Abstract
Using two well-characterized heat stress transcription factors (Hsfs) from tomato (Lycopersicon peruvianum; LpHsfA1 and LpHsfA2), we analyzed the transcriptional activation of the Ha hsp17.6 G1 promoter in sunflower (Helianthus annuus) embryos. In this system, we observed transient promoter activation only with LpHsfA2. In contrast, both factors were able to activate mutant versions of the promoter with improved consensus Hsf-binding sites. Exclusive activation by LpHsfA2 was also observed in yeast (Saccharomyces cerevisiae) without other Hsfs and with a minimal Cyc1 promoter fused to the Ha hsp17.6 G1 heat stress cis-element. Furthermore, the same promoter mutations reproduced the loss of activation selectivity, as observed in sunflower embryos. The results of in vitro binding experiments rule out differential DNA binding of the two factors as the explanation for the observed differential activation capacity. We conclude that the specific sequence of this heat stress cis-element is crucial for Hsf promoter selectivity, and that this selectivity could involve preferential transcriptional activation following DNA binding. In sunflower embryos, we also observed synergistic transcriptional activation by co-expression of LpHsfA1 and LpHsfA2. Mutational analyses of the Ha hsp17.6 G1 promoter, combined with in vitro binding assays, suggest that mixed oligomers of the two factors may be involved in promoter activation. We discuss the relevance of our observations for mechanisms of developmental regulation of plant heat stress protein genes.
During plant zygotic embryogenesis, a subset of the small heat stress proteins (sHSPs) is expressed at normal growth temperatures and in the absence of exogenous stress. It has been noted that the developmental induction of plant sHSPs in seeds involves only certain classes and/or only specific members of a sHSP class (for review, see Waters et al., 1996). Most of the class I sHSP genes that are developmentally regulated in seeds also respond to heat stress conditions not only in vegetative tissues (Coca et al., 1996; Carranco et al., 1997; Almoguera et al., 1998), but also in seeds (Wehmeyer and Vierling, 2000). Studies of developmental regulation of plant sHSP genes may provide the necessary information to generate gain-of-function or loss-of-function mutants with altered expression patterns of sHSPs. These studies may help assess the proposed functions of sHSPs for seed viability, germination, and stress tolerance (for discussion, see Coca et al., 1996; Waters et al., 1996).
Deletion analyses of plant sHSP promoters initially pointed out that the cis-elements necessary for the heat stress response (HSEs) were also required for developmental regulation (Coca et al., 1996; Prändl and Schöffl, 1996). This suggested the involvement of heat stress transcription factors (Hsfs) in both regulatory processes. However, analyses of two developmentally regulated sHSP promoters of sunflower (Helianthus annuus) indicated differences in the transcriptional responses to heat stress or to developmental cues during embryogenesis. Thus, mutagenesis of the complex but imperfect HSE clusters of the sunflower Ha hsp17.7 G4 promoter abolished its heat stress response, but only reduced its activation in desiccating embryos (Almoguera et al., 1998). Another example is the Ha hsp17.6 G1 promoter, which is the only natural example of a sHSP promoter unable to respond to heat stress in sunflower (Carranco et al., 1997), but transcriptionally activated by Hsfs in developing embryos (Carranco et al., 1997, 1999). In this case, we also observed an imperfect, although much simpler, HSE pattern. We proposed that if Hsfs are involved in developmental regulation through this element, the mechanism of promoter activation should be different from that in a heat stress response (Carranco et al., 1997). We recently characterized the expression of chimeric genes with wild-type and null mutant HSE versions of the Ha hsp17.6 G1 promoter in transgenic tobacco (Nicotiana tabacum). These results demonstrated the requirement of Hsfs for the transcriptional activation of this promoter during embryo desiccation. Deletion analyses of 5′-flanking promoter sequences showed that cis-elements immediately adjacent and further upstream of the HSE were also required for this seed-specific activation. Thus, functional interactions of Hsfs with other trans-acting factors could be important for the developmental regulation of the Ha hsp17.6 G1 promoter in seeds (Carranco et al., 1999).
