionary related proteins (Van Damme et al., 1998). Resolution of the three-dimensional structure of the sugar-binding sites from the amaranthins, chitin-binding lectins comprising hevein domains, jacalin-related lectins, legume lectins, monocot Man-binding lectins, and type-2 ribosome-inactivating proteins, demonstrated that each of these families possesses its own typical sugar-binding motif. Moreover, because one can reasonably assume that the Cucurbitaceae phloem lectins also have their own typical three-dimensional structure, it can be concluded that plants developed at least seven distinct structural motifs that are capable of recognizing and binding specific mono- or oligosaccharides.
At present, little is known about the possible origin of the carbohydrate-binding domains of lectins from modern flowering plants. Homologs of plant lectin domains have been identified only for the type-2 ribosome-inactivating proteins and the monocot Man-binding lectins. The β-trefoil structure of the individual domains in the B-chain of type-2 ribosome-inactivating proteins was identified in the Streptomyces olivaceoviridis β-xylanase (Fujimoto et al., 2000), in the lectin of the fungus Rhizoctonia solani (Candy et al., 2001), in murine Man receptor (Liu et al., 2000), in human interleukins-1β and -1α (Finzel et al., 1989; Graves et al., 1990), and in human fibroblast growth factors (Zhu et al., 1991). In accordance with this, the so-called ricin domain most probably arose in prokaryotes and evolved further in a variety of eukaryotic organisms. A homolog of the typical domain of the monocot Man-binding lectins has been identified in comitin, a chimeric Man-specific lectin with actin-binding properties found in Dictyostelium discoideum and in human cells (Jung et al., 1996; Barre et al., 1999). However, at present, the evolutionary link between comitin and the monocot Man-binding lectins is still unclear.
A major reason for the poor insight in the origin and evolution of modern plant lectins is the lack of information about the occurrence and identity of lectins outside flowering plants. There have been a few reports on lectins in gymnosperms and cycads, but in the absence of sequence information, no link can be made with the lectins from flowering plants. The same holds true for a lectin that was isolated from the liverwort Marchantia polymorpha (Adam and Becker, 1993). Because no N-terminal or internal amino acid sequences were determined, it remains to be demonstrated whether this monomeric 16.1-kD lectin with a complex carbohydrate-binding specificity is related to any of the lectin families found in flowering plants.
This paper reports the isolation and characterization of a Man-specific lectin from the thalli of liverwort. Molecular modeling of the liverwort agglutinin (called Marpola) using the deduced amino acid sequence of expressed sequence tags (ESTs) deposited in the databases demonstrated that the liverwort lectin is structurally and evolutionary closely related to the superfamily of monocot Man-binding lectins. To our knowledge, our results demonstrate for the first time the occurrence of a homolog of lectins from flowering plants in a lower plant, and they provide evidence that at least some modern plant lectins evolved from ancestors present in lower plants.
RESULTS
Isolation and Characterization of a Novel Man-Specific Lectin from Thalli of Liverwort
A screening of liverwort for the presence of lectin revealed that thalli from some locations exhibit a reasonably high agglutination activity. Hapten inhibition assays with crude extracts demonstrated that this agglutination activity was completely inhibited by Man/methyl α-d-mannopyranoside, but was insensitive to all other sugars. Therefore, the novel liverwort agglutinin, Marpola, was isolated by affinity chromatography on immobilized Man and was characterized in some detail.
SDS-PAGE demonstrated that Marpola consists of noncovalently linked subunits of approximately 12 kD. Mass spectrometry yielded a more accurate value of 12,841 D. No covalently bound carbohydrate could be detected in purified Marpola, suggesting that the lectin is not glycosylated. Marpola eluted with an apparent molecular mass of approximately 25 kD upon gel filtration, indicating that the native lectin is a homodimer. N-Terminal sequencing yielded the sequence FSNVLLQGSTMFSEQYLAQGPYQFKMQED. A BLAST search revealed that this sequence shares high similarity with the N terminus of the lectins from garlic (Allium sativum; Van Damme et al., 1992) and snowdrop (Galanthus nivalis; Van Damme et al., 1987), suggesting that Marpola is related to the so-called monocot Man-binding lectins.
Agglutination Activity and Carbohydrate-Binding Specificity of Marpola
Marpola readily agglutinates rabbit erythrocytes but is inactive toward human red blood cells, irrespective of their blood group. The minimal concentration required to agglutinate trypsin-treated rabbit erythrocytes was 0.1 μg mL−1. In the same test, the specific agglutination activity of G. nivalis agglutinin (GNA) was 0.5 μg mL−1, indicating that Marpola is a strong agglutinin.
In a first approach to determine the overall carbohydrate specificity of Marpola, the inhibitory effect of a series of simple sugars was tested in hapten inhibition assays of the agglutination of rabbit erythrocytes. As shown in Table I, Marpola was exclusively inhibited by Man and methyl α-d-mannopyranoside, the inhibitory concentration required to cause 50% inhibition (IC50) being 25 and 0.75 mm, respectively. None of the other sugars tested was inhibitory. Besides Man/methyl α-d-mannopyranoside, Marpola was also inhibited by some animal glycoproteins. Asialofetuin was the most potent inhibitor, followed by thyroglobulin (Table I). Though the results shown in Table I are only semiquantitative, they demonstrate that Marpola exhibits an exclusive specificity toward Man and, in this respect, closely resembles GNA and most other monocot Man-binding lectins.
Table I.
Inhibition of the agglutination activitya of Marpola by sugars and glycoproteins
| IC50b | IC50b | |
|---|---|---|
| mm | μg mL−1 | |
| Sugarc | ||
| d-Man | 25 | |
| Methyl α-d-mannopyranoside | 0.75 | |
| Glycoprotein | ||
| Asialofetuin | 50 | |
| Thyroglobulin | 100 | |
| Fetuin | 200 | |
| Mucin | 400 | |
| Asialomucin | >1,000 | |
| Ovomucoid | >1,000 |
Tested with trypsin-treated rabbit erythrocytes at a lectin concentration of 1 μg mL−1.
Concentration required to inhibit 50% of the agglutinating activity.
Ara, cellobiose, 2-deoxyglucose, Fru, d-Fuc, Gal, GalNAc, Glc, GlcNAc, lactose, maltose, melibiose, methylgalactopyranoside, methylglucopyranoside, N-acetylgalactosamine, Rib, l-sorbose, sorbitol, Suc, trehalose, and Xyl (all of the d-configuration except l-Fuc and l-sorbose) were not inhibitory at concentrations <100 mm.
To refine the results obtained with the hapten inhibition assays, the reaction of Marpola with arcelin-1, a plant glycoprotein carrying exclusively high-Man type N-glycans, was analyzed in detail by surface plasmon resonance. Marpola readily interacted with immobilized arcelin-1 (150 resonance units [RU]). However, it should be mentioned that under the same experimental conditions, GNA interacted five to six times more strongly with arcelin-1 (850 RU).
