Abstract
In plants, ammonium released during photorespiration exceeds primary nitrogen assimilation by as much as 10-fold. Analysis of photorespiratory mutants indicates that photorespiratory ammonium released in mitochondria is reassimilated in the chloroplast by a chloroplastic isoenzyme of glutamine synthetase (GS2), the predominant GS isoform in leaves of Solanaceous species including tobacco (Nicotiana tabacum). By contrast, cytosolic GS1 is expressed in the vasculature of several species including tobacco. Here, we report the effects on growth and photorespiration of overexpressing a cytosolic GS1 isoenzyme in leaf mesophyll cells of tobacco. The plants, which ectopically overexpress cytosolic GS1 in leaves, display a light-dependent improved growth phenotype under nitrogen-limiting and nitrogen-non-limiting conditions. Improved growth was evidenced by increases in fresh weight, dry weight, and leaf soluble protein. Because the improved growth phenotype was dependent on light, this suggested that the ectopic expression of cytosolic GS1 in leaves may act via photosynthetic/photorespiratory process. The ectopic overexpression of cytosolic GS1 in tobacco leaves resulted in a 6- to 7-fold decrease in levels of free ammonium in leaves. Thus, the overexpression of cytosolic GS1 in leaf mesophyll cells seems to provide an alternate route to chloroplastic GS2 for the assimilation of photorespiratory ammonium. The cytosolic GS1 transgenic plants also exhibit an increase in the CO2 photorespiratory burst and an increase in levels of photorespiratory intermediates, suggesting changes in photorespiration. Because the GS1 transgenic plants have an unaltered CO2 compensation point, this may reflect an accompanying increase in photosynthetic capacity. Together, these results provide new insights into the possible mechanisms responsible for the improved growth phenotype of cytosolic GS1 overexpressing plants. Our studies provide further support for the notion that the ectopic overexpression of genes for cytosolic GS1 can potentially be used to affect increases in nitrogen use efficiency in transgenic crop plants.
Nitrogen is a costly and rate-limiting element in plant growth. Nitrogenous fertilizer accounts for 40% of costs associated with crops such as corn (Zea mays) and wheat (Triticum aestivum; Sheldrick, 1987). Increasing the efficiency of nitrogen use would be cost-effective and would minimize problems of ground water contamination by excess nitrate application (Sheldrick, 1987). Attempts to select crop plants with enhanced nitrogen use by conventional breeding strategies have been largely unsuccessful because of problems associated with screening large populations for a trait that is difficult to assess under field conditions. Plants do not seem to be limited in their ability to uptake or convert nitrate to ammonium (Crawford et al., 1986), although it does seem that some crop plants may be limited in their ability to incorporate inorganic nitrogen into protein. Gln synthetase (GS; E.C.6.3.1.2) catalyzes the conversion of inorganic nitrogen (ammonium) into organic form (Gln) and, for this reason, is a good candidate to be a critical and possibly rate-limiting enzyme in ammonium assimilation.
Biochemical studies have shown that distinct isoenzymes of GS are located in the chloroplast (GS2) and cytosol (GS1) of numerous plant species (Hirel and Gadal, 1980). In all higher plants examined to date, there is a single nuclear gene for chloroplastic GS2 and multiple homologous but distinct genes for cytosolic GS1 (Tingey and Coruzzi, 1987; Tingey et al., 1987; Sakamoto et al., 1990; Cock et al., 1991; Peterman and Goodman, 1991; Sakakibara et al., 1992; Li et al., 1993; Oliveira et al., 1997; Oliveira and Coruzzi, 1999). The chloroplastic and cytosolic GS isoenzymes seem to serve distinct roles, based on their organ- and cell-specific expression patterns (Edwards et al., 1990; Carvalho et al., 1992; Kamachi et al., 1992). Chloroplastic GS2 is expressed abundantly in leaf mesophyll cells, whereas expression of cytosolic GS1 is detected at low levels in leaves, and it is normally restricted to the phloem (Edwards et al., 1990; Carvalho et al., 1992; Kamachi et al., 1992).
The high-level expression of chloroplastic GS2 in leaf mesophyll cells underscores its role in the reassimilation of photorespiratory ammonium, which is supported by biochemical, genetic, and more recent molecular evidence (Keys et al., 1978; Wallsgrove et al., 1987; Edwards and Coruzzi, 1989; Lea and Forde, 1994; Kozaki and Takeba, 1996; Migge et al., 2000). Reassimilation of photorespiratory ammonium by chloroplast GS2 is crucial to plant growth, as levels of ammonium released during photorespiration may exceed primary nitrogen assimilation by 10-fold (Keys et al., 1978). Barley (Hordeum vulgare) mutants defective in chloroplastic GS2 are unable to reassimilate photorespiratory ammonium and die when grown in air, indicating that chloroplastic GS2 plays a major role in the reassimilation of photorespiratory ammonium in leaf mesophyll cells. It was surprising that these barley mutants in chloroplastic GS2 died when grown under photorespiratory conditions (air), even though leaves contain low levels of cytosolic GS1 (Wallsgrove et al., 1987; Lea and Forde, 1994). The nonoverlapping and cell-specific expression patterns of chloroplastic and cytosolic GS isoenzymes may explain why cytosolic GS1 cannot compensate for the loss of chloroplastic GS2 in leaf mesophyll cells of these barley photorespiratory mutants.
The barley GS mutant studies cited above suggest that there is a subcellular trafficking of photorespiratory ammonium out of the mitochondria and into the chloroplast for reassimilation by chloroplastic GS2. We, therefore, reasoned that the ectopic overexpression of a cytosolic GS1 isoenzyme in the leaf mesophyll cells, where it is not normally expressed at high levels, could potentially provide an additional and/or alternate route to native chloroplastic GS2 in the reassimilation of photorespiratory ammonium. This type of metabolic engineering of cytosolic GS1 could potentially result in an increase in the efficiency of reassimilation of photorespiratory ammonium, leading to increases in nitrogen use efficiency and plant growth. Previous studies showed that overexpression of a gene for chloroplast GS2 from rice in transgenic tobacco (Nicotiana tabacum) increased photorespiratory capacity and resistance to photooxidation, although in this case no effect on growth has been reported (Kozaki and Takeba, 1996).
Several groups have attempted to improve nitrogen assimilation by the overexpression of GS genes with mixed results (Eckes et al., 1989; Hemon et al., 1990; Hirel et al., 1992; Temple et al., 1993; Vincent et al., 1997; Gallardo et al., 1999; Migge et al., 2000; Ortega et al., 2001). For instance, Hirel and co-workers observed accelerated growth rate in transgenic Lotus corniculatus plants, which overexpress a soybean (Glycine max) GS isoenzyme (Vincent et al., 1997). Growth improvements have been reported more recently for poplar (Populus spp.) trees and tobacco plants overexpressing distinct isoforms of GS (Gallardo et al., 1999; Migge et al., 2000; Fuentes et al., 2001). Experimental data available to date have provided evidence that overexpression of GS may affect the modulation/maintenance of photosynthetic rates (Kozaki and Takeba, 1996; Fuentes et al., 2001), and it is a possible mechanism by which GS can improve/accelerate growth in these GS transgenic plants (Fuentes et al., 2001).
