Abstract
The promoter region of the Arabidopsis AtHsp90-1 gene is congested with heat shock elements and stress response elements, as well as with other potential transcriptional binding sites (activating protein 1, CCAAT/enhancer-binding protein element, and metal regulatory element). To determine how the expression of this bona fide AtHsp90-1 gene is regulated, a comprehensive quantitative and qualitative promoter deletion analysis was conducted under various environmental conditions and during development. The promoter induces gene expression at high levels after heat shock and arsenite treatment. However, our results show that the two stress responses may involve common but not necessarily the same regulatory elements. Whereas for heat induction, heat shock elements and stress response elements act cooperatively to promote high levels of gene expression, arsenite induction seems to require the involvement of activating protein 1 regulatory sequences. In stressed transgenic plants harboring the full-length promoter, β-glucuronidase activity was prominent in all tissues. Nevertheless, progressive deletion of the promoter decreases the level of expression under heat shock and restricts it predominantly in the two meristems of the plant. In contrast, under arsenite induction, proximal sequences induce AtHsp90-1 gene expression only in the shoot meristem. Distally located elements negatively regulate AtHsp90-1 gene expression under unstressed conditions, whereas flower-specific regulated expression in mature pollen grains suggests the prominent role of the AtHsp90-1 in pollen development. The results show that the regulation of developmental expression, suppression, or stress induction is mainly due to combinatorial contribution of the cis elements in the promoter region of the AtHsp90-1 gene.
During their lifetime, plant species can be subjected to various stressful environments to which they respond and adapt by means of physiological, developmental, and biochemical changes. One of the most thoroughly characterized is the induction of heat shock proteins (HSPs) when cells or organisms are exposed to supraoptimal temperatures and other types of stresses (for review, see Lindquist and Craig, 1988; Vierling, 1991; Miernyk, 1999). The heat shock response is a universal (Schlessinger et al., 1982; Morimoto and Santoro, 1998) and evolutionarily conserved phenomenon (Schlessinger et al., 1982). However, it is now recognized that the same or closely related proteins are frequently essential components of cells under normal physiological conditions (Boston et al., 1996).
Accumulating evidence reveals that all of the major HSPs serve as molecular chaperones (Georgopoulos and Welch, 1993; Bukau and Horwich, 1998; Pratt et al., 2001). Although the structure and the mechanism of some chaperones such as HSP70, HSP60, and sHSPs have been investigated extensively (Waters et al., 1996; Bukau and Horwich, 1998), the function of HSP90s as molecular chaperones is still controversial. The HSP90s are among the most highly conserved proteins known, with approximately 40% similarity between the prokaryotic 90-kD molecular chaperone, the HtpG, and its eukaryotic counterparts (Csermely et al., 1998). Studies on the chaperone activity of the mammalian HSP90 revealed a cast of the target substrates or client proteins, including members of signal transduction pathways, the cell cycle control machinery, the proteolytic machinery, and other kinds of proteins like nitric oxidase synthase and telomerase (Czar et al., 1997; Nathan et al., 1997; Garcia-Cardena et al., 1998; Holt et al., 1999; Pratt et al., 2001). It has also been proposed that the HSP90 chaperones have other essential, unidentified functions (Nathan et al., 1997).
During the past 10 years, several plant hsp90 genes have been identified and cloned. The proteins are localized in different cell compartments, including the cytoplasm, the endoplasmic reticulum, and chloroplasts (Koning et al., 1992; Takahashi et al., 1992; Marrs et al., 1993; Schröder et al., 1993; Krishna et al., 1995; Schmitz et al., 1996; Milioni and Hatzopoulos, 1997). The corresponding genes were shown to be specifically expressed during embryogenesis, pollen development (Marrs et al., 1993), and seed germination (Reddy et al., 1998) in young and rapidly dividing tissues such as shoot and root apices (Koning et al., 1992) and in flowers (Takahashi et al., 1992; Krishna et al., 1995). In oilseed rape (Brassica napus) and tomato (Lycopersicon esculentum) seedlings, HSP90 protein levels were found to increase by exogenous 24-epibrassinolide application (Dhaubhadel et al., 1999), whereas a glucosinolate-deficient Arabidopsis mutant was shown to be thermosensitive and defective in the cytosolic HSP90 expression after heat stress (Ludwig-Muller et al., 2000). It has also been reported that the hsp90 genes are stimulated by chemical treatments such cadmium or arsenite (Takahashi et al., 1992; Milioni and Hatzopoulos, 1997) and by exogenous treatment with indoleacetic acid or 0.1 m NaCl (Yabe et al., 1994). Also, in rice (Oryza sativa) seedlings, a putative HSP90 protein was shown to accumulate in response to salinity, low temperature stress, and exogenous abscisic acid application (Pareek et al., 1995). The above data suggest that plant HSP90s are encoded by a family of genes that are differentially regulated in response to specific developmental and environmental cues. The Arabidopsis sequencing project recently revealed that the hsp90 family consists of seven members. The AtHsp90-1 through AtHsp90-4 proteins comprise the cytoplasmic subfamily, whereas the AtHsp90-5, AtHsp90-6, and AtHsp90-7 proteins are predicted to be within the plastidial, mitochondrial, and endoplasmic reticulum compartments, respectively (Krishna and Gloor, 2001).
The expression of the heat shock genes is known to be regulated mainly at the transcriptional level. The thermoinducibility of the heat shock genes is attributed to activation of heat shock factors (HSF). HSF act through a highly conserved heat shock promoter element (HSE) that has been defined as adjacent and inverse repeats of the motif 5′-nGAAn-3′ (Amin et al., 1988; Xiao and Lis, 1988; Schöffl et al., 1998). HSEs are the binding sites for the trans-active HSF, and efficient binding requires at least three units. Promoter analyses of individual plant hsp90 genes have indicated the contribution of individual HSEs and their recognition by distinct protein factors during heat shock and development (Yabe et al., 1994; Marrs and Sinibaldi, 1997). In addition, several well-conserved motifs have been identified to have quantitative effects on the expression of certain heat shock genes, i.e. CCAAT-box elements and scaffold-attachment regions (Rieping and Schöffl, 1992; Chinn and Comai, 1996). However, in the promoter region of hsp90 genes, cis-elements that may be important in regulating pathways other than the heat shock response have not been identified as of yet.
The experiments described and the data obtained in the present study explore the contribution of specific regulatory elements in the expression of the Arabidopsis AtHsp90-1 gene under normal conditions, heat stress, or arsenite treatment. We constructed chimeric genes composed of a series of deletions of the AtHsp90-1 promoter and the β-glucuronidase (GUS) gene to quantitatively and qualitatively analyze gene induction in Arabidopsis developing seedlings. In addition, tissue-specific expression was assessed in mature Arabidopsis transgenic plants.