The interaction of Marpola with arcelin-1 was only weakly inhibited by Man, but was very sensitive to methyl α-d-mannopyranoside (Meα-Man) and pNO2-phenyl-α-Man (Table II). DiMan(α1, 4) was virtually inactive, whereas diMan(α1, 2), diMan(α1, 3), and diMan(α1, 6) were approximately twice as active as Man. TriManα1, 3/1, 6 (31% inhibition) and pentaManα1, 3/1, 6 (54% inhibition) yielded a stronger inhibition than any of the dimannosides, which indicates that the Man-binding site of Marpola preferentially accommodates the trimannosidic core of the N-glycans of arcelin-1. Marpola was not inhibited by Gal and Meα-Gal. Glc was also virtually inactive (3% inhibition), but Meα-Glc caused an inhibition of 35% (Table II).
Table II.
Inhibition by simple sugars, sugar derivatives, and oligosaccharides of the interaction of Marpola with arcelin-1
| Inhibitory Sugar | Inhibition of the Interaction between Arcelin-1
and
|
|||
|---|---|---|---|---|
| Marpola
|
GNA
|
|||
| Percentage of inhibitiona | Relative inhibitory potency | Percentage of inhibitiona | Relative inhibitory potency | |
| Man | 8 | 1 | 5 | 1 |
| MeαMan | 65 | 8.12 | 12 | 2.4 |
| pNO2FαMan | 58 | 7.25 | ndb | nd |
| pNO2FβMan | 0 | 0 | nd | nd |
| Gal | 0 | 0 | 1 | 0.2 |
| Methyl α-d-galactopyranoside (MeαGal) | 0 | 0 | 1 | 0.2 |
| p-Nitrophenyl-α-galactopyranoside (pNO2FαGal) | 0 | 0 | nd | nd |
| p-Nitrophenyl-β-galactopyranoside (pNO2FβGal) | 1 | 0.12 | nd | nd |
| Glc | 3 | 0.37 | 2 | 0.4 |
| Methyl α-d-glucopyranoside (MeαGlc) | 35 | 4.37 | 3 | 0.6 |
| diManα1,2 | 19 | 2.37 | 11 | 2.2 |
| diManα1,3 | 21 | 2.62 | 39 | 7.8 |
| diManα1,4 | 1 | 0.12 | 6 | 1.2 |
| diManα1,6 | 16 | 2 | 13 | 2.6 |
| triManα1,3/1,6 | 31 | 3.87 | 60 | 12 |
| pentaManα1,3/1,6 | 54 | 6.75 | 97 | 19.4 |
All sugars were used at a concentration of 10 mm except pNO2FαMan (7 mm) and pNO2-phenyl-β-mannopyranoside (pNO2Fβ-Man, 2.5 mm).
Expressed as the percentage of lectin eluted from the arcelin-1-bound surface (as compared with measurements done in the absence of sugar). Results are the means of duplicate experiments.
nd, Not determined.
Parallel experiments with GNA confirmed the exclusive specificity of GNA toward Man and oligomannosides and indicated that Marpola and GNA respond similarly to Man and the different di- and oligomannosides (Table II). However, the relative inhibitory potency of Meα-Man (over Man) is much higher for Marpola than for GNA.
The data summarized in Table II demonstrate that the Man-binding specificity of Marpola is like that of GNA directed against the trimannosidic core of the N-glycans of arcelin-1. In addition, the strong inhibitory activity of the sugar derivatives Meα-Man (65% inhibition), pNO2Fα-Man (58% inhibition), and Meα-Glc (35% inhibition) suggests that additional hydrophobic interaction(s) in the vicinity of the carbohydrate-binding sites enhance(s) the sugar-binding activity of the lectin.
Marpola Is Structurally and Evolutionarily Related to the Monocot Man-Binding Lectins
Identification of ESTs Encoding Marpola
The predicted relationship between Marpola and the monocot Man-binding lectins could be elaborated in detail by analyzing the sequences of ESTs. A BLAST search yielded two ESTs from immature female sexual organs of liverwort in which an exact match with the N-terminal sequence of Marpola was found at the N terminus of the putative proteins (AU081743 and C96467 encode proteins referred to as Marpola-1 and Marpola-2, respectively; Nagai et al., 1999; Nishiyama et al., 2000). Another EST (C95861) encoding a protein (referred to as Marpola-3) that matched only partially the N terminus of Marpola was also identified. The primary translation products of Marpola-1 and Marpola-2 differ by only one amino acid residue (Ser-72 of Marpola-1 is replaced by Ala-72 in Marpola-2) and hence can be considered virtually identical isoforms. In contrast, the precursor of Marpola-3 shares only 74% sequence identity with that of Marpola-1 and Marpola-2 at the amino acid level, indicating that it is a different isoform.
Analysis of the deduced amino acid sequences encoded by the EST sequences indicated that AU081743 and C96467, as well as C95861, encode a primary translation product of 171 amino acid residues (Fig. 1). AU081743 and C96467 encode polypeptides of 18,501 and 18,485 D, respectively. According to the rules for protein processing of von Heijne (1986), a signal peptide can be cleaved between amino acids 26 and 27, resulting in a polypeptide of 145 amino acids (15,858 and 15,842 D for Marpola-1 and -2, respectively). The length of the predicted signal peptide is in agreement with the fact that the N-terminal sequence of the purified lectin can be aligned starting from residue 27. Because the molecular mass of mature lectin is only 12,841 D, the polypeptide of 145 residues—similar to GNA and other monocot Man-binding lectins—apparently undergoes a post-translational removal of a C-terminal propeptide. According to the differences in Mr between the 145 amino acid polypeptide and the mature lectin, a propeptide of 30 residues is cleaved, resulting in a polypeptide of 12,826 D.
Figure 1.
Comparison of the amino acid sequences of GNA and liverwort lectins Marpola-1 (EST AU081743), Marpola-2 (EST C96467), and Marpola-3 (EST C95861). Deletions (gaps) have been introduced to maximize the homology. Identical residues are boxed.
There is an apparent difference of 16 D between the calculated mass of Marpola-1 and the mass determined by mass spectrometry (15,858 versus 12,841 D, respectively). This difference is possibly due to the oxidation of a Met to the sulfoxide derivative. Sulfenic acid formation from a free Cys residue is unlikely because the free Cys residue of (unreduced) Marpola is readily alkylated [as could be checked by labeling with N-(1-pyrenyl) maleimide (results not shown)]. It is also possible that the observed difference is simply due to an error in the sequence of the deposited EST sequence.
C95861 encodes a polypeptide of 18,508 D with a putative signal peptide cleavage site between residues 26 and 27. Cleavage at this site will result in a peptide of 15,834 D, which most probably will also undergo C-terminal processing.
Molecular Modeling of Marpola
The deduced amino acid sequences of the mature polypeptides encoding Marpola-1 and Marpola-2 share 30% sequence identity and 42% sequence similarity, respectively, with GNA, whereas Marpola-3 shares 32% sequence identity and 43% sequence similarity, respectively, with the snowdrop lectin (Fig. 1). Because Marpola-1 and Marpola-2 are virtually identical, only one of the two sequences (Marpola-1) will be discussed here. In this section, the numbering of the amino acid refers to their position in the mature lectin subunits unless stated otherwise.