Plants contain more than 21 Hsfs that belong to three different classes (Nover et al., 1996, 2001). Although investigations so far have been limited to a few Hsfs, mostly from tomato (Lycopersicon peruvianum), it is clear that they differ with respect to promoter recognition (HSE binding), oligomerization behavior, intracellular localization, and potential as transcription activators (Nover et al., 1996; Boscheinen et al., 1997; Lyck et al., 1997; Bharti et al., 2000; Döring et al., 2000; Heerklotz et al., 2001). The analysis of plant Hsfs is complicated by their capacity to form hetero-oligomers, in contrast to Hsfs in animal systems. In particular, tomato Hsfs A1 and A2 were shown to form stable hetero-oligomers required for efficient nuclear retention of LpHsfA2 (Scharf et al., 1998). The effect is due to shielding of the C-terminal nuclear export signal of LpHsfA2 (Heerklotz et al., 2001). In addition, Hsf hetero-oligomerization might be involved in HSP promoter activation (Scharf et al., 1998). Although there is experimental evidence indicating that plant Hsfs could selectively transactivate promoters with natural (Treuter et al., 1993) or artificial HSE sequences (Bharti et al., 2000), the basis for the observed selectivity is unknown. In particular, it is not clear whether the selectivity involves preferential HSE binding or preferential activation determined by the specific HSE sequences.
In this work, we used the Ha hsp17.6 G1 promoter as a natural model system for analyzing developmental transcriptional activation by Hsfs. Thus, we avoided the complications of more complex HSEs in other promoters and their activation by heat stress. Our starting hypothesis is that the peculiar structure of the HSE in Ha hsp17.6 G1 could confer some selectivity toward promoter regulation in embryos by specific Hsfs or combinations of them. We tested this hypothesis by transient activation assays with two well-characterized plant Hsfs: LpHsfA1 and LpHsfA2 (Scharf et al., 1990; for example, see Boscheinen et al., 1997; Lyck et al., 1997; Döring et al., 2000) using different plant systems and yeast (Saccharomyces cerevisiae). We conclude that the sequence of the HSE in Ha hsp17.6 G1 is crucial for the observed promoter selectivity. Furthermore, we show that this promoter selectivity involves preferential activation by LpHsfA2, rather than preferential binding to the HSE. Mutational analyses of Ha hsp17.6 G1 promoter activation, combined with the results of the in vitro binding assays, suggest that mixed oligomers of LpHsfA1 and LpHsfA2 could be involved in a synergistic transcriptional activation.
RESULTS
LpHsfA2, But Not LpHsfA1, Activated the Ha hsp17.6 G1 Promoter in Sunflower Embryos
We used transient activation assays, performed in particle-bombarded sunflower embryos, to analyze whether the Ha hsp17.6 G1 promoter could be activated at 25°C by two Hsfs from tomato: LpHsfA1 and LpHsfA2 (Scharf et al., 1990). Sunflower embryos were transformed with a luciferase reporter reference plasmid together with the appropriate reporter and effector plasmids (as in Rojas et al., 1999). The effector plasmids encoded LpHsfA1 and LpHsfA2. To investigate the role of the HSE sequences in Ha hsp17.6 G1, we introduced point mutations in crucial nucleotide positions of the HSE motifs (Fig. 1). These mutations should abolish (Mut0, formerly HSEm; Carranco et al., 1999), diminish (Muts 1, 2, 3, and 5; this work), or improve Hsf binding (MutP; this work). All mutants were constructed in the context of a translational fusion that we previously used to analyze this promoter in transgenic tobacco plants (Carranco et al., 1999; Fig. 1).
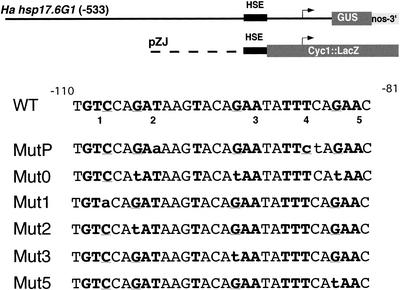
Figure 1.
Chimeric genes with the WT and mutant Ha hsp17.6 G1 HSE sequences. Top, Chimeric gene promoter context for the β-glucuronidase (GUS; −533::GUS, Carranco et al., 1999) and LacZ fusions used in this work. The arrows indicate the Ha hsp17.6 G1 and the Cyc1 promoters. The sequences of the Ha hsp17.6 G1 HSE complex are indicated with a black box. The corresponding, unmodified nucleotide sequences for this HSE (WT) are shown (from −110 to −81 from the transcription initiation site of Ha hsp17.6 G1). The core sequences for the HSE pentanucleotide repeats are marked in bold face with crucial DNA contact positions (G or C) underlined. These core repeats are numbered, below the sequence, from 1 to 5. Nucleotide substitutions in the different mutant HSEs are indicated with lowercase letters. The different mutant sequences were substituted for the WT HSE in each chimeric gene promoter context as described in “Materials and Methods.”