RESULTS
Sequence Analysis of the AtHsp90-1 Promoter
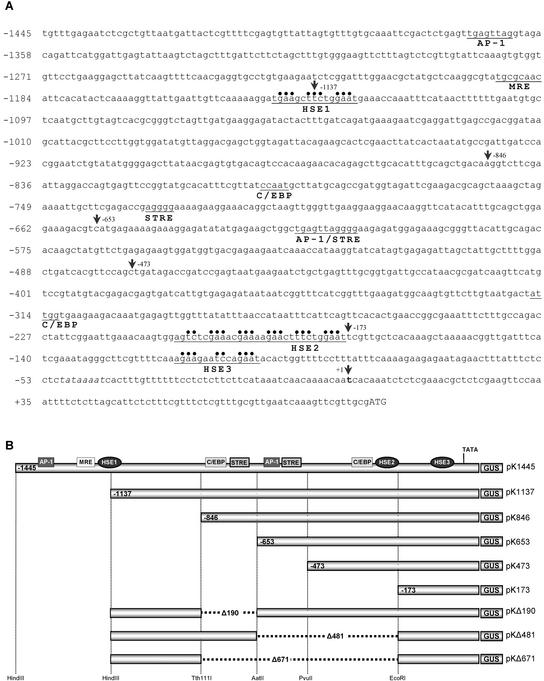
To search the Arabidopsis AtHsp90-1 promoter region for potential binding sites of regulatory transcription factors, we used the MatInspector professional tool (Genomatix) and the transcription factor database (TRANSFAC). Approximately 1,500 bp of the AtHsp90-1 promoter (position −1,445 to +91) was analyzed. This fragment, referred to as full-length promoter (Fig. 1A), contains a sequence of 1,445 bp upstream of the predicted initiation transcription site (Takahashi et al., 1992) and the 5′-untranslated region up to the first codon corresponding to the AtHsp90-1 protein. A putative TATA box (TATAAAAT) is found in an AT-rich region at position −50, upstream of the initiation transcription site. The analysis revealed the presence of several putative transcription factor-binding sites (Fig. 1A). These include consensus sequences for HSE (Wu, 1995), C/EBP (Akira et al., 1990), STRE (Siderius and Mager, 1997), MRE (Culotta and Hamer, 1989), and the animal proto-oncogene AP-1 (Angel et al., 1987).
Figure 1.
A, Sequence of the AtHsp90-1 promoter showing the fusion to the GUS reporter gene and the extent of the promoter deletions. The transcription initiation site is designated in bold letter. The putative TATA box is shown in italics, and the ATG start codon is shown in uppercase. Putative transcriptional cis sequences (HSE, metal regulatory element [MRE], activating protein 1 [AP-1], and CCAAT/enhancer-binding protein element [C/EBP]) are indicated with underlining and are referred to in the text. Dots designate matches to the core consensus GAA/TTC. The vertical arrows show the positions used to generate the promoter-GUS-truncated constructs. Basepair positions are referred to the transcription start point. B, Schematic representation of transformation constructs containing various portions of the 5′-upstream region fused to GUS gene. Horizontal dashed lines indicate internal deletions. The locations of the putative regulatory elements are indicated. The name of each construct is given at the right side.
Transcriptional activation of heat shock genes depends on the interaction of HSFs with highly conserved cis-acting DNA sequences, HSEs, whereas CCAAT-box sequences have been shown to act cooperatively with HSEs to increase promoter activity (Williams and Morimoto, 1990; Rieping and Schöffl, 1992). The upstream region of the AtHsp90-1 gene contains three HSEs conforming to the canonical heat shock consensus of at least three core units of the repeating pentanucleotite sequence 5′-nGAAn-3′ arranged in alternate orientation (Amin et al., 1988; Xiao and Lis, 1988). The most proximal, HSE3, is located 52 nucleotides upstream of the putative TATA box, whereas the most distal HSE1 has been found at position −1,144. HSE1 (tGAAgcTTCtgGAAt) consists of three perfect core units, whereas HSE3 (gGAAgaaTCcaGAAt) consists of two perfect and one imperfect units. HSE2 (agTCtcGAAacGAAaaGAActTTCtgGAAt) is located at position −187 and consists of five perfect and one imperfect core unit of the pentanucleotite consensus. Although the first three core units of this HSE do not follow the general rule of being in alternate orientation, the last three compose a perfect consensus HSF-binding site. It is interesting that the promoter region from point −1,137 to −203 does not contain any sequences matching a consensus HSE. However, two STREs (consensus sequence AGGGG) were identified in this region at position −731 and −612. In contrast to HSEs, STREs are activated not just by heat shock but also by a diverse range of other stress conditions, especially osmotic stress, low pH, and nutrient starvation (Siderius and Mager, 1997). Two perfect CCAAT-boxes, which represent the binding sites for the C/EBP transcription factors, are also present in this region at position −316 and −798 (Fig. 1A).
In animals, AP-1-binding elements have been shown to mediate the induction of the HO-1 gene by CdCl2 (Alam, 1994) and sodium arsenite (Lu et al., 1998), respectively, whereas MREs have been identified in a number of heavy metal-induced promoters including the human and mouse metallothionein genes (Karin et al., 1987; Culotta and Hamer, 1989) and the tomato type II metallothionein-like gene (Whitelaw et al., 1997). Because the heat shock response is known to be mediated by various stress conditions including heavy metals, the 5′-upstream region of AtHsp90-1 was examined to identify possible MREs and AP-1-binding sites, which would suggest the involvement of metals in the regulation of AtHsp90-1 transcription. One MRE-like sequence (TGCGCAAC) matching six of the seven nucleotides of the consensus sequence TGCPuCNC (Culotta and Hamer, 1989) was identified immediately upstream of HSE1 at position −1,192. It is interesting that the AtHsp90-1 promoter contain also two identical octanucleotide sequences (TGAGTTAG), which are highly similar to the animal AP-1 consensus-binding site TGA(G/C) TCAG. The first AP-1 like element is located upstream of HSE1 at position −1,371 and the second is located at position −618. Both putative elements deviate from the animal AP-1 consensus sequence only at position 6 (T instead of C).
AtHsp90-1 Promoter Deletion Constructs and Determination of Transgene Copy Number
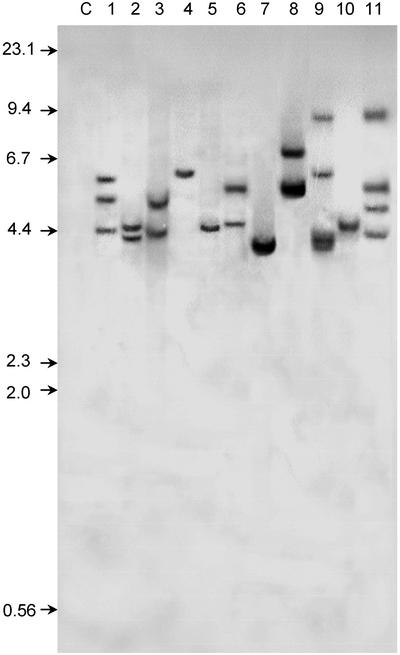
To define the position and function of cis-sequences that regulate the AtHsp90-1 expression, we constructed a series of 5′ and internal deletions of the upstream promoter region and transcriptionally fused them to the GUS reporter gene. Thus, nine constructs (pK1445, pK1137, pK846, pK653, pK473, pK173, pKΔ190, pKΔ481, and pKΔ671) were generated and introduced into Arabidopsis plants via Agrobacterium tumefaciens-mediated transformation (Fig. 1B). In addition, plasmids pBI121 and pBI101.1 were also used to transform Arabidopsis as positive and negative control, respectively. The number of transgene loci was estimated by Southern-blot analysis of HindIII-digested genomic DNA. Five independent T2 transgenic lines from each construct were analyzed and hybridized to a gusA-specific probe. Because HindIII cuts only once into the T-DNA region (except in construct pK1445), each band on the Southern blot most likely represents a single integration event. Figure 2 shows a DNA blot of two independently transformed lines harboring constructs pK173, pK473, pK653, pK846, and pK1137. Three plants (Fig. 2, lane 4, pK473-2; lane 5, pK653-1; and lane 10, pK1137-2) contain one copy of the chimeric construct, four plants (Fig. 2, lane 2, pK173-2; lane 3, pK473-1; lane 6, pK653-2; and most likely lane 7, pK846-1) contain two copies, and two plants (Fig. 2, lane 1, pK173-1; and lane 8, pK846-2) contain three copies of the T-DNA region. A maximum of four copies of the chimeric construct were found in plant pK1137-1 (Fig. 2, lane 9) and in the plant transformed with the pBI121 vector (Fig. 2, lane 11). Similar analysis performed on five independent transgenic plants harboring constructs pK1445, pKΔ190, pKΔ481, and pKΔ671 also revealed a maximum of three to four copies inserted into the plant genome (data not shown).