Herein, we report that transgenic tobacco plants that ectopically overexpress a cytosolic GS1 isoenzyme in leaves have alterations in the photorespiratory pathway. This is evidenced by lower levels of free ammonium, by higher levels of photorespiratory intermediates, and by an increase in the CO2 photorespiratory burst measurements. These GS1 transgenic plants also display an enhanced growth phenotype as quantified by increases in fresh weight, dry weight, and leaf soluble protein. Moreover, these increases are paralleled by corresponding increases in GS activity. These studies provide insights into the mechanism by which overexpression of a cytosolic GS1 isoenzyme may lead to changes in growth and suggest that it may be possible to increase nitrogen use efficiency by the manipulation of genes for specific GS isoenzymes in transgenic crop plants.
RESULTS
Characterization of GS Transgenic Plants
Transgenic lines of tobacco were generated in which a 35S cauliflower mosaic virus promoter was used to drive the ectopic overexpression of pea (Pisum sativum) cDNAs encoding either chloroplastic GS2 or cytosolic GS1 isoenzymes. Two homologous but distinct GS cDNAs encoding cytosolic isoenzymes of GS (80% nucleotide homology and 86% amino acid homology within the coding region) were used; cytosolic GS1 (CytGS1-TR) or cytosolic GS3A (CytGS3A-TR; Tingey et al., 1988). Transgenic lines containing the pea chloroplastic GS2 cDNA were also generated (ChlGS2-TR; Tingey et al., 1988). Controls used in these studies were tobacco plants transformed with an insertless vector (SR1–6). For each construct, multiple independent lines were generated. The results reported below are representative of four CytGS1-TR (three shown below), two CytGS3A-TR (not shown), and nine ChlGS2-TR (one shown below) independent GS transgenic lines, respectively.
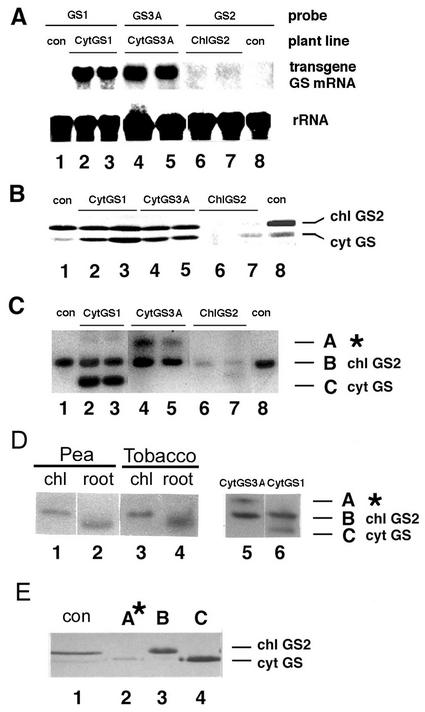
GS expression was examined in transgenic plants at the level of GS mRNA, GS protein, GS holoenzyme, and total GS activity (Figs. 1 and 2). The growth phenotype of two individuals of representative transgenic and control lines are shown side-by-side in Figure 2A. Leaves of CytGS1-TR plants accumulated high levels of mRNA for cytosolic GS1 transgene (Fig. 1A, lanes 2 and 3) and cytosolic GS1 protein (Fig. 1B, lanes 2 and 3). The ectopically expressed pea cytosolic GS1 protein also assembled into a native cytosolic GS1 holoenzyme in leaves (Fig. 1C, lanes 2 and 3, band C). This cytosolic GS1 holoenzyme is normally only detected at significant levels in roots of tobacco (Fig. 1D, lane 4) but not in leaves (Fig. 1C, lane 1). It is noteworthy that the levels of cytosolic GS1 protein present in leaves of control plants detected by western blot (Fig. 1B, lanes 1 and 8) are too low to produce a detectable GS1 holoenzyme band when assayed by enzyme activity staining of extracts run on non-denaturing PAGE (Fig. 1C, lanes 1 and 8). These differences in detection of low levels of native cytosolic GS1 in leaves of tobacco are most likely due to different sensitivities between the two techniques. The increased level of the cytosolic GS1 holoenzyme in leaves of CytGS1-TR plants, resulted in significant increases in levels of total GS activity when compared with controls (Fig. 2, B–D, black circles).
Figure 1.
GS expression profiles in leaves of 35S-GS transgenic tobacco plants. A, GS mRNA was detected by hybridization with full-length cDNA probes for pea cytosolic GS1 (lanes 1–3), pea cytosolic GS3A (lanes 4 and 5), and pea chloroplastic GS2 (lanes 6–8). B, Western-blot analysis with a mixture of antibodies to bean (Phaseolus vulgaris) cytosolic GS1 and tobacco chloroplastic GS2 (Hirel et al., 1984; Lara et al., 1984; Tingey et al., 1988). C, Non-denaturing gel and GS activity stain showing GS holoenzymes A, B, and C. GS holoenzyme A (*) is a nonnative GS isoenzyme detected only in CytGS3A-TR plants. CytGS1-TR and CytGS3A-TR lines contain normal levels of native chloroplastic GS2 (band B). D, Non-denaturing gel and GS activity stain showing a side-by-side comparison between CytGS3A-TR (lane 6) and CytGS1-TR (lane 5) leaf extracts. The cytosolic GS1 holoenzyme (band C), which is detected in leaves of CytGS1-TR plants but not in the control plants, corresponds to the native root-specific tobacco cytosolic GS1 holoenzyme (lanes 4 and 6). Controls: lanes 1 and 2, pea chloroplast and root extracts; lanes 3 and 4, tobacco chloroplast and root extracts. E, Subunit composition of GS holoenzymes. GS holoenzymes A*, B, and C, respectively, were excised from preparative native gels, denatured, separated by PAGE, and detected by western-blot analysis. Crude leaf extract of untransformed tobacco (lane 1), GS holoenzyme A* from CytGS3A-TR (lane 2), GS holoenzyme band B isolated from isolated chloroplasts from untransformed tobacco (lane 3), and GS holoenzyme C from CytGS1-TR (lane 4).
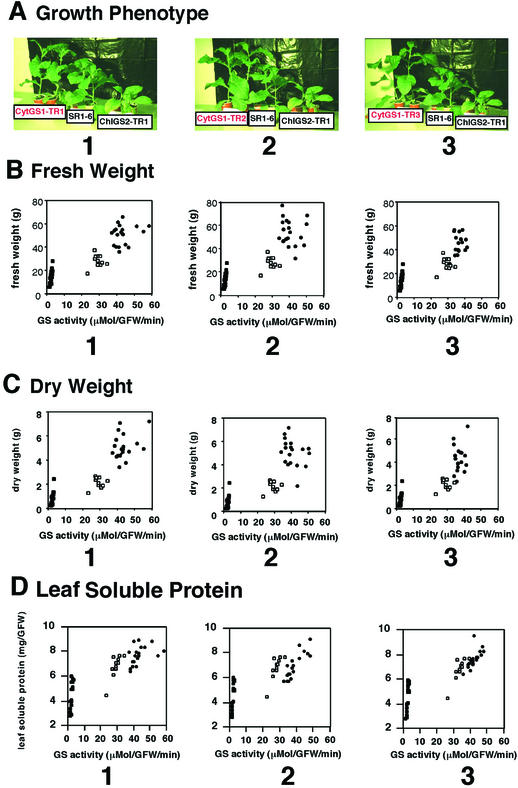
Figure 2.