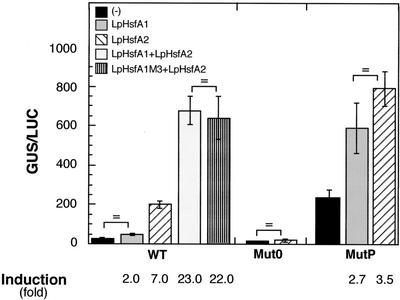
The chimeric gene with the natural promoter and HSE sequences (WT) was significantly activated by LpHsfA2, resulting in induced reporter expression levels about 7-fold higher than in control embryos without LpHsfA2 (Fig. 2, WT; for statistical values of all comparisons mentioned here, see the legend of this figure). This transcriptional activation was fully dependent on the integrity of the HSE sequence, as it was not observed with the Mut0 chimeric gene (Fig. 2, Mut0). In contrast, when LpHsfA1 was used as the effector, basal reporter expression levels were only increased about 2-fold; moreover, this marginal activation was not statistically significant. To confirm that the HSE sequence was a decisive factor for selectivity by LpHSFA2, we analyzed the activation of the chimeric gene with the MutP sequence. In the absence of exogenous Hsfs, the basal level of reporter gene activity was dramatically increased (as expected from the improvement of the HSE consensus sequences and from the likely presence of endogenous Hsfs). We also observed a small (2.7- to 3.5-fold), but statistically significant induction of reporter activity above basal level by either LpHsfA2 or LpHsfA1. Furthermore, induction by LpHsfA1 did not differ from that by LpHsfA2 (Fig. 2, MutP). These results demonstrated, with natural cis-elements and minimal modifications in a native promoter context, the importance of the HSE sequence for the observed Hsf selectivity.
Figure 2.
Transcriptional activation in sunflower embryos. Plant material was particle bombarded with a luciferase reference plasmid, the indicated chimeric GUS reporter gene (WT, Mut0, or MutP), and the Hsf effector plasmid combination shown in the top left corner: (−), no effector plasmid; (+), effector plasmids bombarded simultaneously. We show average reporter GUS activities normalized with luciferase (GUS/LUC, as in Prieto-Dapena et al., 1999), with bars indicating the ses. Data correspond to at least five independent experiments, with each plasmid combination repeated at least 25 times. Nonsignificant differences in reporter gene activities are indicated (P ≥ 0.05). Values for the significant statistical differences mentioned in the text were as follows: for the WT gene with effector plasmid(s) versus without effector plasmid(s), with LpHsfA2, F = 139.6, P = 0.0001; and with LpHsfA1 + LpHsfA2, F = 429.1, P = 0.0001. The induced level obtained with LpHsfA1 + LpHsfA2 did not differ from that of with LpHsfA1 M3 + LpHsfA2, F = 0.25, P = 0.61. In addition, the values obtained with LpHsfA1 + LpHsfA2 were significantly higher than the addition of reporter activities separately obtained with each Hsf (F = 27.35, P = 0.001). For the mutP gene with effector plasmid(s) versus without effector plasmid(s): with LpHsfA2, F = 41.4, P = 0.0001; and with LpHsfA1, F = 9.3, P = 0.021.
Analyses of the activation by LpHsfA1 and LpHsfA2 of other sHSP promoters showed that when simultaneously present, these Hsfs produce higher levels of induced reporter gene activity (for example, the GmHSP17 promoter in tobacco protoplasts; Scharf et al., 1998). This effect occurred at normal growth (25°C) and heat shock temperatures (35°C), and involved nuclear cotransport of both Hsfs. LpHsfA2 is transported to the nucleus with LpHsfA1, and this required a functional nuclear localization signal (NLS) in LpHsfA1. To investigate possible effects of LpHsfA1 on Ha hsp17.6 G1 promoter activation by LpHsfA2, we analyzed the WT gene expression in sunflower embryos cobombarded with effector plasmids for both Hsfs. The functional relevance of the LpHsfA1 NLS sequence was determined by using the previously characterized LpHsfA1M3 mutant (Lyck et al., 1997). We observed that cobombardment of both Hsfs induced the WT promoter about 23-fold above basal levels. This represented a synergistic promoter activation effect by both Hsfs: The activity of the reporter gene after Hsf cobombardment was significantly higher than the cumulative value of the activities observed for each Hsf when assayed separately. Furthermore, LpHsfA2 coexpressed with LpHsfA1 or LpHsfA1M3 showed statistically similar induction levels of reporter gene activity (22- to 23-fold). Thus, the mutated LpHsfA1 NLS was not required for the synergistic activation effect in sunflower embryos. However, LpHsfA1M3, when coexpressed with LpHsfA2 in sunflower leaves, did not increase activation above the level observed with only LpHsfA2. This confirmed that the mutated NLS, in LpHsfA1M3, is functional in sunflower leaves (data not shown).