Figure 2.
Southern-blot analysis of transgenic Arabidopsis lines. Genomic DNA (3 μg per lane) from two independently transformed Arabidopsis plants with pK173 (lanes 1 and 2), pK473 (lanes 3 and 4), pK653 (lanes 5 and 6), pK846 (lanes 7 and 8), pK1137 (lanes 9 and 10), pB121 (lane 11), and from untransformed plants (lane C) was digested with HindIII. DNA restriction fragments were separated on a 0.8% (w/v) agarose gel, transferred to nylon membranes, and hybridized with the 32P-labeled GUS probe. The analysis demonstrated that each of the lines was the result of one to four integration events. Numbers to the left are molecular mass standards in kilobases.
Transcriptional Regulation of the AtHsp90-1 Promoter in Response to Heat Shock
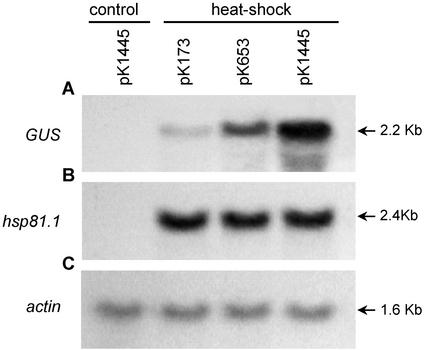
To show that GUS activity is regulated at the transcriptional level and is directly dependent on the length of the AtHsp90-1 promoter or the number of the cis-stress elements within, transgenic plants were monitored for their ability to express the AtHsp90-1::GUS mRNA under normal and heat shock conditions. Ten independent transgenic lines harboring constructs pK1445, pK653, and pK173 were heat shocked, pooled, and total RNA was isolated. As a control, total RNA from pooled, nonheat-shocked pK1445 plants was used. RNA blots were hybridized to gusA- or AtHsp90-1 gene-specific probe. Under normal environmental conditions, the endogenous AtHsp90-1 and pK1445-mRNAs (GUS mRNA) could not be detected. However, a dramatic difference in gene expression was observed when transgenic plants were heat shocked (Fig. 3). The transcript levels of the endogenous AtHsp90-1 gene were strongly increased and were shown to be similar in all three constructs. Nevertheless, AtHsp90-1::GUS transcripts showed a construct-dependent expression pattern in transgenes. GUS mRNA levels decrease 3- or 10-fold in pK653 or pK173 transgenic lines, respectively, when compared with the full-length promoter. Taken together, the above results demonstrate that the expression levels of the transgenes vary between the constructs, showing a promoter-length and therefore a cis-stress element number-dependent (Fig. 1A) pattern, whereas endogenous AtHsp90-1 gene inducibility remains unaffected.
Figure 3.
Heat shock induction of the GUS mRNA levels in transgenic Arabidopsis seedlings was examined by northern blotting. Total RNA (20 μg per lane) was prepared from 5-d-old untreated seedlings harboring the pK1445 construct and seedlings heat-shocked for 1 h at 37°C bearing the pK173, pK653, and pK1445 constructs. Blots were hybridized to the radioactively labeled GUS-coding region and to an AtHsp90-1-specific probe. Equivalent loading of RNA was assessed by actin hybridization. Arrows and numbers indicate the position and size of the transcripts.
Promoter Activity in Control and Heat Shock-Treated Plants
To investigate the contribution of specific regulatory sequences in gene expression under normal and stress conditions, the series of 5′ and internal deletions of the AtHsp90-1 promoter was used (Fig. 1B). The temporal and spatial distribution of AtHsp90-1 promoter-driven gene expression was investigated in in vitro and greenhouse-grown Arabidopsis T2 transformants.
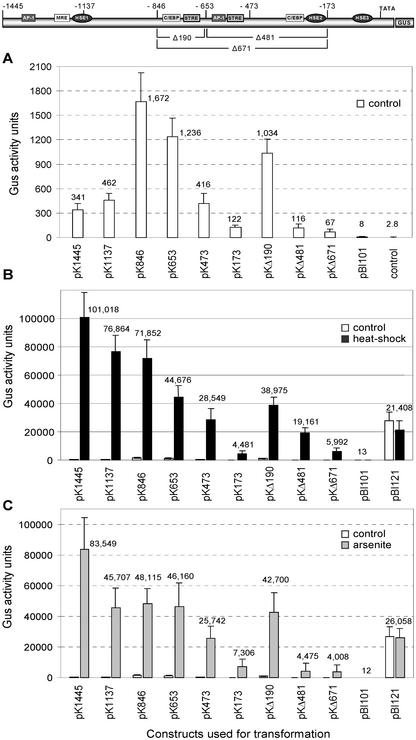
The levels of GUS activity were assayed quantitatively in eight to 12 independently transformed young seedlings. The results showed that plants harboring constructs pK1445 and pK1137 displayed very low GUS activity under normal environmental conditions. However, deletion of the promoter to point −846 resulted in a 5-fold increase in GUS activity (Fig. 4A). Furthermore, under the same conditions, constructs pK653 and pKΔ190 showed a 3-fold increase in expression when compared with the full-length promoter. Further deletion of the promoter to point −473 results in similar expression levels found with the full-length promoter. Arabidopsis plants transformed with the promoterless plasmid pBI101 and untransformed control plants showed negligible activity (Fig. 4A).
Figure 4.
GUS activity in Arabidopsis plants transformed with AtHsp90-1 promoter deletion constructs under physiological (A), heat shock conditions (B), and after treatment with arsenite (C). Tissues were harvested from eight to 12 independent transgenic lines grown in vitro. Fluorometric GUS assays were performed in triplicate and the mean value was calculated for each construct. As a negative control, extracts from nontransgenic seedlings and from seedlings transformed with the promoterless pBI101 vector were assayed. As a positive control, pBI121 transgenic plants were assayed. Error bars represent the se. White bars, Unstressed conditions; black bars, heat shock conditions; gray bars, arsenite treatment. One unit = 1 pm 4-methylumbelliferone (4-MU) produced min−1 mg−1 protein.
We have previously shown substantial elevated transcript levels of the AtHsp90-1 gene from Arabidopsis under heat shock, whereas no signal was detectable in northern blots containing RNA from plants growing at normal conditions (Milioni and Hatzopoulos, 1997). Figure 4B summarizes the fluorometric GUS activity assayed in young heat shock-treated seedlings. Plants carrying the full-length promoter, construct pK1445, showed that expression levels increase 300-fold (101,018 units) after 1 h at 37°C. Deletion of the promoter to point −1137 (pK1137) results in almost 24% reduction of gene expression (from 101,018 to 76,864 units). The deleted region contains one HSE (HSE1), one AP-1-like, and one MRE-like-binding element. As anticipated, deletion of the promoter to point −846 (pK846) had only a minor effect in gene expression (6% reduction) when compared with construct pK1137 because the deleted region does not contain any sequences resembling a consensus HSE or other potential transcription factor-binding sites. Further deletion of the promoter to point −653 (pK653) abolishes one STRE and one CAATT box-binding site, resulting in a decrease in gene expression of about 38% (from 71,852 to 44,676 units) compared with pK846 or by 56% relative to the full-length promoter.