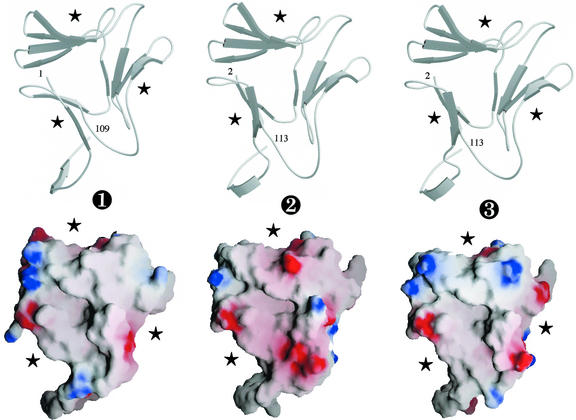
A detailed comparison of the sequences of GNA, Marpola-1, and Marpola-3 indicated that the liverwort lectins have the same structural organization in three subdomains as that found in the GNA monomer (Hester et al., 1995; Fig. 2). Hydrophobic cluster analysis (HCA) confirmed the structural similarity between GNA and Marpola-1 and Marpola-3 (results not shown). All 12 strands of β-sheet in the polypeptide chain of GNA are readily recognized and delineated along the polypeptide chains of both liverwort lectins. The structural similarity between GNA and Marpola is also reflected by the three-dimensional models built from the x-ray coordinates of the GNA monomer. Marpola-1 and Marpola-3 consist of three bundles of antiparallel β-sheet arranged in a β-prism structure (Fig. 3). The three bundles of β-sheet, which correspond to three distinct subdomains I, II, and III, are perpendicularly oriented to the axis of the monomer. They are connected by loops to form a 12-stranded β-barrel harboring three monosaccharide-binding sites located in the clefts formed by the three bundles of β-sheet. The two Cys residues (Cys-30 and Cys-53), which form a disulfide bridge in GNA, are conserved in all three isoforms of Marpola, suggesting that they are like GNA stabilized by an intrachain disulfide bridge. The physicochemical properties of Marpola-1 and Marpola-2 (net charge = −3, calculated pI = 5.15) resemble those of GNA (net charge = −4, calculated pI = 4.87). Marpola-3 (net charge = −1, calculated pI = 6.18) is a less acidic protein than GNA and Marpola-1. Marpola-1 is more electronegatively charged at its surface than Marpola-3 and GNA (Fig. 3).
Figure 2.
Alignment of the amino acid sequence stretches corresponding to subdomains I, II, and III of GNA with those of Marpola-1 (I′, II′, and III′) and Marpola-3 (I′′, II′′, and III′′). The conserved amino acid residues forming the Man-binding sites of GNA and the corresponding residues of the liverwort lectins are in bold. The conserved Val residue participating to the Man-binding sites of GNA is in bold and is underlined.
Figure 3.
Top, Ribbon diagrams of the modeled three-dimensional models of Marpola-1 (➋) and Marpola-3 (➌) compared with the three-dimensional structure of the GNA monomer (➊). The strands of β-sheet (indicated by gray arrows) associate in three four-stranded bundles to form the β-prism fold. Asterisks indicate the location of the functional carbohydrate-binding sites. Figures are rendered with MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merritt and Bacon, 1997). Bottom, Representation of the electrostatic surface potentials of GNA (➊), Marpola-1 (➋), and Marpola-3 (➌) by GRASP (Nicholls et al., 1991). The negative potential was colored red and displayed at −5 kT level, and the positive potential was colored blue and displayed at +5 kT level (1 kT = 0.6 kcal). Neutral surfaces are in white. Stars indicate the location of the functional Man-binding sites.
Molecular Modeling of the Sugar-Binding Sites of Marpola
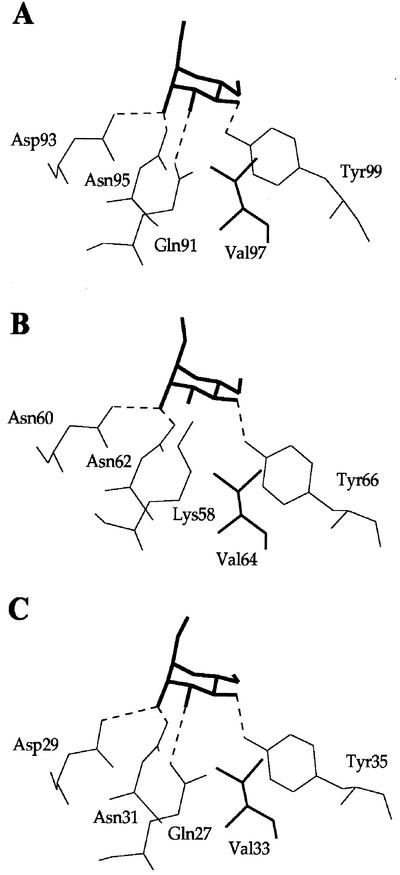
All the amino acid residues that form the three Man-binding sites of subdomains I (Gln-89, Asp-91, Asn-93, and Tyr-97), II (Gln-57, Asp-59, Asn-61, and Tyr-65), and III (Gln-26, Asp-28, Asn-30, and Tyr-34) of GNA (Hester et al., 1995) are strictly conserved in Marpola-3. In addition, the conserved Val residues (Val-95, Val-63, and Val-32) that participate in the binding of Man via a hydrophobic interaction are also conserved in Marpola-3 (Fig. 2). Docking experiments performed with Man suggested that all the Man-binding sites of Marpola-3 are fully functional because they can accommodate a Man unit through a network of four hydrogen bonds similar to that determined in the GNA-methyl α-d-mannoside complex (Hester et al., 1995). In a similar manner, all residues forming the Man-binding sites of subdomains I and III of Marpola-1 and Marpola-2 are conserved, and are, as suggested from docking experiments, fully functional because they can accommodate Man (Fig. 4, A and C). However, the Man-binding site located in subdomain II of Marpola-1 and Marpola-2 is suspected to be inactive due to the replacement of Gln-57 (of GNA), which is essential for the proper binding of Man by Lys-58. This presumed lack of activity of the Man-binding site of subdomain II of Marpola-1 and Marpola-2 is not unique. Previous studies have demonstrated that some monocot Man-binding lectins possess only one or two instead of three active Man-binding sites per domain (Barre et al., 1996).
Figure 4.
Docking of Man (bold) into the sites of subdomains I (A), II (B), and III (C) of Marpola-1. A conserved Val residue (thick line), responsible for a hydrophobic interaction with the pyranose ring of Man, occurs in all the sites. The replacement of Gln-57 of GNA by Lys-58 in the site of subdomain II creates a steric clash (1.2 Å) with O3 of Man that prevents the binding of the sugar into the site.