Selective Activation by LpHsfA2 through the WT HSE in Yeast
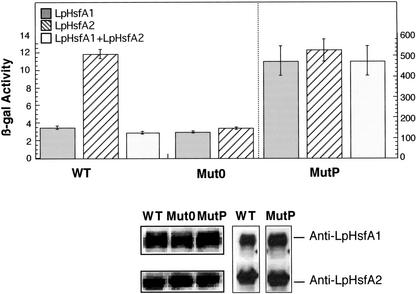
The same natural and mutant Ha hsp17.6 G1 HSE sequences analyzed in sunflower embryos were transcriptionally fused with a minimal Cyc1 promoter and lacZ reporter gene (pZJ derived; see Fig. 1 and Slater and Craig, 1987), and their expression was analyzed in RSY4-derived yeast strains with LpHsfA1 or LpHsfA2 as the only Hsf present (Boscheinen et al., 1997). Using yeast containing the null mutant HSE plasmid, we observed a low-level β-galactosidase reporter activity with the LpHsfA1 or LpHsfA2 strains (Fig. 3, Mut0). We considered this level of reporter gene activation as the basal transcription level from the Cyc1 promoter. In yeast cells with the WT HSE reporter plasmid, we observed a significant transcriptional induction above this basal level, but only with the LpHsfA2 strain (Fig. 3, WT). This is similar to what was observed in sunflower embryos with the native Ha hsp17.6 G1 promoter (Fig. 2, WT). We also tested the MutP plasmid and found activation by LpHsfA2 or LpHsfA1, as was the case with sunflower embryos. The observed β-galactosidase activity was similar for both Hsfs and was much higher than with the WT plasmid (Fig. 3, compare MutP and WT and note the difference in scale between left and right panels). We conclude that in absence of other endogenous Hsfs, the HSE sequences used were crucial for promoter activation and discrimination by Hsfs in yeast. However, the yeast system could not reproduce other results obtained in the sunflower system. For example, we tested the WT HSE plasmid in a yeast strain containing LpHsfA1 and LpHsfA2, and we did not observe reporter gene induction above the levels obtained with LpHsfA1. In addition, using the MutP HSE plasmid, similar reporter gene activities were observed in strains, irrespective of whether LpHsfA1, LpHsfA2, or both factors were present (Fig. 3).
Figure 3.
Transcriptional activation in yeast. Reporter β-galactosidase activity measured in yeast strains transformed with plasmids containing the indicated HSE (WT, Mut0, or MutP) is shown. These RSY4 strains (Boscheinen et al., 1997) contained one (LpHsfA1 or LpHsfA2) or two (LpHsfA1 + LpHsfA2) tomato Hsfs. The values for two independent transformants per plasmid combination were averaged, and the ses are represented with bars. Bottom, The expression level of the Hsfs determined by western analysis of protein extracts from each yeast strain using the immune sera indicated to the right. Left, Data for the single Hsf strains with each reporter plasmid (LpHFSA1 above, LpHsfA2 below); right, data for the double Hsf strain (with the WT or MutP plasmid); in this case, LpHsfA1 and LpHsfA2 were sequentially detected using the same protein extracts.