Consistent with the above observation, transgenic plants harboring the internal deletion construct pKΔ190, which also lacks these STRE and CAATT box-binding sites, showed similar expression levels (Fig. 4B). Deletion of the promoter to point −473 (pK473) results in a further reduction in gene expression by 36% (from 44,676 to 28,549 units) relative to pK653 or by 76% relative to the full-length promoter. The deleted region contains one AP-1 like-binding site and one STRE element positioned next to each other. Further reduction of the promoter size to 173 bp (construct pK173) results in abolishing HSE2 and the upstreamlocated CAATT box element at position −316, resulting in a dramatic decrease in gene expression by 84% (from 28,549 to 4,481 units). Transgenic plants harboring only 173 bp of the AtHsp90-1 promoter show relatively low expression levels (about 4%) of the reporter gene when compared with the full-length promoter. Even so, this is a 37-fold increase in GUS activity compared with the nontreated control plants. As anticipated, seedlings transformed with construct pKΔ671 showed similar expression levels to those transformed with construct pK173. However, the presence of the STRE and the CCAAT-box element (at position −731 and −798, respectively) in construct pKΔ481 results in at least 3-fold increase in GUS activity compared with construct pKΔ671 (Fig. 4B).
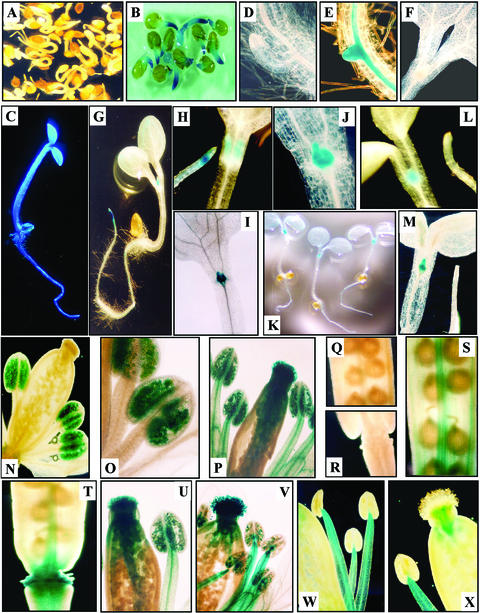
Histochemical analysis of seedlings and mature plants harboring construct pK1445, cultivated under control conditions, revealed no detectable GUS activity, with the exception of mature pollen grains (Fig. 5, N and O). In contrast, high levels of AtHsp90-1 promoter activity were detected in all tissues of heat shock-treated seedlings mature plants, as evidenced by strong blue GUS staining (Fig. 5, C, P, S, and T). Reporter gene activity was prominent in the root meristematic region of germinated seeds (Fig. 5B). In 5-d-old seedlings, preferential high levels of GUS activity were observed in the vascular system of the root and in the emerging secondary root primordia 15 min after staining (Fig. 5E). Progressive staining of the cortex and the epidermis of the root was obvious after 1 to 2 h (Fig. 5C). Plants carrying progressive deletions of the promoter showed in general a reduction in GUS staining pattern in most tissues (data not shown). However, construct pK173 (containing only HSE3) showed significant levels of GUS activity localized predominantly in the shoot and root meristematic zones (Fig. 5, G–J). In mature stressed plants, high levels of expression were seen in almost all parts of the developing flower, including the stigma, the anther, and the filaments (Fig. 5P). Nevertheless, the analysis revealed an unexpected differential expression pattern in the stamen of the developing flower. Progressive deletion of the promoter results in a respective decrease in GUS expression in the pistil (stigma, style, and ovary) and in the pollen grains of the anther (Fig. 5, U–X). Hence, plants harboring constructs pK173 show very low GUS activity in the style of the pistil and almost no activity in the stigma, the ovary, and the pollen grains of the anther (Fig. 5, W–X). However, it is interesting that GUS activity remains the same in the filament of the anther in all constructs (Fig. 5, P and U–W).
Figure 5.
Reporter gene expression patterns. Histochemical analysis of GUS activity during vegetative and reproductive growth of transgenic plants. Arabidopsis plants transformed with pK1445 (A–E) and pK173 (F–M) constructs. A, Nontreated germinating seedlings. B, Heat-shocked germinating seedlings. C, Heat-shocked 5-d-old transgenic seedling. D and E, Nontreated and heat-shocked seedling showing a meristematic region initiating a lateral root. F, Close-up on shoot meristematic zone in 5-d-old unstressed plants bearing the pK173 construct. G through J, Activity in shoot and root meristematic zones in 5-d-old heat-shocked seedlings. I and J, Close-up on shoot meristematic zone in 5-d-old heat-shocked transgenic seedlings. K, Seedlings 5 d after germination transformed with pK173 construct and treated with arsenite. L and M, Close-up on shoot and root meristematic regions in 5-d-old arsenite-treated transgenic seedlings. N through P, Mature flowers from unstressed (N and O) and heat-shocked transgenic plants (P) transformed with construct pK1445. Q through T, Detail of the upper part of early developing siliques from unstressed plants (Q and R) and from heat-shocked plants (S and T). U through X, Developing flowers of heat-shocked plants transformed with the pK846 (U) and pK473 (V) constructs. W and X, GUS activity is restricted in filaments and the style in heat-shocked transgenic plants bearing construct pK173.
Promoter Activity in Arsenite-Treated Plants
Heavy metal toxicity is well known to trigger HSP induction. In several species, for example, cadmium induces the synthesis of a considerable number of stress proteins with a molecular mass ranging from 10 to 70 kD. However, little is known about the induction of AtHsp90 genes by arsenite in plants. To carry out a comparative analysis of the AtHsp90-1 promoter to the heat shock response, we investigated the expression of the GUS reporter gene after exposing the plants to 10 mm arsenite for 6 h. Depending on the construct, eight to 12 independent transformants were obtained and examined for GUS activity by quantitative assays and histochemical staining.
Figure 4C summarizes the expression levels measured in all deletion constructs, showing that the AtHsp90-1 promoter responds tremendously to arsenite. Transgenic plants harboring the full-length promoter construct (pK1445) showed a 245-fold (83,500 units) increase in gene expression when compared with the untreated control plants. Deleting the promoter region −1,445 to −1,137 (construct pK1137) significantly affects gene expression because GUS activity drops by 45% (from 83,500 to 45,707 units). The deleted region contains, apart from HSE1, one putative AP-1-like and one MRE-like-binding site. This decline in gene expression is considerably higher than the one observed with heat shock (21%). Deletion of the promoter to point −846 does not affect gene expression, similar to the heat shock treatment. However, it is surprising that GUS activity does not decrease even after deleting the promoter to point −653, which abolishes the CCAAT-box and STRE-binding sites located at position −798 and −731, respectively. In agreement with the above observation, transgenic plants harboring construct pKΔ190 (lacking also the above CCAAT-box and STRE) showed comparable expression levels. By deleting the promoter up to −473, gene expression declines further by 44% (from 46,160 to 25,742 units). It is interesting that this significant reduction in gene expression is due to the deletion of the 180-bp AatII/PvuII fragment, which contains the STRE and AP-1-like-binding sites at position −612 and −618, respectively. Gene expression levels decrease further by 28% (from 25,742 to 7,306 units) when the promoter is deleted up to point −173. This decline in gene expression is due to the deletion of the 300-bp PvuII/EcoRI fragment containing the CCAAT-box and HSE2-binding sites at position −316 and −183, respectively. However, arsenite-treated plants harboring construct pK173 demonstrate a 60-fold induction in gene expression when compared with the untreated control plants. Construct pKΔ481 and pKΔ671 showed similar GUS expression levels, which are comparable with the levels of construct pK173. pKΔ481 and pKΔ671 lack the region containing the HSE2, the STRE, and the AP-1-like-binding site at position −187, −612, and −618, respectively. It is interesting that the presence of CCAAT-box and STRE elements (at position −798 and −731, respectively) in construct pKΔ481 does not affect the level of induction (Figs. 1B and 3B).