Qualitative and quantitative growth phenotype of GS transgenic plants. A, Plants from the control line (SR1–6) and the cosuppressed chloroplastic GS2 (ChlGS2-TR) line are shown next to three independent lines of cytosolic GS1 overexpressors: CytGS1-TR1 (1), CytGS1-TR2 (2), and CytGS1-TR3 (3). The same ameliorated growth phenotype was also observed in another independent CytGS1-TR line, CytGS1-TR4 (not shown). B through D, Growth analysis of cytosolic GS1 overexpressor lines (●) CytGS1-TR1 (1), CytGS1-TR2 (2), and CytGS1-TR3 (3). Also represented are the control tobacco line (SR1–6, □) and the cosuppressed chloroplastic GS2 line (ChlGS2-TR, ▪). The growth assays were performed in 19 plants for the CytGS1-TR or ChlGS2-TR lines and 10 plants for the SR1–6 line. All plants were analyzed individually for total plant fresh weight (B), dry weight (C), and soluble protein (D) as a function of total leaf GS specific activity (Shapiro and Stadtman, 1971). The plants were grown and assayed as described in “Materials and Methods.”
Plants overexpressing a distinct pea cytosolic GS isoenzyme named GS3A (CytGS3A-TR) showed increases in levels of GS3A mRNA (Fig. 1A, lanes 4 and 5) and GS3A protein (Fig. 1B, lanes 4 and 5). However, the GS3A protein assembled into a nonnative-sized GS holoenzyme (Fig. 1C, lanes 4 and 5 and band A*), as demonstrated by its anomalous migration pattern when compared with either pea or tobacco native isoforms from chloroplasts and roots (Fig. 1D). To determine the subunit composition of the GS holoenzymes in the CytGS3A-TR plants, bands A*, B, and C were excised from preparative gels, and the GS subunit peptides were detected by western-blot analysis (Fig. 1E). GS activity bands A* and C are composed exclusively of GS polypeptides (Fig. 1E, lanes 2 and 4). This discounted the possibility that the larger GS activity band A* was the result of the assembly of GS3A subunits expressed ectopically in leaf mesophyll cells with endogenous prechloroplastic GS2 subunits containing an unprocessed chloroplastic transit peptide. Therefore, because the anomalous migrating GS3A holoenzyme was shown to be composed of normal-sized cytosolic GS3A polypeptides (Fig. 1E), one formal possibility is that the larger GS activity band A* in the CytGS3A-TR plants could result from a post-translation modification by the association of this GS holoenzyme with another uncharacterized protein. Evidence for the association of cytosolic GS with other associated proteins has previously been suggested by other studies (Temple et al., 1993; Ortega et al., 2001). Therefore, the unusual migration of the cytosolic GS3A holoenzyme in the CytGS3A-TR plants may reflect changes in conformation and/or additional GS-associated proteins. These CytGS3A-TR plants, which had the anomalous GS holoenzyme, exhibited only modest changes in total GS enzyme activity and growth when compared with controls (not shown). These results with the CytGS3A-TR lines are reminiscent of previous reports in which posttranslational modification of a transgenic cytosolic GS protein was suggested to be associated with the lack of increase in GS enzyme activity and/or ameliorated plant growth in the transgenic GS lines (Eckes et al., 1989; Hemon et al., 1990; Hirel et al., 1992; Temple et al., 1993; Vincent et al., 1997).
All transgenic lines engineered to overexpress pea chloroplastic GS2 (ChlGS2-TR) showed a cosuppressed phenotype. Cosuppression was manifested by no expression of transgene GS2 mRNA (Fig. 1A, lanes 6 and 7) and by a dramatic reduction in levels of native tobacco GS protein and holoenzyme for chloroplastic GS2 and cytosolic GS1 (Fig. 1, B and C, lanes 6 and 7). The cosuppression effect on ChlGS2-TR was very consistent and was observed in 23 independent transformants using two different constructs (not shown). It is noteworthy that the pea GS2 transgene was able to suppress expression of genes for chloroplastic GS2 and cytosolic GS1 of tobacco. This is consistent with the relatively high identity between the GS genes of these two plant species (76%–88% amino acid homology; Tingey and Coruzzi, 1987). There are other examples where one member of a gene family can cause cosuppression of other gene family members with significant homology (e.g. ACC synthase; Que et al., 1998). Because the actual mechanism(s) underlying the phenomenon of cosuppression in plants is not totally understood (Vaucheret et al., 1998), the cause for the observed cosuppression of both GS isoenzymes in the ChlGS2-TR plants can only be conjectured.
Transgenic GS Lines Show a Correlation between GS Activity and Fresh Weight, Dry Weight, and Leaf Soluble Protein
We monitored the above transgenic GS lines for growth phenotypes (Fig. 2A, 1–3), and observed a correlation between the levels of GS enzyme activity and plant fresh weight, dry weight, and leaf soluble protein (Fig. 2, B–D). Analysis of at least three independent lines for each construct consistently showed that the transgenic lines transformed with the pea cytosolic GS1 cDNA (CytGS1-TR1, CytGS1-TR2, and CytGS1-TR3; black circles), showed the highest levels of GS activity and the highest increases in plant fresh weight, dry weight, and leaf soluble protein compared with controls (open squares; Fig. 2, B–D). These increases in fresh weight, dry weight, and leaf soluble protein exhibited by the CytGS1-TR plants were most pronounced at early stages of development (Figs. 3 and 4), but also persisted in older plants (Fig. 2A) and in flowering plants (50–60 d old; not shown). The improved growth phenotype of transgenic lines transformed with the pea cytosolic GS1 cDNA was observed in soil-germinated seedlings (Fig. 3) and in plants cultured in media, before transfer to soil (Fig. 2A). Lines transformed with the gene encoding a distinct cytosolic GS gene (CytGS3A) showed only modest increases in GS activity and correspondingly modest increases in fresh weight, dry weight, and leaf soluble protein when compared with the control (not shown). All lines containing the chloroplast GS2 gene (ChlGS2-TR lines) were co-suppressed, and the growth of these lines was characterized by extensive leaf chlorosis (Fig. 2A, 1–3) and by reductions in growth, fresh weight and dry weight (Fig. 2, B–D). The chlorotic phenotype of the cosuppressed ChlGS2-TR plants was relieved when plants were grown in an atmosphere of elevated CO2 (0.8%–1.2%) to suppress photorespiration or when plants were supplemented with exogenous Gln (not shown). As such, these GS cosuppressed transformants resembled the GS2-deficient photorespiratory mutants of barley (Wallsgrove et al., 1987; Lea and Forde, 1994). Previous studies showed that the barley GS2 mutants could also survive if photorespiration was suppressed (by 1% [v/v] CO2) or if supplemented with Gln (Blackwell et al., 1987). These results indicate that chloroplastic GS2 mutants (and the cosuppressed GS transgenic plants described herein) die from the depletion of amino donors from the pool of organic nitrogen, caused by their inability to reassimilate photorespiratory ammonium (Blackwell et al., 1987).