LpHsfA1 Efficiently Binds to the WT HSE in Vitro
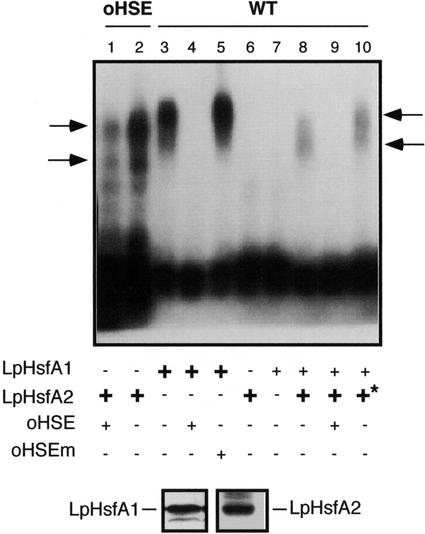
Results presented so far established that the WT HSE conferred selective chimeric gene transcriptional activation by LpHsfA2. This HSE would not allow the activation of the natural or chimeric promoters by LpHsfA1 in the absence (Fig. 3) or in the likely presence of other endogenous plant Hsfs (Fig. 2). The mechanism of Hsf discrimination could involve their differential binding to the WT HSE and/or differential promoter activation subsequent to DNA binding. These two possibilities are not mutually exclusive. We investigated the relevance of Hsf binding by performing in vitro gel retardation experiments using yeast extracts containing LpHsfA2 or LpHsfA1 and appropriate oligonucleotide-labeled DNA probes. The presence of comparable amounts of LpHsfA2 and LpHsfA1 in the protein extracts was verified by western-immunoblot detection using specific antibodies (Fig. 4, bottom). To our surprise, we could easily detect binding of LpHsfA1 to the WT probe, whereas LpHsfA2 did not bind to this probe, even with the highest amount of total extract protein allowed in our experimental conditions (Fig. 4, compare the retarded bands in lanes 3 and 5 with lane 6, and data not shown). We also observed similar differential binding results using other binding conditions (data not shown). This clearly indicated that the binding behavior of each Hsf corresponded to their respective affinity for the DNA probe rather than to the particular binding conditions in the experiments shown in Figure 4. The specificity of the detected Hsf::DNA complexes was verified by competition with two unlabeled oligonucleotides, one with the synthetic consensus HSE sequences (oHSE) and a mutant version that is no longer bound by Hsfs (oHSEm). We only observed competition using the oHSE competitor (Fig. 4, compare lanes 4 and 5). We also verified that the LpHsfA2 extract contained a functional HSE-binding Hsf by observing the corresponding specific retarded complexes using oHSE as a labeled DNA probe (Fig. 4, lanes 1 and 2). As expected from the smaller Mr of LpHsfA2, these complexes migrated in the gel faster than those seen with LpHsfA1 (Fig. 4, compare lanes 2 and 3). The observation of two separate retarded bands with the oHSE probe was also expected as this HSE, with five GAA/TTC consensus repeats, would be able to simultaneously bind two Hsf oligomers. We conclude that binding of LpHsfA1 to the WT HSE is very efficient, at least in vitro. Thus, the observed failure of promoter activation by LpHsfA1 through this HSE would not be explained by inefficient DNA binding of the Hsf. Most likely, LpHsfA1 binds the WT HSE under the experimental conditions used for Figures 2 and 3, but is unable to activate transcription. In a converse manner, it is clear that even if we failed to detect in vitro binding of LpHsfA2 to the WT HSE, LpHsfA2 bound to WT HSE in absence of other Hsfs was able to activate transcription (Fig. 3).
Figure 4.
In vitro binding experiments. Two labeled DNA fragments that contained synthetic (oHSE) or natural HSE sequences from Ha hsp17.6 G1 (WT) were subjected to mobility shift assays in agarose gels. Binding reactions contained the probe indicated on top (0.13 ng of labeled DNA) and the components were summarized at the bottom, including unlabeled oligonucleotide fragments used as specific or unspecific competitors (oHSE and oHSEm, respectively; see “Material and Methods”), as well as different amounts of protein extracts from yeast strains containing LpHsfA1 or LpHsfA2. Optimal and limiting amounts of total protein used for each extract were experimentally determined (see “Results”): LpHsfA1, limiting = 10 μg (+), and optimal = 20 μg (+). In the case of LpHsfA2, 40 μg (+) was used as the optimal amount of total protein. Higher amounts of this extract were used in some reactions (+*, 70 μg). Ticks to the left mark the position of the LpHsfA2 homo-oligomeric complexes. The arrows to the right mark the position of the different mixed complexes observed with increasing amounts of the LpHsfA2 extract (lane 8, 40 μg of protein, and lane 10, 70 μg of protein). Bottom, Western immunodetection of LpHsfA1 and LpHsfA2 using 25 μg of total protein from the respective extracts used in the binding reactions.