In all constructs (except pK173 and pKΔ671), histochemical staining of germinating and young seedlings treated with arsenite revealed a similar pattern to that seen with the heat shock-treated plants (data not shown). However, construct pK173 and pKΔ671 showed an “arsenite-specific” differential expression pattern in the two meristems of the plants. Whereas in these constructs heat shock induces GUS expression in both meristems (Fig. 5, G–J), arsenite remarkably triggers expression only in the shoot and not in the root meristem (Fig. 5, K–M).
DISCUSSION
We have previously shown (Milioni and Hatzopoulos, 1997) that the AtHsp90-1 gene was highly heat-inducible in Arabidopsis plants, whereas the transcripts were undetectable in the absence of any stress condition. As a step forward in understanding regulatory mechanisms controlling AtHsp90-1 gene expression, we have examined the temporal and spatial expression of the AtHsp90-1 during development and following heat or arsenite stress. A series of promoter deletion constructs were generated and transcriptionally fused to the GUS reporter gene to quantitatively and qualitatively monitor gene expression. Our results demonstrate that the AtHsp90-1 promoter from Arabidopsis is highly induced by heat and arsenite treatment, involving presumably a number of regulatory sequences such as HSEs, STREs, AP-1, or MRE-binding sites. However, analysis of the deletion constructs indicates that the two pathways may involve common but not necessarily the same regulatory sequences. Furthermore, the two responses (heat shock and arsenite) most likely implicate additional regulatory elements (MRE and CCAAT-box) and/or tissue-specific components.
Computational analysis of the AtHsp90-1 promoter revealed several cis-regulatory elements known to be involved in a number of stress responses in different organisms. These include consensus sequences for HSE, STRE, and MRE, as well as CCAAT-box and AP-1-binding elements (Fig. 1). This is the first report, to our knowledge, of an AtHsp90 promoter presumably involving a number of different stress regulatory sequences for gene induction under different environmental conditions. It is worth mentioning that G-box-like motifs (CACGTG) and scaffold-attachment regions, which are required for expression of genes induced by stress, were not identified in the promoter of the AtHsp90-1 gene.
In the absence of any stress condition, the AtHsp90-1 gene is barely expressed (Yabe et al., 1994; Milioni and Hatzopoulos, 1997). Consistent with these reports, the full-length AtHsp90-1 promoter displays relatively low GUS activity levels (Fig. 4A). However, the 5-fold increase in GUS activity of construct pK846 indicates the existence of upstream regulatory elements that suppress gene expression in vivo. Although the 846 bp of the promoter region could interact with regulatory proteins to form appropriate complexes for the induction of the AtHsp90-1 gene, the existence of further upstream sequences (−1,445 to −846) seems to mask this induction under normal environmental conditions. To envisage such a phenomenon in vivo, one possibility could be the folding of the DNA in chromatin in such a way that brings upstream sequences (−1,445 to −846) close to the basal transcriptional apparatus.
Heat shock results in a tremendous 300-fold increase in the reporter gene expression. It is known that transcriptional activation of heat shock genes depends on the interaction of HSFs with highly conserved cis-acting DNA sequences, the HSEs. All HSEs contain multiple units of the repeating 5-bp consensus sequence 5′-nGAAn-3′ arranged in head-to-head or head-to-tail orientation (Amin et al., 1988; Xiao and Lis, 1988). Although at least three units are thought to be required for heat inducible expression, the degree of homology of each pentameric unit to the consensus motif can vary. Mutational analysis of plant heat shock elements revealed that the G/C bp (G and complementary C) at position one of the unit is more important than the A/T base in the third position (Barros et al., 1992). In addition to HSEs, a number of sequence motifs were found to have quantitative effects on the expression of certain heat shock genes. An interaction of C/EBP and HSF, bound to their respective cis elements, has been postulated to be required for maximum stress-induced transcription from human hsp70 promoters (Williams and Morimoto, 1990). In plants, there is evidence for the involvement of CCAAT-box elements, HSEs, and scaffold-attachment regions in stress-induced transcription (Rieping and Schöffl, 1992; Schöffl et al., 1993). Furthermore, STREs are known to activate transcription in response to a variety of stress conditions, especially heat (Siderius and Mager, 1997). In the AtHsp90 -1 promoter, HSE1 and HSE2 represent a perfect HSF-binding site, whereas HSE3 deviates from the consensus sequence only at the second core unit (nATCn instead of nTTCn) (Fig. 1). However, in the absence of any other putative transcription factor-binding site, HSE3 (construct pK173) is able to drive a 37-fold increase in gene expression under heat stress. The presence of HSE2, HSE3, and the upstream located CCAAT-box at position −316 (construct pK473) results in a 69-fold increase of GUS activity, indicating an additive effect of the two HSEs and/or the CCAAT-box element in gene expression.
Despite its very distal position (−1,144), HSE1 represents a perfect consensus HSF-binding site and seems to be required for full promoter activity. However, the involvement of other sequences (MRE- and/or AP-1-like elements), located upstream of HSE1, to assist the enhancement of gene expression cannot be excluded and remains to be tested. It is interesting that a promoter region from −1,137 to −203 does not contain any sequences resembling an HSF-binding consensus sequence. Nevertheless, region −846 to −653 and region −653 to −473 contain one STRE-binding site (Fig. 1). Taking in account that the CCAAT-box element itself does not respond to heat, the significant decline in gene expression in construct pK653 and pK473 could be due to the deletion of these STREs. The AP-1 element, located at position −612 (pK653), may also be involved in enhancing gene expression. Consistent with the above results, construct pKΔ190 showed similar expression levels to construct pK653, indicating that sequences within the Tth111I/AatII fragment (presumably the STRE) are positive determinants of gene expression following heat stress. Hence, full-length AtHsp90-1 promoter activity requires the presence of all cis elements, HSEs, STREs, AP-1-like, and CCAAT-boxes, indicating a synergistically mode of action in promoting high levels of gene expression.
The heat shock response and the arsenite-induced stress share many features at the molecular level. Both phenomena induce HSPs ranging from the very small αB-crystallin to the larger HSPs, such as HSP105. The central component of the heat shock response is oxidative stress, which in fact is also a typical arsenite-related effect (Bernstam and Nriagu, 2000). These stimuli lead to up-regulation of HSF phosphorylation and hence HSP induction. However, it is suggested that the pathways of HSF phosphorylation induced by heat or arsenite are different, implying distinct mechanisms of transcriptional control (Elia et al., 1996). AP-1-binding elements have been implicated in CdCl2 and arsenite induction (Alam, 1994; Lu et al., 1998), whereas STREs and AP-1 elements are involved in responses to a range of stresses in yeast (Saccharomyces cerevisiae; Ruis and Schuller, 1995). Furthermore, MREs have been identified in a number of heavy metal-induced promoters such as the human and mouse metallothionein genes (Karin et al., 1987; Culotta and Hamer, 1989), the tomato type II metallothionein-like gene (Whitelaw et al., 1997), and the mouse and chicken heme oxygenase genes (Alam, 1994; Lu et al., 1998). However, the involvement of AP-1- and/or MRE-binding sites in the expression of an hsp90 gene has not yet been shown in plants.