To explain the strongly enhanced affinity of Marpola for methyl and p-nitrophenyl groups, the overall surface hydrophobicity and the hydrophobic environment of the carbohydrate-binding sites of subdomains I and III of Marpola-1 (the binding-site of subdomain II was omitted because it was predicted as being inactive) were calculated. The overall exposed surface area occupied by nonpolar residues is higher in Marpola-1 than in GNA (23% versus 18.7%). Furthermore the hydrophobic environment of the carbohydrate-binding site of subdomain I is more extended in Marpola-1 than in GNA (results not shown). No obvious difference in hydrophobicity was found between the binding sites of subdomain III of Marpola-1 and GNA (results not shown). It is possible that the (slightly) higher hydrophobic character of Marpola, especially in the vicinity of the carbohydrate-binding site of subdomain I, which is the most reactive binding site in GNA (Hester et al., 1995), can account for the enhanced affinity of Marpola for methylated or p-nitrophenylated sugar derivatives.
Evolutionary Relationship between Marpola and the Superfamily of Monocot Man-Binding Lectins
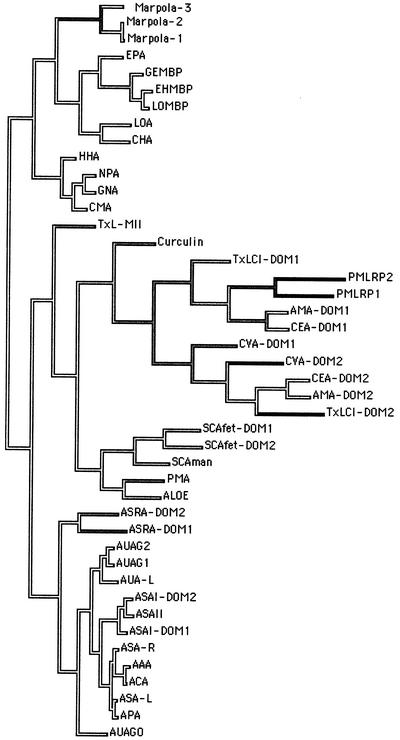
To trace the evolutionary relationships of Marpola, a phylogenetic tree based on a distance matrix was built from the sequences of the liverwort lectins and all other available sequences of monocot Man-binding lectins. As shown in Figure 5, Marpola is most closely related to the Orchidaceae lectins. It is very striking that the orchid lectins are more closely related to a lectin from a liverwort than to the Amaryllidaceae lectins. The fact that the orchid lectins are the closest relatives of Marpola suggests that of all modern monocot Man-binding lectins the Orchidaceae lectins are probably the most “primitive” (in the sense that they share the highest similarity with homologs from a lower plant). A detailed discussion of the rest of the dendrogram has been elaborated in a previous paper (Van Damme et al., 2000).
Figure 5.
Phylogenetic tree built from the amino acid sequences of the liverwort lectins (Marpola-1, Marpola-2, and Marpola-3) and other structurally related monocot Man-binding lectins. Codes are as follows: AAA, Allium ascalonicum agglutinin; ACA, shallot (Allium cepa) agglutinin; APA, leek (Allium porrum) agglutinin; ASAI-DOM1 and ASAI-DOM2, domains 1 and 2 of garlic agglutinin; ASAII, garlic agglutinin II; ASA-l, garlic leaf lectin; ASA-R, garlic root lectin; ASRA-DOM1 and ASRA-DOM2, domains 1 and 2 of putative garlic lectin-related protein; AUAG0, lectin polypeptides of Allium ursinum agglutinin II; AUAG1 and AUAG2, lectin polypeptides composing A. ursinum agglutinin I; AUA-l, A. ursinum leaf lectin; ALOE, Aloe arborescens agglutinin; AMA-DOM1 and AMA-DOM2, domains 1 and 2 of lily (Arum maculatum) agglutinin; CEA-DOM1 and CEA-DOM2, domains 1 and 2 of taro (Colocasia esculenta) agglutinin; CHA, Cymbidium hybrid agglutinin; CMA, Clivia miniata agglutinin; Curculin, sweet-tasting protein from Curculigo latifolia; CVA-DOM1 and CVA-DOM2, domains 1 and 2 of Crocus vernus agglutinin; EHMBP, Epipactis helleborine monomeric Man-binding protein; EPA, E. helleborine agglutinin; GEMBP, Gastrodia elata Man-binding protein; HHA, Hippeastrum hybrid agglutinin; LOA, Listera ovata agglutinin; LOMBP, monomeric Man-binding protein of L. ovata; NPA, daffodil (Narcissus pseudonarcissus) agglutinin; PMA, Polygonatum multiflorum agglutinin; PMLRP1 and PMLRP2, domains 1 and 2 of putative P. multiflorum lectin-related protein; SCAfet-DOM1 and SCAfet-DOM2, domains 1 and 2 of Scilla campanulata fetuin-binding agglutinin; SCAman, S. campanulata Man-binding agglutinin; TxLCI-DOM1 and TxLCI-DOM2, domains 1 and 2 of tulip (Tulipa spp.) lectin TxLCI; TxL-MII, tulip lectin MII. Branches of the tree are shaded according to the amount of amino acid changes.
DISCUSSION
Screening of liverwort revealed the occurrence of a novel Man-specific lectin, which clearly differs from a previously described liverwort lectin (Adam and Becker, 1993) with respect to its specificity and molecular structure. Biochemical analyses of the purified protein combined with sequence analysis of existing ESTs and molecular modeling allowed us, for the first time, to characterize in detail a lectin from a lower plant. In addition, the availability of sequence data and models of the three-dimensional structure of Marpola enabled us to establish for the first time the structural and evolutionary relationships between a lectin from a lower plant and a well-characterized family of lectins from flowering plants. The high sequence identity and structural similarity leave no doubt that Marpola is closely related to the family of monocot Man-binding lectins, which hitherto have exclusively been found in a subgroup of flowering plants comprising the families Alliaceae, Amaryllidaceae, Araceae, Bromeliaceae, Iridaceae, Liliaceae, and Orchidaceae. This unexpected finding not only demonstrates that at least some families of modern plant lectins evolved from ancestral homologs present in ancient lower plants, but it also allows us to construct a refined model of the molecular evolution of the monocot Man-binding lectins. The identification of a homolog of the monocot Man-binding lectins in a lower plant is important in view of the possible origin of the GNA domain present in modern flowering plants. Hitherto, the GNA domain was identified in the monocot Man-binding lectins (Van Damme et al., 1998) and in comitin, an actin-binding protein found in the slime mold D. discoideum as well as in mammals (Barre et al., 1999).