LpHsfA1-Aided Binding of LpHsfA2 to the WT HSE
We examined whether LpHsfA1 could facilitate the binding of LpHsfA2 to the WT HSE. We reduced the amount of the LpHsfA1 extract so that LpHsfA1 reached a limiting concentration in the binding reaction. Under these conditions, LpHsfA1 by itself did not bind to the WT HSE probe (Fig. 4, lane 7). We then added the LpHsfA2 extract to the binding reactions with LpHsfA1 at the limiting concentration. We used two LpHsfA2 extract concentrations that were not sufficient by themselves to detect Hsf binding (Fig. 4, lane 6 and data not shown). Under such conditions, we observed retarded bands that corresponded to specific Hsf::HSE complexes that could be competed with unlabeled oHSE (Fig. 4, compare lanes 8 and 9). We conclude that LpHsfA1 facilitates the binding of LpHsfA2 to the WT HSE by increasing the DNA affinity of LpHsfA2 or the stability of the Hsf::HSE complex. The mobility of the retarded complexes observed was dependent on the amount of LpHsfA2 in the binding reaction. At the lowest LpHsfA2 concentration, a single broad band was apparent (Fig. 4, lane 8). The mobility of this band was between those of the two bands corresponding to one (Fig. 4, lane 2, bottom band) or two (Fig. 4, lane 2, top band) HSE-bound LpHsfA2 homo-oligomeric complexes. At the highest LpHsfA2 concentration, the observed band shifted to a more retarded position (Fig. 4, lane 10). This position was slightly more retarded than that of the double LpHsfA2 homo-oligomeric complex (Fig. 4, compare lanes 2, 8, and 10). We conclude that, as expected from the sequence (Fig. 1), the WT HSE in Ha hsp17.6 G1 can simultaneously bind two Hsf oligomeric complexes. Most likely, the bands of intermediate mobility observed with mixed LpHsfA1 and LpHsfA2 extracts correspond to one (Fig. 4, lane 8) or two (Fig. 4, lane 10) HSE-bound Hsf hetero-oligomers.
Mutational Analyses of Ha hsp17.6 G1 Promoter Activation by the Hsfs in Protoplasts
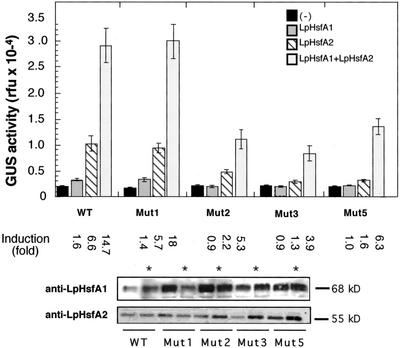
To further investigate the mechanisms of WT promoter activation by LpHsfA2 (with and without LpHsfA1), we analyzed the additional HSE mutants 1, 2, 3, and 5. In these mutants, which have crucial nucleotide substitutions in each of the four consensus-matching GAA/TTC sequences, the number of active HSE modules was reduced from four to three (see Fig. 1). Thus, we obtained simplified versions of the natural HSE to which the efficient binding of more than a single Hsf oligomer at a time should not be possible (based on previous observations with yeast or Drosophila melanogaster Hsfs and similar HSEs; Amin et al., 1988; Perisic et al., 1989; Xiao et al., 1991; Bonner et al., 1994; Fernandes et al., 1995). To allow the analyses of the numerous combinations of chimeric genes and Hsfs, we used tobacco mesophyll protoplasts as a transient expression system (Scharf et al., 1998). We first verified that the results that we obtained for the WT chimeric gene in sunflower embryos (Fig. 2) could be reproduced in the tobacco protoplast system. We observed that in tobacco protoplasts, the WT gene could be clearly induced by LpHsfA2 (statistical values for all comparisons in the legend of Fig. 5), but only marginally induced with LpHsfA1 (Fig. 5). The very low induction levels (1.6-fold) observed with LpHsfA1 were not different from those observed with the null HSE mutant (Mut0; data not shown), and this is likely to be HSE-independent, unspecific promoter activation. In the tobacco protoplast system, coexpression of LpHsfA1 and LpHsfA2 also showed the synergistic and not additive activation of the WT promoter (Fig. 5). We conclude that the tobacco protoplast system reproduced our observations in sunflower embryos (compare Figs. 2 and 5). We next analyzed the additional HSE mutants. The results that we obtained (shown also in Fig. 5) can be summarized as follows: Mut1 was fully functional and behaved exactly as the WT gene with respect to activation by the Hsfs; and, the rest of the mutants (Mut 2, 3, and 5) were similar to each other and showed reduced activation by LpHsfA2 (without or with LpHsfA1) compared with WT; moreover, all these mutants showed synergistic promoter activation by the two Hsfs (statistical comparisons in the legend of Fig. 5). These results would support the suggestion that LpHsfA1, together with LpHsfA2, could bind and activate transcription on the Ha hsp17.6 G1 HSE as hetero-oligomeric complexes.
Figure 5.