Our results indicate the involvement of additional distinct regulatory elements, apart from the HSEs, in mediating the arsenite-related response. The full-length promoter of the AtHsp90-1 gene is highly respondent to arsenite (Fig. 4C). GUS activity decreases by 45% when the region −1,445 to −1,137 (containing, apart from HSE1, one AP-1-like and one MRE-like-binding element) is deleted (Fig. 4). It is interesting that this decline in gene expression is about 21% higher than the one observed with heat shock. In animal systems, Lu et al. (1998) have shown that sodium arsenite treatment increases nuclear protein binding to an AP-1 element. Therefore, it is possible that the considerable decline of gene expression in construct pK1137 is due to the combined deletion of HSE1 and the AP-1 like sequence. Furthermore, the upstream located MRE-like-binding site, in complex with its corresponding factor, may also be involved in a “crosstalk” interaction with HSF- and/or AP-1 like-binding factor. Because the imperfect heat shock element HSE3 by itself or HSE3 and HSE2 contribute to arsenite induction, it is expected that the canonical HSE1 should also contribute. In a similar manner, because the AP-1 element at position −612 contributes to arsenite induction (see later), it is highly plausible that the other AP-1 elements present in the promoter region −1,445 to −1,137 should also contribute. Whether the AP-1 or the MRE-like element or both contribute more to arsenite treatment than that of the HSE1 (all present in −1,445 to −1,137 promoter region) is unknown. However, the functionality of these sequences enhancing or regulating differential gene expression under arsenite treatment remains to be tested. Because deletion of the STRE and CCAAT-box-binding site at position −731 and/or −793, respectively, in two independent constructs (pK653 and pKΔ190) does not affect gene expression in arsenite-treated plants, we therefore assume that the STRE and C/EBP elements present in the promoter region −653 to −473 and −473 to −173, respectively, do not contribute to arsenite induction. Deletion of the AP-1-like-binding site at position −612 (construct pK473) strongly reduces expression by 44% (Fig. 4C). Taken together, these results indicate that the HSEs, the AP-1 elements, and probably the MRE in the AtHsp90-1 promoter are presumably positive determinants of gene expression under arsenite treatment.
GUS staining of unstressed mature plants transformed with the full-length promoter revealed detectable levels of expression only in pollen grains of the anthers (Fig. 5, N and O). This observation is consistent with previous findings in maize (Zea mays; Marrs et al., 1993; Magnard et al., 1996), indicating the significance of chaperones and in particular a prominent role of the AtHsp90-1 in Arabidopsis pollen development. After heat shock, GUS staining was prominent in all parts of the developing flower (Fig. 5P). However, the deletion of the promoter toward 3′ showed an interesting tissue-specific expression pattern. Although progressive deletion results in a respective decline in GUS expression in the pistil and the pollen grains of the anther, the expression in the filaments of the stamen remains unaffected. This observation is more profound in transformed plants carrying 473- and 173-bp upstream promoter sequences (Fig. 5, V and W). Taken together, the above results indicate that “filament-specific” sequences are most likely located proximally within the 173 bp of the promoter and that distal pollen-specific sequences are necessary for the expression of the AtHsp90-1 gene in pollen grains irrespective of the heat shock. On the other hand, GUS staining was prominent in all tissues in heat-shocked transgenic seedlings carrying the pK1445 construct (Fig. 5, C and P). Progressive deletions from the 5′ end of the promoter resulted in respectively lower expression levels (data not shown). Although GUS activity dropped to undetectable levels in most tissues, pK173 transgenic seedlings (containing only HSE1) showed relatively high levels of GUS activity in the shoot and the root apical meristems. Because meristematic cells are known to have the highest rate of cell divisions, it is reasonable to assume that heat stress may be most detrimental to rapidly dividing cells. Thus, the accumulation of GUS protein in these most vital parts of the seedling may reflect the significant role of AtHsp90-1 as a chaperone.
GUS staining of young seedlings and mature plants treated with arsenite revealed a similar pattern to that seen with the heat shock-treated plants (data not shown). Nevertheless, sequences within the 173-bp promoter region, bearing HSE3, direct differential gene expression under arsenite treatment. As mentioned above, heat shock treatment results in high GUS expression in both meristems of the plant. However, arsenite directs gene expression specifically in the shoot and not in the root meristem (Fig. 5, K–M). This result indicates that additional unidentified regulatory elements located within the 300-bp PvuII/EcoRI region are necessary in driving “root-specific” gene expression under arsenite treatment.
Therefore, our results indicate that the combinatorial contribution of a number of different cis elements in the promoter region of the AtHsp90-1 gene is important in specifying suppression, developmental, or tissue-specific expression and stress induction. In this context, knowledge of the AtHsp90-1 promoter elements and their associated regulatory proteins may eventually lead to a better understanding of the regulatory mechanisms controlling AtHsp90 gene expression under various environmental conditions.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Landsberg erecta) plants were used in all transformation experiments. Wild-type and transgenic plants were grown under standard conditions at 22°C in 70% humidity with a light/dark cycle of 16 h/8 h. Seeds from individual transgenic plants were imbibed at 4°C overnight, and were surface sterilized for 2 min with 70% (v/v) ethanol and for 5 min with 15% (v/v) sodium hypochlorite containing 0.1% (v/v) Tween 20. After several washing steps with sterile deionized water, seeds were germinated on Murashige and Skoog medium containing 50 mg L−1 kanamycin and 200 mg L−1 cefotaxime under the same growth conditions. Transgenic plants were transferred to soil for further development.
Plasmid Construction and Plant Transformation
A 1.9-kb SacI genomic fragment containing approximately 1,800 bp of the regulatory sequence (AJ010947) and 100 bp of the 5′ coding region of the Arabidopsis AtHsp90-1 gene (Milioni and Hatzopoulos, 1997) was cloned into the SacI site of the pUC19 vector. This plasmid was used to remove the initiation of translation start codon (ATG) from the native AtHsp90-1 gene by exonuclease III digest using the SalI/SphI restriction sites. After SI nuclease treatment and recircularization of the plasmids, the starting point of the deletions was determined by dideoxy-nucleotide sequencing using the Sequenase 2.0 sequencing kit. Furthermore, approximately 1,500 bp of the promoter region of the AtHsp90-1 gene was sequenced. Routine DNA manipulations were carried out (Sambrook et al., 1989) using pUC19 (New England Biolabs, Beverly, MA) and pBluescript SK (Stratagene, La Jolla, CA) as intermediate vectors to obtain appropriate fragment lengths of the promoter and convenient restriction sites for directional cloning into the pBI121 (CLONTECH, Palo Alto, CA) binary vector (details available upon request). 5′ end deletions were made by digesting the promoter with different restriction enzymes (partial HindIII digest for deletion to point −1,445, HindIII digest for deletion to point −1,137, Tth111I/PstI digest for deletion to point −846, AatII/PstI digest for deletion to point −653, PvuI/BamHI digest for deletion to point −473, and EcoRI digest for deletion to point −173). Furthermore, three internal deletions of 190, 481, and 671 bp were obtained by using appropriate restriction enzyme combinations. Constructs generated in the pUC19 or pBluescript SK background were cloned upstream of the GUS reporter gene of the pBI121 binary vector by replacing the 35S-cauliflower mosaic virus promoter. In this way, the constructs pK1445, pK1137, pK846, pK653, pK473, pK173, pKΔ190, pKΔ481, and pKΔ671 were generated (Fig. 1B). Plasmids pBI121 and pBI101.1 (CLONTECH) were used as positive and negative controls, respectively. The binary vector constructs were introduced into the Agrobacterium tumefaciens strain C58C1::pGV2260 by the direct transfer method (An et al., 1988). Arabidopsis (Landsberg erecta) plants were transformed by using the in planta A. tumefaciens infiltration method as described (Bechtold et al., 1993).