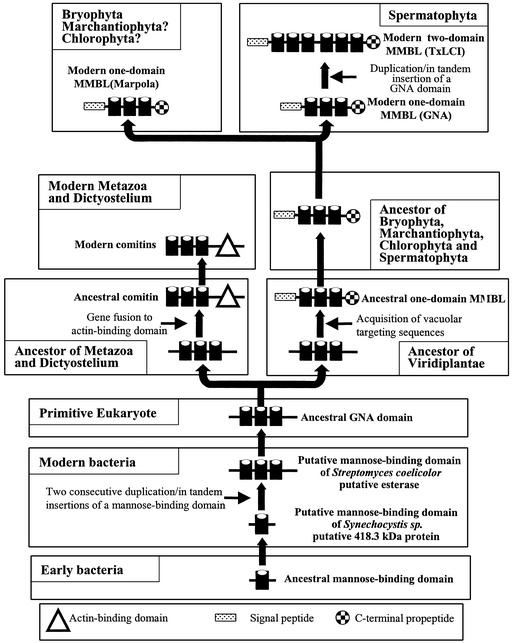
Comitin is a chimeric cytoplasmic Man-binding protein consisting of an N-terminal Man-binding domain and a C-terminal actin-binding domain (Jung et al., 1996). This protein simultaneously binds to Man residues on the surface of vesicle membranes and to actin, and hence is capable of linking these membranes to the cytoskeleton (Jung et al., 1996). Because no comitin-like protein has been found in plants and no sequences encoding putative homologs have been identified (e.g. in the completed genome of Arabidopsis), there is apparently no direct evolutionary link between the monocot Man-binding lectins and comitin. To explain the enigmatic relationship between comitin and the monocot Man-binding lectins on the one hand, and the limited taxonomic distribution of these lectins on the other hand, it was suggested that the common ancestor of the Alliaceae, Amaryllidaceae, Araceae, Bromeliaceae, Iridaceae, Liliaceae, and Orchidaceae lectins acquired the Man-binding domain of comitin through a horizontal gene transfer (Van Damme et al., 1998). It is evident that this idea has to be abandoned because the identification of Marpola leaves no doubt that the modern monocot Man-binding lectins evolved from a lectin that already existed in the common ancestor of lower and higher plants. The apparent occurrence in organisms as diverse as slime molds, mammals, liverworts, and flowering plants indicates that a common ancestor of the GNA/comitin domain already existed in the predecessor of all higher eukaryotes. Though the origin of the ancestral GNA/comitin domain itself is still unclear, the occurrence of three homologous internal repeats of approximately 35 amino acid residues indicates that two consecutive duplication/in tandem insertions of an ancestral gene encoding a shorter polypeptide chain of approximately 35 amino acid residues gave rise to the original ancestor of the GNA/comitin domain (Van Damme et al., 1998). Evidence for the occurrence of a hypothetical Man-binding domain equivalent to a single GNA subdomain has recently been obtained from the deduced sequences of a Synechocystis sp. (strain pcc 6803) putative 418.3-kD protein (accession no. BAA17165) and a Streptomyces hygroscopicus hypothetical protein (accession no. T30223). In addition, a putative Streptomyces coelicolor putative esterase (accession no. CAC44518) has been identified containing a domain consisting of three in tandem-arrayed equivalents of the hypothetical Man-binding domain of these Synechocystis sp. (strain PCC 6803) and S. hygroscopicus hypothetical proteins.
These findings not only suggest a bacterial origin of the GNA subdomain, but they also indicate that the in tandem duplication/insertion of the original Man-binding domain already took place in prokaryotes. Therefore, it is very likely that the evolution of the monocot Man-binding lectins started in a prokaryote (Fig. 6). Taking into consideration the high sequence similarity between the GNA subdomains of the modern bacterial and eukaryotic proteins, one can reasonably assume that they all evolved from an ancestral Man-binding domain of approximately 40 amino acid residues (equivalent to a single GNA subdomain) that already existed in an early prokaryote before the first eukaryotes originated. How this ancestral Man-binding domain possibly gave rise to the modern monocot Man-binding lectins is schematically represented in Figure 6. It should be mentioned here that the scheme given in Figure 6 applies exclusively to the monocot Man-binding lectins and cannot be extrapolated to other families of plant lectins. Moreover, it should also be emphasized that there is no evolutionary relationship between the monocot Man-binding lectins and any other plant lectin family. However, it is interesting that despite the obvious absence of sequence similarity, the β-prism fold of the monocot Man-binding lectins resembles the β-prism structure of the jacalin-related lectins, most of which exhibit an exclusive specificity toward Man and oligomannosides (Bourne et al., 1999). This close structural relationship between both lectin families illustrates that two unrelated sequences eventually gave rise to a structurally similar Man-binding motif through a convergent evolution.
Figure 6.
Model of the molecular evolution of the superfamily of monocot Man-binding lectins. An ancestral prokaryotic Man-binding domain of approximately 40 amino acid residues evolved into a modern prokaryotic Man-binding domain that can be considered the ancestor of all modern GNA subdomains. Two consecutive duplication/in tandem insertions of this Man-binding domain gave rise to a prokaryotic domain equivalent to the modern GNA domain. In the ancestor of the modern metazoa and slime molds, the ancestral GNA domain fused to an actin binding to yield a comitin. Fusion of the ancestral GNA domain to vacuolar targeting sequences in an early ancestor of the Viridiplantae resulted in an extra-cytoplasmic protein similar to the modern one-domain monocot Man-binding lectins. This ancestral one-domain monocot Man-binding lectin served as the direct ancestor of all modern one-domain and two-domain monocot Man-binding lectins.
Marpola is, of all modern monocot Man-binding lectins, probably the closest relative of the common ancestor of this lectin family. In accordance with this, one can reasonably expect that the liverwort lectin fulfills a role that closely resembles that of the “original” monocot Man-binding lectins. Therefore, the discovery of Marpola may, in the long term, help to elucidate the function of these lectins. At present, little is known about the regulation of the expression of Marpola. A screening of liverwort thalli from different locations for the presence of lectin indicated that some, but not all, thalli contained an agglutinin. The factor(s) governing the expression of the lectin are not known, but there are indications that genetic and environmental factors play a role. Checking of samples from different patches at the same location revealed that lectin-positive thalli occurred only in some patches, whereas thalli from patches in the direct vicinity reacted negatively in the agglutination test. More extensive testing indicated that within a single patch, virtually all thalli reacted similarly. The occurrence of lectin is not linked to the sex of the plants because agglutinating activity was found in male and female thalli. Regular checking of samples from a single patch revealed that the lectin was only present during certain periods.
Semiquantitative assays based on agglutination tests indicated that the lectin content remains low. The titer of the sap squeezed from the thalli never exceeded 100 (corresponding to a Marpola concentration of approximately 10 μg mL−1), indicating that the lectin concentration remains below 10 μg g−1 tissue (fresh weight), and accordingly, Marpola is a minor protein in the thalli. This (maximal) concentration is very low when compared with the expression level of most monocot Man-binding lectins from flowering plants. For example, typical values for leaf lectins from Orchidaceae, Amaryllidaceae, and Alliaceae are in the range between 0.1 and 0.5 mg g−1 tissue (fresh weight). In general, these lectins can be distinguished as discrete polypeptide bands upon SDS-PAGE of crude leaf extracts. Expression levels of monocot Man-binding lectins in vegetative storage organs usually vary between 1 and 10 mg g−1 tissue (fresh weight). Many of these lectins are the most predominant proteins, representing up to 50% of the total protein (e.g. in garlic cloves). Therefore, it can be concluded that the expression level of Marpola in the liverwort thalli is considerably lower than that of the monocot Man-binding lectins from flowering plants. Another important difference concerns the temporal regulation of the expression. In flowering plants, the leaf lectins are constitutively expressed. The same holds true for the storage protein-like lectins, but in this case, the concentration depends on the developmental stage of the vegetative storage tissues. In contrast, Marpola is not constitutively expressed, but is synthesized only under certain yet unknown conditions.