Figure 3.
Qualitative growth phenotype of soil-grown GS transgenic plants. Control line (SR1–6; A), CytGS1-TR1 (B), CytGS1-TR2 (C), and CytGS1-TR3 (D) were germinated and grown for 28 d in soil as described in “Materials and Methods.”
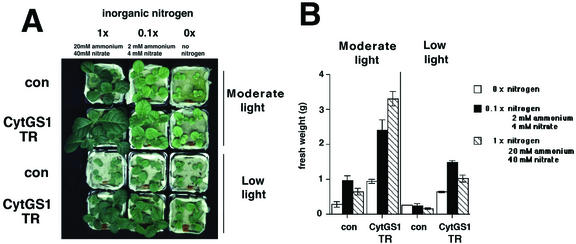
Figure 4.
Effect of light on growth of GS transgenic plants grown under different nitrogen regimes. Plants were incubated in a normal day/night cycle either under high light (moderate PFD, 200 μmol cm−2 s−1) or low light (low PFD, 50 μmol cm−2 s−1) and subirrigated with ammonium-free/nitrate-free liquid Murashige and Skoog medium containing 0× nitrogen (no nitrogen supplementation), 0.1× nitrogen (4 mm nitrate/2 mm ammonium), or 1× nitrogen (40 mm nitrate/20 mm ammonium). A, Qualitative growth phenotype. B, Fresh weight (n = 4, mean ± se) from plants in A. The plants for this experiment were grown as described in “Materials and Methods.”
Transgenic Plants That Ectopically Overexpress Cytosolic GS1 Display a Light-Dependent, Improved Growth Phenotype under Nitrogen-Limiting and Nitrogen-Non-Limiting Conditions
To determine the possible mechanisms underlying the enhanced growth of transgenic plants overexpressing cytosolic GS1 (CytGS1-TR), we examined whether this improved growth was related to the concentration of exogenously supplied inorganic nitrogen or by light (Fig. 4). The CytGS1-TR plants showed increases in fresh weight under nitrogen-limiting and nitrogen-non-limiting conditions when compared with plants grown at lower PFDs (Fig. 4, A and B). The effects of light and inorganic nitrogen were additive, because the growth of CytGS1-TR plants was maximal under conditions of high inorganic nitrogen (40 mm nitrate and 20 mm ammonium) and “moderate light” (moderate PFD, 200 μmol cm−2 s−1). It is noteworthy that even under conditions of no exogenous nitrogen application (0× nitrogen), the CytGS1-TR plants still show a growth advantage compared with control plants (Fig. 4). This suggests that the observed growth advantage of the GS transgenics may relate to increased efficiencies in use of internal stores of nitrogen such as the reassimilation of “recycled” ammonium released during photorespiration (see below).
Transgenic Plants That Ectopically Overexpress Cytosolic GS1 Display Increased Photorespiratory CO2 Burst
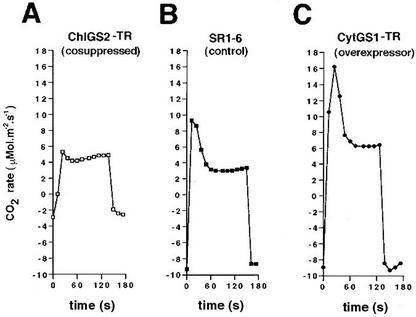
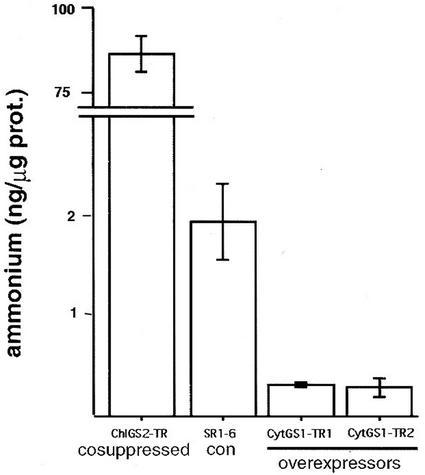
Because primary nitrogen assimilation, photorespiration, and the reassimilation of photorespiratory ammonium are all light-dependent processes (Blackwell et al., 1987; Wallsgrove et al., 1987; Lea and Forde, 1994; Kozaki and Takeba, 1996), we next tested whether photorespiration was affected in the CytGS1-TR transgenic plants by measuring the postillumination photorespiratory CO2 burst. Several independent lines of evidence suggest a direct correlation between increased levels of cytosolic GS1 overexpression in the CytGS1-TR plants and increased rates of photorespiration. First, gas exchange experiments revealed that postillumination photorespiratory CO2 evolution was increased in the overexpressing CytGS1-TR and decreased in the ChlGS2-TR-cosuppressed plants when compared with the controls (Fig. 5; Table I). Second, levels of amino acids known to be involved in the photorespiratory cycle were elevated in the leaves of CytGS1-TR transgenic plants. CytGS1-TR plants showed a 3.5-fold increase in the Ser/Gly ratios (669.0 ± 86.6 Ser/71.3 ± 4.6 Gly) when compared with the SR1–6 controls (465.7 ± 4.4 Ser/175.7 ± 3.2 Gly) and a 2-fold increase in Glu levels (719.5 ± 27.8) when compared with the SR1–6 controls (338.8 ± 3.7), as measured in picomoles per milligram fresh weight (±se, n = 3 individual plants). Third, the increased photorespiratory rates in the CytGS1-TR plants correlated with a 6.3- to 7-fold reduction in the total levels of free ammonium when compared with the SR1–6 controls (Fig. 6). This reduction in levels of ammonium was related to the level of GS expression, because transgenic plants that are cosuppressed for GS activity display the opposite phenotype (i.e. 44-fold increase in the levels ammonium; Fig. 6). These results collectively provide three independent measures suggesting that the CytGS1-TR plants have changes associated with photorespiration: (a) increased postillumination CO2 evolution, (b) increased levels of photorespiratory amino acids, and (c) decreases in free ammonium. These correlated changes support the notion that ectopic overexpression of cytosolic GS1 in the cytoplasm of leaf mesophyll cells leads to increases in the levels of photorespiration in the transgenic GS1 plants. Although these measures indicate increased photorespiratory rate, the CO2 compensation point in the GS1-TR plants was unchanged from wild type (not shown). Because the CO2 compensation point is the point at which CO2 consumption by photosynthesis equals the rate of CO2 evolution by photorespiration, this suggests that the changes in photorespiration were most likely accompanied by commensurate changes in photosynthesis.