Effect of the HSE mutations on synergistic activation of the Ha hsp17.6 G1 promoter by LpHsfA2 and LpHsfA1. Bars represent average GUS activity determined in tobacco protoplasts transformed with different combinations of reporter and effector plasmids. Results correspond to three independent experiments for each plasmid combination, with GUS activity measured in triplicate in each experimental repetition. The chimeric genes, indicated by WT, Mut1, Mut2, Mut3, and Mut5, correspond to the natural and mutant HSE denominations of the GUS fusions in Figure 1. Each reporter gene was transformed without (−) and with the effector plasmids for the Hsfs indicated in the upper right corner. Induction is the ratio between activities obtained with and without effector plasmid(s). Statistical values for relevant comparisons of observed induced activities: WT with LpHsfA2 versus no effector plasmid (F = 119.6, P = 0.001). WT with LpHsfA1 + LpHsfA2 versus the addition of the activities separately obtained with each Hsf (F = 19.6, P = 0.0001). WT versus Mut1 with LpHsfA1 + LpHsfA2 (F = 0.047, P = 0.83). WT versus Mut2 (F = 33.7, P = 0.0001), Mut3 (F = 58.7, P = 0.0001), or Mut5 (F = 20.6, P = 0.0001), in all cases with LpHsfA1 + LpHsfA2. Statistical significance as in the legend of Figure 2. Panels below depict the verification, by western immunodetection, of expression levels for LpHsfA1 and LpHsfA2 in each experimental combination. For combinations simultaneously expressing LpHsfA1 and LpHsfA2, the same protein samples were used for sequential detection of both Hsfs (marked with asterisks).
DISCUSSION
Promoter discrimination by different Hsfs might explain the differential transcription of sHSP genes during plant embryogenesis. Discrimination may also be crucial during other developmental processes, and might require additional transcription factors (for discussion, see Carranco et al., 1999). It is conceivable that the sequences of plant Hsfs have diversified during evolution in parallel with that of the HSE elements in Hsf-regulated promoters. Such diversification would result in activation or repression of specific promoters by given Hsf(s) (Nover et al., 1996, 2001). This hypothesis would be consistent with published observations of differential activation of chimeric genes containing particular HSE sequences (Treuter et al., 1993; Bharti et al., 2000). However, these previous observations did not indicate the mechanism(s) involved in promoter discrimination. These mechanisms could involve differential HSE binding (for example, as observed with animal Hsfs; Kroeger et al., 1993, 1994) and/or steps in the Hsf-mediated transcriptional activation that could occur before or after DNA binding. By analyzing the activation of the Ha hsp17.6 G1 promoter, we observe discrimination by Hsfs that depends on the HSE sequence, but does not appear to involve preferential Hsf binding to the HSE. We also show that in plant cells, the Ha hsp17.6 G1 promoter is transiently activated by hetero-oligomers of LpHsfA1 and LpHsfA2.
HSE Discrimination without Preferential Hsf Binding
Perhaps the most crucial observation reported here is that LpHsfA1 cannot activate the Ha hsp17.6 G1 promoter (Figs. 2, 3, and 5), although in vitro, it binds efficiently to the HSE from this promoter (Fig. 4). The failure to activate the promoter was surprising given the precedents in the literature. Thus, LpHsfA1 is a well-characterized factor that has been found to activate a large number of different promoters (with natural or synthetic HSEs) in homologous or heterologous transient expression systems under normal and heat shock temperatures (Treuter et al., 1993; Boscheinen et al., 1997; Scharf et al., 1998; Bharti et al., 2000; Döring et al., 2000). Our results with LpHsfA1 and the converse results observed with LpHsfA2 denote a case of promoter discrimination that involves a peculiar low-consensus HSE in the native promoter context. We conclude that the HSE sequence in the Ha hsp17.6 G1 promoter is crucial for discrimination by LpHsfA2. Such discrimination would most likely occur in vivo following DNA binding. Structural Hsf differences involved in promoter discrimination could be intrinsic to LpHsfA2 (i.e. presence of specific protein domains and AHA motifs; for example, see Döring et al., 2000), but combined with a dynamic interaction with the HSE sequence. An example would be, leading to specific conformational changes. This would be similar to what has been observed for the single Hsf (ScHsf1) in yeast and the HSEs of the CUP1 promoter. It was proposed that the transcriptional activation of different yeast promoters occurred through the preferential use of one of the two activation domains of ScHsf1 (N- or C-terminal), depending on the HSE sequences present (Santoro et al., 1998). In systems with multiple Hsfs, as in plants, different proteins would have adapted their structure (including divergence of the activation and other domains) for efficient activation through specific HSE sequences. Our results show that LpHsfA2 has special functional characteristics, as it is able to transactivate though imperfect HSE sequences in the natural context of the Ha hsp17.6 G1 promoter. The homologous Hsf(s) involved in the developmental regulation of Ha hsp17.6 G1 in seeds is (are) likely to share with LpHsfA2 some structural and functional characteristics. This factor(s) is also expected to functionally interact with the additional embryo-specific factors revealed by our previous analyses of chimeric gene expression in transgenic plants (Carranco et al., 1999).