MATERIALS AND METHODS
Chemicals
Man, Gal, Glc, Meα-Man, pNO2Fα-Man, pNO2Fβ-Man, Meα-Gal, pNO2Fα-Gal, pNO2Fβ-Gal, and Meα-Glc were purchased from Sigma (St. Louis). The oligomannosides diManα1, 2; diManα1, 3; diManα1, 4; diManα1, 6; triManα1, 3/1, 6; and pentaManα1, 3/1, 6 were purchased from Dextra Laboratories (Reading, UK). Sensor chips (CM 5), 10 mm HEPES, 150 mm NaCl containing 0.05% (v/v) BIAcore surfactant P20, and 3.0 mm EDTA, pH 7.4 (HBS), and all the chemicals required for the activation of the carboxymethylated Dextran and the immobilization of arcelin-1 [100 mm N-hydroxysuccinimide, 400 mm N-ethyl-N′-(3-dimethylaminopropyl) carbodiimide hydro-chloride, and 1 m ethanolamine hydrochloride adjusted to pH 8.5 with NaOH] were obtained from Amersham Biosciences AB (Uppsala).
Arcelin-1 was isolated from seeds of Phaseolus vulgaris cv RAZ2 (Fabre et al., 1998). The Man-binding GNA was purified from snowdrop (Galanthus nivalis) bulbs as previously described (Van Damme et al., 1987).
Plant Material
Thalli of liverwort (Marchantia polymorpha; with gemma cups) were collected locally. Because the lectin content of the thalli differs strongly from patch to patch, the presence of the lectin was checked with a simple agglutination assay. Thalli were collected only from those patches in which most of the thalli showed a strong agglutinating activity. After collection, thalli were intensively washed with tap water to remove debris and soil particles.
Isolation of the Liverwort Agglutinin (Marpola)
Fresh thalli (200 g) were transferred to 1 L of a solution of 0.1% (w/v) unbuffered ascorbic acid and were homogenized with a Waring blender. The homogenate was filtered through cheesecloth and centrifuged (3,000g for 5 min). The supernatant was decanted, CaCl2 was added (1.5 g L−1), and the pH was raised to 9.0 by the addition of 1 n NaOH. After standing in the cold (2°C) for 1 h, the precipitate was removed by centrifugation (3,000g for 10 min). The cleared extract was filtered through filter paper (3MM, Whatman, Clifton, NJ), adjusted to pH 3.0 with 1 n HCl, and loaded on a column of S Fast Flow (Amersham Biosciences AB; 2.6 cm × 5 cm; 50-mL bed volume) equilibrated with 20 mm acetic acid. The column was subsequently washed with 500 mL of formate buffer (20 mm Na-formate, pH 3.8), and the bound proteins were eluted with 250 mL of 1 m NaCl in 0.1 m Tris-HCl (pH 7.8). To this partially purified protein fraction solid ammonium sulfate was added to reach a final concentration of 1 m, and the protein mixture was loaded on a column of Man-Sepharose 4B (2.6 cm × 5 cm; 25-mL bed volume) equilibrated with 1 m ammonium sulfate. The column was washed with 1 m ammonium sulfate until the A280 fell below 0.01, and the bound proteins were eluted with 100 mL of a solution of 20 mm unbuffered Tris. After desorption, the pH of the eluate was lowered to 7.5 with 1 n acetic acid, and solid NaCl was added to a final concentration of 0.2 m. The partially purified lectin fraction was centrifuged (20,000g for 10 min) and loaded on a small column (1.6 cm × 2 cm; 4-mL bed volume) of Man-Sepharose 4B. The column was washed with 0.2 m NaCl until the A280 fell below 0.01, and the bound lectin was desorbed with 20 mL of 0.1 m Man in 0.2 m NaCl. To concentrate the affinity-purified lectin, the eluate was diluted with an equal volume of a solution of 2 m ammonium sulfate and was applied on a column (1 cm × 2 cm; 1.5-mL bed volume) of Phenyl-Sepharose (Amersham Biosciences AB) equilibrated with 1 m ammonium sulfate. After washing the column with 1 m ammonium sulfate, the lectin was desorbed with 2 mL of 20 mm Tris-HCl (pH 8.7), dialyzed against appropriate buffers, and stored at −20°C until use. An approximate 0.5 mg of total affinity-purified Marpola was obtained starting from 200 g of fresh weight.
Analytical Techniques
Purified proteins were analyzed by SDS-PAGE using 12.5% to 25% (w/v) acrylamide gradient gels as described by Laemmli (1970). Analytical gel filtration was performed on a Superose 12 column (Amersham Biosciences AB) using phosphate-buffered saline (PBS) containing 0.2 m Man (to avoid possible interactions of the lectins with the matrix) as running buffer. The well-characterized lectins from Galanthus nivalis (50 kD; Van Damme et al., 1987) and garlic (Allium sativum; 25 kD; Van Damme et al., 1992) were used as molecular mass markers. Total neutral sugar was determined by the phenol/H2SO4 method with d-Glc as standard (Dubois et al., 1956).
Purified Marpola was labeled with fluorescent N-(1-pyrenyl) maleimide as described by Bhattacharyya and Roy (1993), and was analyzed by SDS-PAGE. Labeling was visualized on a UV transilluminator. GNA was included as a control.
For N-terminal amino acid sequencing, purified proteins were separated by SDS-PAGE and electroblotted on a polyvinylidene difluoride membrane. Polypeptides were excised from the blots and were sequenced on a protein sequencer (model 477A/120A or Procise 491 cLC; Applied Biosystems, Foster City, CA). Prior to mass spectrometry, proteins were desalted on a C4-ZipTip (Millipore, Bedford, MA). Proteins were dissolved in 50% (v/v) water/50% (v/v) acetonitrile containing 0.1% (w/v) acetic acid and were injected at 5 μL min−1 on an electrospray ion trap mass spectrometer (Esquire-LC-MS; Bruker Daltonic, Bremen, Germany). About 300 spectra were averaged, resulting in an accuracy of ±0.01% for proteins with a relative molecular mass of 10,000 D.
Hemagglutination and Hapten Inhibition Assays
Agglutination assays were carried out in small glass tubes in a final volume of 50 μL containing 40 μL of a 1% (v/v) suspension of red blood cells and 10 μL of extracts or lectin solutions. To determine the specific agglutination activity, the lectin was serially diluted with 2-fold increments. Agglutination was assessed visually after 1 h at room temperature. Rabbit erythrocytes were treated with trypsin as described previously (Van Damme et al., 1987).
The overall carbohydrate-binding specificity of the lectin was determined by hapten inhibition of the agglutination of trypsinized rabbit erythrocytes. To 10 μL of a solution of Marpola (1 μg mL−1 in PBS), 10-μL aliquots of solutions of sugars (0.5 m in PBS) or glycoproteins (5 mg mL−1 in PBS) were added. After preincubation for 1 h at 25°C, 30 μL of a 1% (v/v) suspension of trypsinized rabbit erythrocytes was added, and the agglutination was evaluated after 1 h. To determine the inhibitory potency of the most active monosaccharides and glycoproteins, the assays were repeated with serially diluted stock solutions of sugars and glycoproteins. The IC50 of the agglutination of trypsin-treated rabbit erythrocytes was determined visually.