Figure 5.
Levels of photorespiration correlate with GS expression in transgenic plants. Detached leaves of the cosuppressed chloroplastic GS2 line (ChlGS2-TR1, □), the control tobacco line (SR1–6, ▪), and a cytosolic GS1 overexpressor line (CytGS1-TR1, ●) were initially illuminated (1,000 μmol cm−2 s−1) for 1 h and subsequently exposed to dark by blocking the light source for a period of 2 min. The composition of the gas entering the chamber was 79 μL CO2 L−1 (PPM), 21% (v/v) O2, and balanced nitrogen. Total gas flow was approximately 1 L min−1. The temperature was kept at 28°C to 29°C for dark and light conditions. The rate of CO2 exchange was measured at 12-s intervals. The measurements were done in two individual plants from each transgenic line analyzed. A representative result is shown.
Table I.
Determination of CO2 evolution in detached leaves of tobacco plants
| Time | ChlGS2-TR | SR1-6 | CytGS1-TR |
|---|---|---|---|
| s | μmol m−2 s−1 | ||
| 0 | −2.895 | −9.300 | −8.932 |
| 12 | 0 | 9.300 | 10.586 |
| 24 | 5.308 | 8.636 | 16.242 |
| 36 | 4.504 | 5.646 | 12.570 |
| 48 | 4.182 | 3.820 | 7.675 |
| 60 | 4.182 | 3.155 | 6.881 |
| 72 | 4.343 | 3.023 | 6.318 |
| 90 | 4.504 | 2.989 | 6.285 |
| 102 | 4.665 | 2.989 | 6.285 |
| 114 | 4.826 | 3.023 | 6.285 |
| 126 | 4.826 | 3.155 | 6.451 |
| 138 | 4.890 | 3.255 | −8.435 |
| 150 | −1.930 | 3.355 | −9295 |
| 162 | −2.413 | −8.636 | −8.932 |
| 174 | −2.574 | −8.636 | −8.435 |
Results shown are a representative one from measurements done in two individual plants from each transgenic line analyzed.
Figure 6.
Correlation between the levels of ammonium and expression of GS in tobacco transgenic plants. The plants were incubated under moderate light (moderate PFD, 200 μmol cm−2 s−1) subirrigated with 0.5× Hoagland for 20 to 30 d. Ammonium was determined from leaf extracts of the tobacco transgenic lines as indicated. Results are in nanograms of NH4+ per microgram of protein ± se, n = 3 individual plants.
DISCUSSION
Here, we report the positive effects on plant growth related to the ectopic overexpression of a pea gene for cytosolic GS1 in transgenic tobacco (CytGS1-TR lines). To begin to uncover the mechanisms by which the ectopic expression of cytosolic GS1 leads to improved growth, we examined the effects of light, nitrogen, and photorespiration in the GS transgenic plants. Analysis of GS levels in transgenic and control lines indicated a correlation between levels of GS activity and fresh weight, dry weight, and leaf soluble protein. The CytGS1-TR plants display increases in photorespiration, as judged by three measurements: (a) an increased pool size of photorespiratory metabolites, (b) an increased CO2 burst, and (c) an accompanying decrease in the levels of free ammonium. These results suggest that cytosolic GS1 is not solely operating to suppress the negative effects of photorespiration (e.g. nitrogen drain). Instead, because photorespiratory rates are actually elevated in the CytGS1-TR lines, these results suggest that GS may be a critical enzyme linking photosynthesis with photorespiration (via ammonium assimilation). This role for GS is supported by previous studies on the overexpression of chloroplastic GS2 in tobacco (Kozaki and Takeba, 1996), where increased levels of chloroplastic GS2 in tobacco led to increased photorespiration and resistance to photooxidation. In our studies, overexpression of cytosolic GS1 isoenzyme in tobacco leaves in the CytGS1-TR lines led to increased photorespiratory rates as indicated by an increased CO2 burst and by increased levels of photorespiratory intermediates. However, the CO2 compensation point (71) of the CytGS1-TR is unchanged compared with control plants (data not shown). The CO2 compensation point is the concentration of CO2 at which the rate of CO2 evolution from photorespiration equals the rate of CO2 assimilated via photosynthesis at a given O2 level (Tolbert, 1997). The fact that the CO2 compensation point is unchanged in the CytGS1-TR lines suggests that the increase in photorespiration in CytGS1-TR plants is most likely accompanied by a concomitant increase in photosynthesis. This conclusion is supported by the recent findings of Fuentes et al. (2001). The Fuentes et al. (2001) study has demonstrated that tobacco plants overexpressing the alfalfa GS1 gene under control of the 35S promoter display growth advantage when compared with the controls. The authors concluded that this was due to the ability of these transgenic plants to maintain normal photosynthetic rates even under nitrogen limiting conditions. The observations of these authors are complementary with our own observations that the growth phenotype of the CytGS1-TR plants is positively affected by light and is accompanied by changes in photorespiration.
We cannot rule out the possibility that the observed increase in growth and photosynthesis/photorespiration may be an indirect effect of the increase in leaf soluble protein observed in the CytGS1-TR plants. However, other genetic and biochemical evidence support a direct correlation between changes in GS expression, levels of photorespiration, and photosynthetic rates (Blackwell et al., 1987; Wallsgrove et al., 1987; Edwards and Coruzzi, 1989; Hausler et al., 1994a, 1994b). Photorespiratory mutants in GS2 display a decline in photorespiration and in photosynthetic carbon fixation (Blackwell et al., 1987; Wallsgrove et al., 1987; Edwards and Coruzzi, 1989; Hausler et al., 1994b). These photorespiratory GS mutants also show a 2- to 50-fold increase in ammonium accumulation (Wallsgrove et al., 1987; Hausler et al., 1994a). In the barley GS2 mutants, the failure to reassimilate photorespiratory ammonium into Gln also resulted in a 5-fold decrease in the Ser/Gly ratio (Hausler et al., 1994a) and a reduction of photorespiratory amino acids (Blackwell et al., 1987; Hausler et al., 1994a). The CytGS1-TR tobacco plants that ectopically overexpress cytosolic GS1 described herein, show the exact opposite phenotypes compared with the barley GS2 mutants, deficient in GS activity. The CytGS1-TR plants display enhanced photorespiration, an increase in the Ser/Gly ratio (3.5-fold), and a dramatic reduction in the levels of free ammonium. In addition, the levels of Glu (the product of photorespiratory ammonium assimilation) were also increased in CytGS1-TR plants when compared with controls (2-fold; not shown).