Heat Stress and Arsenite Treatment
Transgenic T2 plants were germinated on Murashige and Skoog medium plates containing 50 mg L−1 kanamycin. Five-day-old seedlings and flowering plants were heat shocked for 1 h at 37°C. Five-day-old seedlings were incubated at 22°C for 6 h in liquid Murashige and Skoog medium containing 10 mm Na2HAsO4.7H2O. After each treatment, the material was frozen in liquid nitrogen and kept at −80°C until further use or was treated to histochemical GUS staining.
Southern- and Northern-Blot Analysis
Genomic DNA, isolated from T2 Arabidopsis plants using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA), was digested with the restriction enzyme HindIII and fractionated on a 0.8% (w/v) agarose gel (3 μg per lane). DNA denaturation, transfer onto Hybond N+ nylon membrane (Amersham Biosciences, Piscataway, NJ), and UV-cross-linking were performed as described (Sambrook et al., 1989). Hybridization was carried out with the [α-32P]-labeled gusA-specific probe under high stringency conditions at 65°C (Church and Gilbert, 1984). Total RNA was isolated from control and transgenic plants using a modified phenol-chloroform extraction procedure. One gram of frozen tissues was ground in liquid nitrogen, resuspended in 2 mL of homogenization buffer (100 mm Tris-HCl, pH 9, and 5% [w/v] SDS) and 2 mL of phenol, mixed well, and centrifuged for 5 min at 10,000g. The aqueous phase was removed, and was extracted twice with phenol/chloroform/isoamyl alcohol (25:24:1) and once with chloroform/isoamyl alcohol (24:1). Nucleic acids were precipitated with 2 volumes of ethanol and one-tenth volume 3 m sodium acetate, and were resuspended in sterile deionized water. RNA concentration was determined spectrophotometrically and was verified by ethidium bromide staining on agarose gels. The RNAs (20 μg per lane) were electrophoresed on 1.4% (w/v) denaturing formaldehyde-agarose gels and transferred without any further treatment onto Hybond N+ nylon membranes. After immobilization by UV cross-linking, the blots were hybridized with the [α-32P]-labeled gusA-specific probe or with [α-32P]-labeled 0.7-kb HindIII/EcoRI fragment specific for the AtHsp90-1 gene under high stringency conditions at 65°C (Church and Gilbert, 1984). As a control, actin gene from pea (Pisum sativum) was hybridized to northern blot.
Fluorometric and Histochemical GUS assays
Quantitative GUS assays were carried out essentially as described by Jefferson et al. (1987) on T2 transgenic plants. Young seedlings were homogenized in 50 μL of ice-cold phosphate buffer (50 mm sodium phosphate, pH 7, 40 mm 2-mercaptoethanol, and 10 mm Na2EDTA). Samples were centrifuged for 5 min at 4°C and GUS activity was measured using standard conditions and buffers containing 4-methylumbelliferyl-β-d-glucuronide (Sigma, St. Louis) with a fluorometer (LS50B; PerkinElmer Instruments, Norwalk, CT). Standard curves were prepared with 4-MU (Sigma). Specific GUS activity is shown in units of nanomoles 4-MU produced per milligram of protein per minute. All measurements were repeated three times on eight to 12 independently transformed plants from each construct.
Histochemical staining for GUS activity was performed in seedlings and flower parts at different stages of plant development using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-gluc) as a substrate (Jefferson et al., 1987). Tissues were stained for 2 h (or less if otherwise stated) at 37°C in X-gluc reaction buffer (50 mm sodium phosphate buffer, pH 7.2, 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, and 2 mm X-gluc), dehydrated by series of ethanol washes, and kept in 3.7% (w/v) formaldehyde, 50% (w/v) ethanol, and 5% (w/v) acetic acid at 4°C before being photographed.
Footnotes
This work was supported by the General Secretariat of Research and Technology, Greece (grant no. 91/910 to P.H.). K.H. and S.R. were supported by State Foundation Scholarships, Greece.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.004044.
LITERATURE CITED
- Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for Il-6 expression (Nf-Il6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam J. Multiple elements within the 5′ distal enhancer of the mouse heme oxygenase-1 gene mediate induction by heavy metals. J Biol Chem. 1994;269:25049–25056. [PubMed] [Google Scholar]
- Amin J, Ananthan J, Voellmy R. Key features of heat-shock regulatory elements. Mol Cell Biol. 1988;8:3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Edbert PR, Mitra A, Ha SB. Binary vectors. In: Gelvin SB, Shilperoort RA, Verma DPS, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. pp. 1–9. [Google Scholar]
- Angel P, Imagawa M, Chiu R, Stein B, Imbra RJ, Rahmsdorf HJ, Jonat C, Herrlich P, Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- Barros MD, Czarnecka E, Gurley WB. Mutational analysis of a plant heat-shock element. Plant Mol Biol. 1992;19:665–675. doi: 10.1007/BF00026792. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Comptes Rendus Acad Sci Ser III-Sci Vie-Life Sci. 1993;316:1194–1199. [Google Scholar]
- Bernstam L, Nriagu J. Molecular aspects of arsenic stress. J Toxicol Environ Health Crit Rev. 2000;3:293–322. doi: 10.1080/109374000436355. [DOI] [PubMed] [Google Scholar]
- Boston RS, Viitanen PV, Vierling E. Molecular chaperones and protein folding in plants. Plant Mol Biol. 1996;32:191–222. doi: 10.1007/BF00039383. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Chinn AM, Comai L. The heat shock cognate 80 gene of tomato is flanked by matrix attachment regions. Plant Mol Biol. 1996;32:959–968. doi: 10.1007/BF00020492. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, Schnaider T, Soti C, Prohaszka Z, Nardai G. The 90-kDa molecular chaperone family: structure, function, and clinical applications: a comprehensive review. Pharmacol Ther. 1998;79:129–168. doi: 10.1016/s0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- Culotta VC, Hamer DH. Fine mapping of a mouse metallothionein gene metal response element. Mol Cell Biol. 1989;9:1376–1380. doi: 10.1128/mcb.9.3.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czar MJ, Galigniana MD, Silverstein AM, Pratt WB. Geldanamycin, a heat shock protein 90-binding steroid-dependent translocation of the glucocorticoid receptor from the cytoplasm to the nucleus. Biochemistry. 1997;36:7776–7785. doi: 10.1021/bi970648x. [DOI] [PubMed] [Google Scholar]
- Dhaubhadel S, Chaudhary S, Dobinson KF, Krishna P. Treatment with 24-epibrassinolide, a brassinosteroid, increases the basic thermotolerance of Brassica napus and tomato seedlings. Plant Mol Biol. 1999;40:333–342. doi: 10.1023/a:1006283015582. [DOI] [PubMed] [Google Scholar]
- Elia G, DeMarco A, Rossi A, Santoro MG. Inhibition of HSP70 expression by calcium ionophore A23187 in human cells: an effect independent of the acquisition of DNA-binding activity by the heat shock transcription factor. J Biol Chem. 1996;271:16111–16118. doi: 10.1074/jbc.271.27.16111. [DOI] [PubMed] [Google Scholar]
- Garcia-Cardena G, Fan R, Shah V, Sorrentino R, Cirino G, Papapetropoulos A, Sessa WC. Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature. 1998;392:821–824. doi: 10.1038/33934. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C, Welch WJ. Role of the major heat-shock proteins as molecular chaperones. Annu Rev Cell Biol. 1993;9:601–634. doi: 10.1146/annurev.cb.09.110193.003125. [DOI] [PubMed] [Google Scholar]
- Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, Trager JB, Morin GB, Toft DO, Shay JW et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Haslinger A, Heguy A, Dietlin T, Cooke T. Metal-responsive elements act as positive modulators of human metallothionein-IIA enhancer activity. Mol Cell Biol. 1987;7:606–613. doi: 10.1128/mcb.7.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning AJ, Rose R, Comai L. Developmental expression of tomato heat-shock cognate protein 80. Plant Physiol. 1992;100:801–811. doi: 10.1104/pp.100.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P, Gloor G. The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperon. 2001;6:238–246. doi: 10.1379/1466-1268(2001)006<0238:thfopi>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P, Sacco M, Cherutti JF, Hill S. Cold-induced accumulation of hsp90 transcripts in Brassica napus. Plant Physiol. 1995;107:915–923. doi: 10.1104/pp.107.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Lu TH, Lambrecht RW, Pepe J, Shan Y, Kim T, Bonkovsky HL. Molecular cloning, characterization, and expression of the chicken heme oxygenase-1 gene in transfected primary cultures of chick embryo liver cells. Gene. 1998;207:177–186. doi: 10.1016/s0378-1119(97)00623-9. [DOI] [PubMed] [Google Scholar]
- Ludwig-Muller J, Krishna P, Forreiter C. A glucosinolate mutant of Arabidopsis is thermosensitive and defective in cytosolic hsp90 expression after heat stress. Plant Physiol. 2000;123:949–958. doi: 10.1104/pp.123.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnard JL, Vergine P, Dumas C. Complexity and genetic variability of heat-shock protein expression in isolated maize microspores. Plant Physiol. 1996;111:1085–1096. doi: 10.1104/pp.111.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs KA, Casey ES, Capitant SA, Bouchard RA, Dietrich PS, Mettler IJ, Sinibaldi RM. Characterization of two maize hsp90 heat-shock protein genes and expression during heat-shock, embryogenesis, and pollen development. Dev Genet. 1993;14:27–41. doi: 10.1002/dvg.1020140105. [DOI] [PubMed] [Google Scholar]
- Marrs KA, Sinibaldi RM. Deletion analysis of the maize hsp82, hsp81, and hsp17.9 promoters in maize and transgenic tobacco: contributions of individual heat shock elements and recognition by distinct protein factors during both heat shock and development. Maydica. 1997;42:211–226. [Google Scholar]
- Miernyk JA. Protein folding in the plant cell. Plant Physiol. 1999;121:695–703. doi: 10.1104/pp.121.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milioni D, Hatzopoulos P. Genomic organization of hsp90 gene family in Arabidopsis. Plant Mol Biol. 1997;35:955–961. doi: 10.1023/a:1005874521528. [DOI] [PubMed] [Google Scholar]
- Morimoto RI, Santoro MG. Stress-inducible responses and heat shock proteins: new pharmacologic targets for cytoprotection. Nat Biotechnol. 1998;16:833–838. doi: 10.1038/nbt0998-833. [DOI] [PubMed] [Google Scholar]
- Nathan DF, Vos MH, Lindquist S. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci USA. 1997;94:12949–12956. doi: 10.1073/pnas.94.24.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek A, Singla SL, Grover A. Immunological evidence for accumulation of two high-molecular-weight (104 and 90 kDa) HSPs in response to different stresses in rice and in response to high temperature stress in diverse plant genera. Plant Mol Biol. 1995;29:293–301. doi: 10.1007/BF00043653. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Krishna P, Olsen LJ. Hsp90-binding immunophilins in plants: the protein movers. Trends Plant Sci. 2001;6:54–58. doi: 10.1016/s1360-1385(00)01843-4. [DOI] [PubMed] [Google Scholar]
- Reddy RK, Chaudhary S, Patil P, Krishna P. The 90-kDa heat shock protein (hsp90) is expressed throughout Brassica napus seed development and germination. Plant Sci. 1998;131:131–137. [Google Scholar]
- Rieping M, Schöffl F. Synergistic effect of upstream sequences, CCAAT box elements, and HSE sequences for enhanced expression of chimeric heat-shock genes in transgenic tobacco. Mol Gen Genet. 1992;231:226–232. doi: 10.1007/BF00279795. [DOI] [PubMed] [Google Scholar]
- Ruis H, Schuller C. Stress signaling in yeast. Bioessays. 1995;17:959–965. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schlessinger J, Ashburner N, Tissieres A. Heat-Shock: from Bacteria to Man. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Schmitz G, Schmidt M, Feierabend J. Characterization of a plastid-specific HSP90 homologue: identification of a cDNA sequence, phylogenetic descendence and analysis of its mRNA and protein expression. Plant Mol Biol. 1996;30:479–492. doi: 10.1007/BF00049326. [DOI] [PubMed] [Google Scholar]
- Schöffl F, Prandl R, Reindl A. Regulation of the heat-shock response. Plant Physiol. 1998;117:1135–1141. doi: 10.1104/pp.117.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöffl F, Schröder G, Kliem M, Rieping M. A SAR sequence containing 395 bp fragment mediates enhanced, gene-dosage-correlated expression of a chimeric heat shock gene in transgenic tobacco plants. Transgenic Res. 1993;2:93–100. doi: 10.1007/BF01969382. [DOI] [PubMed] [Google Scholar]
- Schröder G, Beck M, Eichel J, Vetter HP, Schröder J. HSP90 homologue from Madagascar periwinkle (Catharanthus roseus): cDNA sequence, regulation of protein expression and location in the endoplasmic reticulum. Plant Mol Biol. 1993;23:583–594. doi: 10.1007/BF00019305. [DOI] [PubMed] [Google Scholar]
- Siderius M, Mager WH. General stress response: in search of a common denominator. In: Hohmann S, Mager WH, editors. Yeast Stress Responses. Heidelberg: Springer-Verlag; 1997. pp. 213–230. [Google Scholar]
- Takahashi T, Naito S, Komeda Y. Isolation and analysis of the expression of two genes for the 81-kilodalton heat-shock proteins from Arabidopsis. Plant Physiol. 1992;99:383–390. doi: 10.1104/pp.99.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierling E. The roles of heat-shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:579–620. [Google Scholar]
- Waters ER, Lee GJ, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot. 1996;47:325–338. [Google Scholar]
- Whitelaw CA, LeHuquet JA, Thurman DA, Tomsett AB. The isolation and characterization of type II metallothionein-like genes from tomato (Lycopersicon esculentum L.) Plant Mol Biol. 1997;33:503–511. doi: 10.1023/a:1005769121822. [DOI] [PubMed] [Google Scholar]
- Williams GT, Morimoto RI. Maximal stress-induced transcription from the human hsp70 promoter requires interactions with the basal promoter elements independent of rotational alignment. Mol Cell Biol. 1990;10:3125–3136. doi: 10.1128/mcb.10.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- Xiao H, Lis JT. Germline transformation used to define key features of heat-shock response elements. Science. 1988;239:1139–1142. doi: 10.1126/science.3125608. [DOI] [PubMed] [Google Scholar]
- Yabe N, Takahashi T, Komeda Y. Analysis of tissue-specific expression of Arabidopsis thaliana HSP90-family gene HSP81. Plant Cell Physiol. 1994;35:1207–1219. doi: 10.1093/oxfordjournals.pcp.a078715. [DOI] [PubMed] [Google Scholar]