Biosensor Measurements
Analysis of the specific interaction of Marpola with the high-Man N-glycans of arcelin-1 (which contains predominantly oligosaccharide chains of the high-Man type) was performed by surface plasmon resonance (SPR) using a biosensor BIAcore 2000 (Amersham Biosciences AB). Parallel experiments were done with GNA because the snowdrop lectin is considered the prototype of the monocot Man-binding lectins (Shibuya et al., 1988) and hence serves as a suitable reference to trace similarities/differences between Marpola and the classic monocot Man-binding lectins.
For immobilization on the sensor chips CM 5, arcelin-1 was used at a concentration of 1 mg mL−1 in 5 mm sodium acetate buffer (pH 4.0). Based on the change of SPR response (expressed in RU) as a result of the immobilization on the carboxymethylated Dextran layer covering the sensor chip, the surface concentration of arcelin-1 was estimated at 10 ng mm−2 of Dextran.
Marpola and GNA, used at concentrations of 175 and 100 μg mL−1, respectively, in HBS (pH 7.4), were injected for 5 min onto the glycoprotein-bound surface of the sensor chip at a flow rate of 5 μL min−1. The change of the SPR response was monitored at 25°C for 9.3 min. The same glycoprotein sensor chip surfaces were used repeatedly after removing the remaining immobilized Marpola by two successive washes with 10 mm HCl.
Inhibition by monosaccharides, monosaccharide derivatives, and oligomannosides was performed by injecting sugars at a concentration of 10 mm in HBS (pH 7.4; except for pNO2Fα-Man and pNO2Fβ-Man, which were used at concentrations of 7 and 2.5 mm, respectively) at the beginning of the dissociation phase for 5 min at a flow rate of 5 μL min−1, and the change of the SPR response was monitored at 25°C for 9.3 min.
Molecular Modeling
The program SEQVU (J. Gardner, The Garvan Institute of Medical Research, Sydney, Australia) was used to compare the amino acid sequences of GNA and liverwort lectins. Multiple amino acid sequence alignments based on CLUSTAL W (Thompson et al., 1994) were carried out using SEQPUP (D.G. Gilbert, Indiana University, Bloomington) and were modified manually to build the phylogenetic tree. MACCLADE (Maddison and Maddison, 1992) was used to build a parsimony phylogenetic tree relating the liverwort lectin to other lectins.
HCA (Gaboriaud et al., 1987; Lemesle-Varloot et al., 1990) was performed to delineate the structurally conserved β-sheets along the amino acid sequences of Marpola by homology to the GNA used as a model. HCA plots were generated using the program HCA-Plot2 (Doriane, Le Chesnay, France).
Molecular modeling of Marpola was carried out on a workstation (O2 R10000) using the programs INSIGHTII, HOMOLOGY, and DISCOVER (Accelrys Inc., San Diego). The atomic coordinates of GNA (Hester et al., 1995) were used to build the three-dimensional model of the liverwort lectins. Steric conflicts resulting from the replacement or the deletion of some residues in the liverwort lectins were corrected during the model-building procedure using the rotamer library (Ponder and Richards, 1987) and the search algorithm implemented in the HOMOLOGY program (Mas et al., 1992) to maintain proper side chain orientation. Energy minimization and relaxation of the loop regions was carried out by several cycles of steepest descent and conjugate gradient using the cvff forcefield of Discover. The program TURBOFRODO (Bio-Graphics, Marseille, France) was run on the O2 workstation (Silicon Graphics) to draw the Ramachandran plot of the modeled lectins and to perform the superposition of the models and the docking of Man into their binding sites. The lowest apparent binding energy (Ebind, expressed in kilocalories per mole) compatible with the four hydrogen bonds (considering Van de Waals interactions and strong [2.5 Å < dist(D-A) <3.1 Å and 120° < ang(D-H-A)] and weak [2.5 Å < dist(D-A) <3.5 Å and 105° < ang(D-H-A) <120°] hydrogen bonds; with D: donor, A: acceptor, and H: hydrogen) found in the GNA-Man complex (Hester et al., 1995; Hester and Wright, 1996; Wright and Hester, 1996) was calculated with the cvff forcefield and was used to anchor the pyranose ring of Man into the binding sites of lectins. The programs MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merritt and Bacon, 1997) were used to draw the figures.
Electrostatic potentials were calculated and displayed with GRASP using the parse3 parameters (Nicholls et al., 1991). The solvent probe radius used for molecular surfaces was 1.4 Å, and a standard 2.0 Å-Stern layer was used to exclude ions from the molecular surface (Gilson and Honing, 1987). The inner and outer dielectric constants applied to the protein and the solvent were fixed at 4.0 and 80.0, respectively, and the calculations were performed keeping a salt concentration of 0.145 m NaCl. No even distribution of the net negative charge of the carboxylic group of negatively charged residues was assigned between their two oxygen atoms prior to the calculations. The surfaces occupied by hydrophobic (Ala, Leu, Ile, Val, and Met) and aromatic (Phe, Trp, and Trp) residues on the solvent accessible surface of the modeled lectins were calculated with the GRASP program.
Footnotes
This work was supported in part by the Catholic University of Leuven (grant no. OT/98/17), by Centre National de la Recherche Scientifique (grants to A.B. and P.R.), and by the Fund for Scientific Research-Flanders (Belgium; grant no. G.0113.01). P.P. is a Postdoctoral Fellow of this fund.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010959.