Our growth assays suggest that enhanced photorespiratory rates combined with increased reassimilation of photorespiratory ammonium in the CytGS1-TR plants (7-fold reduction) have beneficial effects on plant growth. Migge et al. (2000) have overexpressed a plastidic form of Gln synthetase (GS2) in leaves of tobacco, which led to a 3.7-fold reduction in the leaf ammonium pool with parallel effects on growth. The increased ammonium assimilation observed in our study (7-fold) may be due to the fact that the CytGS1-TR plants ectopically overexpress a cytosolic GS1 isoform in tobacco leaf mesophyll cells, where it is normally not expressed at high levels. The ectopic expression of cytosolic GS1 in leaf mesophyll cells may provide a complementary and/or alternative route to chloroplastic GS2 for the reassimilation of photorespiratory ammonium. Because nitrogen flux through the photorespiratory pathway is 10-fold higher than primary N-assimilation, the enhanced reassimilation of photorespiratory ammonium could lead to enhanced nitrogen use efficiency. Mechanistically, the improved growth phenotype observed in the CytGS1-TR plants may be a consequence of increased photosynthesis/photorespiration, combined with enhanced nitrogen efficiency. These findings for cytosolic GS1 seem to be generally applicable to other C3 plants, because preliminary results from our laboratory indicate that a similar improved growth phenotype also occurs in Arabidopsis plants overexpressing the pea cytosolic GS1 gene (not shown).
The overexpression of GS genes has been attempted before by several groups with mixed results (Eckes et al., 1989; Hemon et al., 1990; Hirel et al., 1992; Temple et al., 1993; Vincent et al., 1997; Ortega et al., 2001). For instance, Hirel and co-workers have observed accelerated growth rate in transgenic L. corniculatus plants that overexpress a soybean cytosolic GS isoenzyme. Those plants also displayed increases in some amino acids. However, that report does not indicate a correlation between plant dry/fresh weight and GS activity (Vincent et al., 1997). Previous studies also showed that overexpression of a gene for chloroplast GS2 from rice in transgenic tobacco led to increased levels of photorespiration and resistance to photooxidation, although no accompanying increase in growth or yield was reported (Kozaki and Takeba, 1996). It is unlikely that photoprotection plays a major role in the improved growth phenotype in the CytGS1-TR plants reported herein, because of the moderate PFD used in our experiments (200 μmol cm−2 s−1). In more recent reports, overexpression of distinct GS isoenzymes has been associated with improvement of plant growth in two other species, including poplar, supporting the generality of this approach. However, in those studies, no studies were performed to gain insight into the mechanisms underlying such growth improvement (Gallardo et al., 1999; Migge et al., 2000). The recent study by Fuentes et al. (2001) has demonstrated that tobacco plants overexpressing the alfalfa GS1 gene under control of the 35S promoter display growth advantage when compared with the controls, and they cite increases in photosynthetic rate as a possible mechanism. The improvement in plant growth for the CytGS1-TR plants reported herein most likely results from a combination of factors including: (a) ectopic overexpression of a cytosolic GS1 isoenzyme in leaf mesophyll cells of a species where it is normally expressed at low levels (e.g. Solanaceous species); (b) a threshold level of transgene expression; (c) a cytosolic GS1 isoenzyme that assembles into a native holoenzyme in the host plant system; and (d) an appropriate plant background (e.g. plants with low levels of native cytosolic GS1 in leaves or C3 plants).
The CytGS1-TR plants described herein exhibit increases in biomass (dry weight) at all stages of growth tested, up to flowering (50–60 d old; not shown). This increase may reflect an accelerated growth rate (Vincent et al., 1997) and/or an increase in total biomass. Either trait could have important agronomic applications. The physiological parameters relevant to seed yield and seed-nitrogen content include not only the efficiency of nitrogen assimilation or reassimilation in vegetative tissues, but also the remobilization of nitrogen reserves at the onset of bolting and flowering. Whether the increases in dry weight and soluble protein observed in transgenic lines overexpressing cytosolic GS1 will also lead to a significant improvement in seed yield or seed quality is an important question that remains to be answered in future studies of these and other transgenic lines currently under investigation in our laboratory.
MATERIALS AND METHODS
Plasmids and Plant Transformation
The plant expression vector and the cDNAs corresponding to the pea (Pisum sativum) genes GS1, GS2, and GS3A have been described elsewhere (Tingey and Coruzzi, 1987; Tingey et al., 1988; Brears et al., 1993). Transfer of constructs to the Agrobacterium tumefaciens strain LBA4404 and tobacco (Nicotiana tabacum line SR1) transformation was as described (Bevan, 1984; Horsch et al., 1985; Brears et al., 1993). All experiments described below were performed with T3 and T4 generation transgenic plants.
Plant Growth Conditions. Growth Assays for Plants Germinated on Medium
Plants were germinated on Murashige and Skoog/kanamycin medium under a light irradiance of 90 μmol cm−2 s−1 generated by a mixture of fluorescent, incandescent, high-pressure sodium, and metal halide lights. After 14 to 18 d, kanamycin-resistant seedlings were transferred to white sand. The plants were further grown for 20 to 42 d (depending on the experiment) and subirrigated with 0.5× Hoagland (0.6 mm ammonium and 7 mm nitrate) in a 16-h-light/8-h-dark cycle. Fresh weight and dry weight determinations were from the whole plant. Dry weight was determined after incubation of the plant at 65°C for 72 h. Soluble protein was calculated by measuring the total soluble protein from approximately 1 g of leaf tissue (Bradford, 1976). All protein measurements were conducted either in fresh harvested tissue ground immediately after excision or from leaves quickly deep-frozen in liquid nitrogen and kept at −80°C until the assay. Material for all biochemical determinations (including protein measurement) was collected from plants in mid-light cycle. The transgenic lines used in these experiments have not been analyzed for transgene copy number or homozygosity. Therefore, to compensate for possible variations within individuals of each line, a large number of individuals were analyzed (19 individuals/line).
Growth Assays for Plants Germinated on Soil
Plants were germinated on soil under a light irradiance of 60 to 90 μmol cm−2 s−1 generated by a mixture of fluorescent and incandescent lights. The plants were grown for 28 d subirrigated with 0.5× Hoagland (0.6 mm ammonium and 7 mm nitrate) in a 16-h-light/8-h-dark cycle.
Light and Inorganic Nitrogen Dependence
Plants were germinated on Murashige and Skoog/kanamycin medium under a light irradiance of 60 μmol cm−2 s−1 provided by incandescent and hi-gro fluorescent lights. After 14 to 18 d, the kanamycin-resistant seedlings were transferred to white sand. Plants were subirrigated with ammonium-free/nitrate-free Murashige and Skoog liquid medium containing 0× nitrogen (no nitrogen supplementation), 0.1× nitrogen (4 mm nitrate/2 mm ammonium), or 1× nitrogen (40 mm nitrate/20 mm ammonium), further subdivided into two sets, incubated under moderate light (moderate PFD, 200 μmol cm−2 s−1) or low light (low PFD, 50 μmol cm−2 s−1), and further grown for 20 to 30 d in a 16-h-light/8-h-dark cycle.