LITERATURE CITED
- Adam K-P, Becker H. A lectin from the liverwort Marchantia polymorphaL. Experientia. 1993;49:1098–1100. doi: 10.1007/BF01929921. [DOI] [PubMed] [Google Scholar]
- Barre A, Van Damme EJM, Peumans WJ, Rougé P. Structure-function relationship of monocot mannose-binding lectins. Plant Physiol. 1996;112:1531–1540. doi: 10.1104/pp.112.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre A, Van Damme EJM, Peumans WJ, Rougé P. Homology modelling of the endogenous lectin comitin: structural basis for its mannose-binding specificity. Plant Mol Biol. 1999;39:969–978. doi: 10.1023/a:1006133527621. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya T, Roy SA. Fluorescence spectroscopic study of substrate-induced conformational changes in glutaminyl-tRNA synthetase. Biochemistry. 1993;32:9268–9273. doi: 10.1021/bi00087a002. [DOI] [PubMed] [Google Scholar]
- Bourne Y, Zamboni V, Barre A, Peumans WJ, Van Damme EJM, Rougé P. Crystal structure of Helianthus tuberosuslectin, a widespread scaffold for mannose-binding lectins. Struct Fold Des. 1999;7:1473–1482. doi: 10.1016/s0969-2126(00)88338-0. [DOI] [PubMed] [Google Scholar]
- Candy L, Peumans WJ, Menu-Bouaouiche L, Houlès Astoul C, Van Damme J, Van Damme EJM, Erard M, Rougé P. The Gal/GalNAc-specific lectin from the plant pathogenic basidiomycete Rhizoctonia solani is a member of the ricin-B family. Biochem Biophys Res Commun. 2001;282:655–661. doi: 10.1006/bbrc.2001.4626. [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugar and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- Fabre C, Causse H, Mourey L, Koninkx J, Rivière M, Hendriks H, Puzo G, Samama JP, Rougé P. Characterization and sugar-binding properties of arcelin-1, an insecticidal lectin-like protein isolated from kidney bean (Phaseolus vulgarisL. cv raz-2) seeds. Biochem J. 1998;329:551–560. doi: 10.1042/bj3290551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzel BC, Clancy LL, Holland DR, Muchmore SW, Watenpaugh KD, Einspahr HM. Crystal structure of recombinant human interleukin-1β at 2.0 Å resolution. J Mol Biol. 1989;209:779–791. doi: 10.1016/0022-2836(89)90606-2. [DOI] [PubMed] [Google Scholar]
- Fujimoto Z, Kuno A, Kaneko S, Yoshida S, Kobayashi H, Kusakabe I, Mizuno H. Crystal structure of Streptomyces olivaceoviridisE-86 β-xylanase containing xylan-binding domain. J Mol Biol. 2000;300:575–585. doi: 10.1006/jmbi.2000.3877. [DOI] [PubMed] [Google Scholar]
- Gaboriaud C, Bissery V, Benchetrit T, Mornon JP. Hydrophobic cluster analysis: an efficient new way to compare and analyze amino acid sequences. FEBS Lett. 1987;224:149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Gilson MK, Honing BH. Calculation of electrostatic potential in an enzyme active site. Nature. 1987;330:84–86. doi: 10.1038/330084a0. [DOI] [PubMed] [Google Scholar]
- Graves BJ, Hatada MH, Hendrickson WA, Miller JK, Madison VS, Satow Y. Structure of interleukin-1α at 2.7 Å resolution. Biochemistry. 1990;29:2679–2684. doi: 10.1021/bi00463a009. [DOI] [PubMed] [Google Scholar]
- Hester G, Kaku H, Goldstein IJ, Wright CS. Structure of mannose-specific snowdrop (Galanthus nivalis) lectin is representative of a new plant lectin family. Nat Struct Biol. 1995;2:472–479. doi: 10.1038/nsb0695-472. [DOI] [PubMed] [Google Scholar]
- Hester G, Wright CS. The mannose-specific bulb lectin from Galanthus nivalis(snowdrop) binds mono- and dimannosides at distinct sites: structure analysis of refined complexes at 2.3 Å and 3.0 Å resolution. J Mol Biol. 1996;262:516–531. doi: 10.1006/jmbi.1996.0532. [DOI] [PubMed] [Google Scholar]
- Jung E, Fucinin P, Stewart M, Noegel AA, Schleicher M. Linking microfilaments to intracellular membranes: the actin-binding and vesicle associated protein comitin exhibits a mannose-specific lectin activity. EMBO J. 1996;15:1238–1246. [PMC free article] [PubMed] [Google Scholar]
- Kraulis PJ. Molscript: a program to produce both detailed and schematic plots of protein structures. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lemesle-Varloot L, Henrissat B, Gaboriaud C, Bissery V, Morgat A, Mornon JP. Hydrophobic cluster analysis: procedure to derive structural and functional information from 2-D representation of protein sequences. Biochimie. 1990;72:555–574. doi: 10.1016/0300-9084(90)90120-6. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chirino AJ, Misulovin Z, Leteux C, Feizi T, Nussenzweig MC, Bjorkman PJ. Crystal structure of the cysteine-rich domain of mannose receptor complexed with a sulfated carbohydrate ligand. J Exp Med. 2000;7:1105–1115. doi: 10.1084/jem.191.7.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. MacClade: Analysis of Phylogeny and Character Evolution, Version 3.0. Sunderland, MA: Sinauer Associates; 1992. [Google Scholar]
- Mas MT, Smith KC, Yarmush DL, Aisaka K, Fine RM. Modeling the anti-CEA antibody combining site by homology and conformational search. Proteins Struct Func Genet. 1992;14:483–498. doi: 10.1002/prot.340140409. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ. Raster3D photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct Func Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Nagai J, Yamato KT, Sakaida M, Yoda H, Fukuzawa H, Ohyama K. Expressed sequence tags from immature female sexual organ of a liverwort, Marchantia polymorpha. DNA Res. 1999;6:1–11. doi: 10.1093/dnares/6.1.1. [DOI] [PubMed] [Google Scholar]
- Nishiyama R, Yamato KT, Miura K, Sakaida M, Okada S, Kono K, Takahama M, Sone T, Takenaka M, Fukuzawa H et al. Comparison of expressed sequence tags from male and female sexual organs of Marchantia polymorpha. DNA Res. 2000;7:165–174. doi: 10.1093/dnares/7.3.165. [DOI] [PubMed] [Google Scholar]
- Ponder JW, Richards FM. Tertiary templates for proteins: use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Shibuya N, Goldstein IJ, Van Damme EJM, Peumans WJ. Binding properties of a mannose-specific lectin from the snowdrop (Galanthus nivalis) bulb. J Biol Chem. 1988;263:728–734. [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJM, Allen AK, Peumans WJ. Isolation and characterization of a lectin with exclusive specificity towards mannose from snowdrop (Galanthus nivalis) bulbs. FEBS Lett. 1987;215:140–144. [Google Scholar]
- Van Damme EJM, Smeets K, Torrekens S, Van Leuven F, Goldstein IJ, Peumans WJ. The closely related homomeric and heterodimeric mannose-binding lectins from garlic are encoded by one-domain and two-domain lectin genes, respectively. Eur J Biochem. 1992;206:413–420. doi: 10.1111/j.1432-1033.1992.tb16941.x. [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Peumans WJ, Barre A, Rougé P. Plant lectins: a composite of several distinct families of structurally and evolutionary related proteins with diverse biological roles. Crit Rev Plant Sci. 1998;17:575–692. [Google Scholar]
- Van Damme EJM, Houles Astoul C, Barre A, Rougé P, Peumans WJ. Cloning and characterization of a monocot mannose-binding lectin from Crocus vernus(family Iridaceae) Eur J Biochem. 2000;267:5067–5077. doi: 10.1046/j.1432-1327.2000.01563.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;11:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CS, Hester G. The 2.0 Å structure of a cross-linked complex between snowdrop lectin and a branched mannopentaose: evidence for two unique binding modes. Structure. 1996;11:1339–1352. doi: 10.1016/s0969-2126(96)00141-4. [DOI] [PubMed] [Google Scholar]
- Zhu X, Komiya H, Chririno A, Faham S, Fox GM, Arakawa T, Hsu BT, Rees DC. Three-dimensional structures of acidic and basic fibroblast growth factors. Science. 1991;251:90–93. doi: 10.1126/science.1702556. [DOI] [PubMed] [Google Scholar]