Ammonium Determination
Plants were germinated in Murashige and Skoog/kanamycin medium, transferred to white sand, and subirrigated with 0.5× Hoagland as above. Thereafter, the plants were incubated under moderate light (moderate PFD, 200 μmol cm−2 s−1) for 20 to 30 d.
Postillumination Photorespiratory CO2 Evolution Experiments
Plants were germinated and grown as above except that in this case the plants were transferred to soil and subirrigated with 0.5× Hoagland. Thereafter plants were grown in a greenhouse and subirrigated with 0.5× Hoagland for 20 to 25 d.
Measurement of Photorespiration
The levels of photorespiration were estimated by postillumination photorespiratory CO2 evolution (Decker, 1955; Peterson, 1983) using an infra-red CO2 gas analyzer (LI-COR, Lincoln, NE).
HPLC Analysis of Free Amino Acids
HPLC analysis was performed as previously described (Brears et al., 1993) with minor modifications. In brief, leaf samples were harvested and quickly frozen in liquid nitrogen until the moment of the assay. Thereafter, the leaf samples were frozen-ground and mixed in an ice-cold buffer containing 50 mm Tris-HCl, pH 8.0, 10 mm imidazole, and 0.5% (w/v) β-mercaptoethanol quickly followed by extraction with 200 μL of methanol:chloroform (6:2.5, v/v). HPLC analysis of amino acid was performed using a supelcosil LC-18 reversed-phase analytical column (25-cm × 4.6-mm i.d., particle size 5 μm; Supelco Inc., Bellefonte, PA). The mobile phase consisted of a gradient of 26 mm phosphate buffer, pH 7.5 (buffer A), with increasing concentrations of 72% (v/v) methanol in water (buffer B). The column eluate was read by a LS30 luminescence spectrometer (PerkinElmer, South Plainfield, NJ) and recorded in a ChromJet integrator (ThermoSeparations, Bergenfield, NJ). The amino acid analog nor-Val was used as an internal standard.
Determination of Gln Synthetase Activity and Free Ammonium Levels
GS enzyme activity analysis was essentially as described (Shapiro and Stadtman, 1971). Ammonium was extracted by grinding liquid nitrogen-frozen plant tissue samples with a mortar in cold GS assay buffer (50 mm Tris-HCl, pH 8.0, 10 mm imidazole, and 0.5% [w/v] β-mercaptoethanol). Samples were kept on ice until assay that was performed immediately after grinding with a kit (Boehringer Mannheim, Mannheim, Germany) following instructions from the manufacturer. Material for all determinations was collected from plants in mid-light cycle.
ACKNOWLEDGMENTS
We thank Dr. Israel Zelitch for advice on the photorespiration aspects of these studies and Richard Peterson for the setup in the gas exchange experiments. We thank Rosana Melo-Oliveira for insightful advice; Joanna Wysocka-Diller and Joshua Layne for critical reading of the manuscript; and Paula Gonzales, Ravi Mistri, Dimitrios Bliagos, and Christopher Liu for help with several technical aspects of this work. We also thank William F. Thompson for the gift of the pea rRNA gene probe and Bertrand Hirel and Miguel Lara for providing GS antibodies.
Footnotes
This research was supported by the National Institutes of Health (grant no. GM 32877) and by a New York University Technology Transfer grant (to G.M.C.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.020013.
LITERATURE CITED
- Bevan M. Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell RD, Murray AJS, Lea PJ. Inhibition of photosynthesis in barley with decreased levels of chloroplastic glutamine synthetase activity. J Exp Bot. 1987;38:1799–1809. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brears T, Liu C, Knight T, Coruzzi G. Ectopic overexpression of asparagine synthetase in transgenic tobacco. Plant Physiol. 1993;103:1285–1290. doi: 10.1104/pp.103.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho H, Pereira S, Sunkel C, Salema R. Detection of cytosolic glutamine synthetase in leaves of Nicotiana tabacum L. by immunocytochemical methods. Plant Physiol. 1992;100:1591–1594. doi: 10.1104/pp.100.3.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock JM, Brock IW, Watson AT, Swarup R, Morby AP, Cullimore JV. Regulation of glutamine synthetase genes in leaves of Phaseolus vulgaris. Plant Mol Biol. 1991;17:761–771. doi: 10.1007/BF00037059. [DOI] [PubMed] [Google Scholar]
- Crawford NM, Campbell WH, Davis RW. Nitrate reductase from squash: cDNA cloning and nitrate regulation. Proc Natl Acad Sci USA. 1986;83:8073–8076. doi: 10.1073/pnas.83.21.8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker JP. A rapid post-illumination deceleration of respiration in green leaves. Plant Physiol. 1955;30:82–84. doi: 10.1104/pp.30.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckes P, Schmitt P, Daub W, Wengenmayer F. Overproduction of alfalfa glutamine synthetase in transgenic tobacco plants. Mol Gen Genet. 1989;217:263–268. doi: 10.1007/BF02464891. [DOI] [PubMed] [Google Scholar]
- Edwards JW, Coruzzi GM. Photorespiration and light act in concert to regulate the expression of the nuclear gene for chloroplast glutamine synthetase. Plant Cell. 1989;1:241–248. doi: 10.1105/tpc.1.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JW, Walker EL, Coruzzi GM. Cell-specific expression in transgenic plants reveals nonoverlapping roles for chloroplast and cytosolic glutamine synthetase. Proc Natl Acad Sci USA. 1990;87:3459–3463. doi: 10.1073/pnas.87.9.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes SI, Allen DJ, Ortiz-Lopez A, Hernández G. Over-expression of cytosolic glutamine synthetase increases photosynthesis and growth at low nitrogen concentrations. J Exp Bot. 2001;52:1071–1081. doi: 10.1093/jexbot/52.358.1071. [DOI] [PubMed] [Google Scholar]
- Gallardo F, Fu J, Canton FR, Garcia-Gutierez A, Canovas FM, Kirby EG. Expression of a conifer glutamine synthetase gene in transgenic poplar. Planta. 1999;210:19–26. doi: 10.1007/s004250050649. [DOI] [PubMed] [Google Scholar]
- Hausler RE, Blackwell RD, Lea PJ, Leegwood RC. Control of photosynthesis in barley leaves with reduced activities of glutamine synthetase and glutamate synthase: I. Plant characteristics and changes in nitrate, ammonium and amino acids. Planta. 1994a;194:406–417. [Google Scholar]
- Hausler RE, Lea PJ, Leegwood RC. Control of photosynthesis in barley leaves with reduced activities of glutamine synthetase and glutamate synthase: II. Control of electron transport and CO2 assimilation. Planta. 1994b;194:418–435. [Google Scholar]
- Hemon P, Robbins M, Cullimore J. Targeting of glutamine synthetase to the mitochondria of transgenic tobacco. Plant Mol Biol. 1990;15:895–904. doi: 10.1007/BF00039428. [DOI] [PubMed] [Google Scholar]
- Hirel B, Gadal P. Glutamine synthetase in rice. Plant Physiol. 1980;66:619–623. doi: 10.1104/pp.66.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel B, Marsolier M, Hoarau A, Hoarau J, Brangeon J, Schafer R, Verma DPS. Forcing expression of a soybean root glutamine synthetase gene in tobacco leaves induces a native gene encoding cytosolic enzyme. Plant Mol Biol. 1992;20:207–218. doi: 10.1007/BF00014489. [DOI] [PubMed] [Google Scholar]
- Hirel B, Weatherly C, Cretin C, Bergounioux C, Gadal P. Multiple subunit composition of chloroplastic glutamine synthetase of Nicotiana tabacum. Plant Physiol. 1984;74:448–450. doi: 10.1104/pp.74.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry JW, Hoffman NL, Eicholtz D, Rogers SG, Fraley RJ. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Kamachi K, Yamaya T, Hayakawa T, Mae T, Ojima K. Vascular bundle-specific localization of cytosolic glutamine synthetase in rice leaves. Plant Physiol. 1992;99:1481–1486. doi: 10.1104/pp.99.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys AJ, Bird IF, Cornelius MJ, Lea PJ, Wallsgrove RM, Miflin BJ. Photorespiratory nitrogen cycle. Nature. 1978;275:741–743. [Google Scholar]
- Kozaki A, Takeba G. Photorespiration protects C3 plants from photooxidation. Nature. 1996;384:557–560. [Google Scholar]
- Lara M, Porta H, Padilla J, Folch J, Sanchez F. Heterogeneity of glutamine synthetase polypeptides in Phaseolus vulgaris. Plant Physiol. 1984;76:1019–1023. doi: 10.1104/pp.76.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea PJ, Forde BG. The use of mutants and transgenic plants to study amino acid metabolism. Plant Cell Environ. 1994;17:541–556. [Google Scholar]
- Li M-G, Villemur R, Hussey PJ, Silflow CD, Gantt JS, Snustad DP. Differential expression of six glutamine synthetase genes in Zea mays. Plant Mol Biol. 1993;23:401–407. doi: 10.1007/BF00029015. [DOI] [PubMed] [Google Scholar]
- Migge A, Carrayol E, Hirel B, Becker TW. Leaf-specific overexpression of plastidic glutamine synthetase stimulates growth of transgenic tobacco seedlings. Planta. 2000;210:252–260. doi: 10.1007/PL00008132. [DOI] [PubMed] [Google Scholar]
- Oliveira IC, Coruzzi G. Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis thaliana. Plant Physiol. 1999;121:301–309. doi: 10.1104/pp.121.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira IC, Coschigano K, Lam HM, Melo-Oliveira R, Coruzzi G. Molecular-genetic dissection and metabolic engineering of nitrogen assimilation in plants. Plant Physiol Biochem. 1997;35:185–198. [Google Scholar]
- Ortega JL, Temple SJ, Sengupta-Gopalan C. Constitutive overexpression of cytosolic glutamine synthetase (GS1) gene in transgenic alfalfa demonstrates that GS1 may be regulated at the level of RNA stability and protein turnover. Plant Physiol. 2001;126:109–121. doi: 10.1104/pp.126.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman TK, Goodman HM. The glutamine synthetase gene family of Arabidopsis thaliana: light-regulation and differential expression in leaves, roots and seeds. Mol Gen Genet. 1991;230:145–154. doi: 10.1007/BF00290662. [DOI] [PubMed] [Google Scholar]
- Peterson RB. Estimation of photorespiration based on the initial rate of postillumination CO2 release: II. Effects of O2, CO2, and temperature. Plant Physiol. 1983;73:983–988. doi: 10.1104/pp.73.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Q, Wang H, Jorgensen R. Distinct patterns of pigment suppression are produced by allelic sensine and antisense chalcone synthase transgenics in petunia flowers. Plant J. 1998;13:401–409. [Google Scholar]
- Sakakibara H, Kawabata S, Takahashi H, Hase T, Sugiyama T. Molecular cloning of the family of glutamine synthetase genes from maize: expression of genes for glutamine synthetase and ferredoxin-dependent glutamate synthase in photosynthetic and non-photosynthetic tissues. Plant Cell Physiol. 1992;33:49–58. [Google Scholar]
- Sakamoto A, Takeba G, Shibata D, Tanaka K. Phytochrome-mediated activation of the gene for cytosolic glutamine-synthetase (GS1) during imbibition of photosensitive lettuce seeds. Plant Mol Biol. 1990;15:317–323. doi: 10.1007/BF00036917. [DOI] [PubMed] [Google Scholar]
- Shapiro BM, Stadtman ER. Glutamine synthetase (Escherichia coli) Methods Enzymol. 1971;17A:910–922. doi: 10.1016/s0076-6879(85)13032-6. [DOI] [PubMed] [Google Scholar]
- Sheldrick WF. World Nitrogen Survey. Technical paper no. 59, Washington, DC: World Bank; 1987. [Google Scholar]
- Temple S, Knight T, Unkefer P, Sengupta-Gopalan C. Modulation of glutamine synthetase gene expression in tobacco by the introduction of an alfalfa glutamine synthetase gene in sense and antisense orientation: molecular and biochemical analysis. Mol Gen Genet. 1993;236:315–325. doi: 10.1007/BF00277128. [DOI] [PubMed] [Google Scholar]
- Tingey SV, Coruzzi GM. Glutamine synthetase of Nicotiana plumbaginifolia: cloning and in vivo expression. Plant Physiol. 1987;84:366–373. doi: 10.1104/pp.84.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingey SV, Tsai F-Y, Edwards JW, Walker EL, Coruzzi GM. Chloroplast and cytosolic glutamine synthetase are encoded by homologous nuclear genes which are differentially expressed in vivo. J Biol Chem. 1988;263:9651–9657. [PubMed] [Google Scholar]
- Tingey SV, Walker EL, Coruzzi GM. Glutamine synthetase genes of pea encode distinct polypeptides which are differentially expressed in leaves, roots and nodules. EMBO J. 1987;6:1–9. doi: 10.1002/j.1460-2075.1987.tb04710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert NE. The C2 oxidative photosynthetic carbon cycle. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:1–25. doi: 10.1146/annurev.arplant.48.1.1. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel J-B, Mourrain P, Palauqui J-C, Vernhettes S. Transgene-induced gene silencing in plants. Plant J. 1998;16:651–659. doi: 10.1046/j.1365-313x.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- Vincent R, Fraisier V, Chaillou S, Limani AM, Deleens E, Phillipson B, Douat C, Boutin J-P, Hirel B. Overexpression of a soybean gene encoding cytosolic glutamine synthetase in shoots of transgenic Lotus corniculatus L. plants triggers changes in ammonium assimilation and plant development. Planta. 1997;201:424–433. doi: 10.1007/s004250050085. [DOI] [PubMed] [Google Scholar]
- Wallsgrove RM, Turner JC, Hall NP, Kendally AC, Bright SWJ. Barley mutants lacking chloroplast glutamine synthetase-biochemical and genetic analysis. Plant Physiol. 1987;83:155–158. doi: 10.1104/pp.83.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]