Synergistic Activation by LpHsfA1 + LpHsfA2. Involvement of Hetero-Oligomers?
LpHsfA1 and LpHsfA2 synergistically activated the Ha hsp17.6 G1 promoter when expressed simultaneously in plant cells (Figs. 2 and 5). Based on the DNA-binding and activation results obtained with the Ha hsp17.6 G1 promoter and each Hsf, LpHsfA1 should have acted as a competitive inhibitor of transcriptional activation by LpHsfA2. LpHsfA1 inhibited activation by LpHsfA2, but only in yeast (see Fig. 3, WT); thus, the synergistic activation observed in plant cells is paradoxical. An explanation for this observation would be that LpHsfA1 helps LpHsfA2 without binding the WT HSE in vivo. This might be the case, for example, if LpHsfA1 only contributed to the nuclear cotransport of LpHsfA2, as previously observed in tomato and tobacco cells (Scharf et al., 1998). In that case, only LpHsfA2 would bind and activate the Ha hsp17.6 G1 promoter. Our results are not consistent with this simple explanation. First, we observed similar synergistic effects using LpHsfA1 or the LpHsfA1 M3 mutation. This suggests that if cotransport were involved, its mechanism would differ from that in whole leaves and leaf protoplasts, where the M3 mutation was shown to be inactive (Scharf et al., 1998; data not shown for sunflower leaves). Second, we showed that LpHsfA1 facilitated the in vitro binding of LpHsfA2 to the WT HSE. Thus, functional interactions between both Hsfs could involve other aspects beside their previously described nuclear cotransport (Scharf et al., 1998).
The inability to reproduce the synergistic interaction between LpHsfA1 and LpHsfA2 for transcriptional activation through the Ha hsp17.6 G1 HSE in yeast (Fig. 3) is interesting and would be consistent with the involvement of additional factors. These factors could bind DNA in sequences not included in the pZJ construct (see Figs. 1 and 3) and/or they may not be conserved in yeast cells. Our previous work has identified potential binding sites for these unidentified factors in upstream promoter sequences (Carranco et al., 1999). Recent work has allowed these cis-elements to be further defined, and this may lead to cloning these factors by one-hybrid interaction (unpublished data from J. Jordano's lab).
Our experimental observations would support a model in which LpHsfA1 facilitates the binding of LpHsfA2 to the WT HSE in the Ha hsp17.6 G1 promoter, most likely through hetero-oligomerization. LpHsfA1/LpHsfA2 hetero-oligomers would have an increased binding affinity for this HSE compared with LpHsfA2, at least in vitro, due to the higher binding affinity showed by LpHsfA1 (Fig. 4). Such hetero-oligomers might bind to the HSE and, by incorporating other structural characteristics of LpHsfA2, would be able to efficiently activate the Ha hsp17.6 G1 promoter. The demonstration of the physical interaction of LpHsfA1 and LpHsfA2 supports the formation of hetero-oligomers prior to DNA binding (Scharf et al., 1998). Our model goes beyond DNA binding and is backed by a second crucial observation: that the sequence of the mutant HSEs used in Figure 5 allowed synergistic activation by LpHsfA1 and LpHsfA2. These sequences would not support the efficient binding of more than one Hsf oligomer at a time, based on previous observations for yeast and animal Hsfs (Amin et al., 1988; Perisic et al., 1989; Xiao et al., 1991; Bonner et al., 1994). Even if LpHsfA2 or LpHSFA1 were to bind the HSEs as monomers, we still should have observed that LpHsfA1 would repress the transcriptional activation by LpHsfA2 in cobombardment experiments given the DNA-binding affinities (Fig. 4) and transcriptional activation capacities of both Hsfs (Figs. 2, 3, and 5). Our observation of functional interaction among LpHsfA1 and LpHsfA2 could be relevant for the developmental activation of the Ha hsp17.6 G1 and other similar HSP promoters. This observation indicates potential synergistic activation of such promoters by Class A Hsf hetero-oligomers. However, our experimental conditions may not mimic the natural situation in embryos. For example, the nuclear abundance of LpHsfA1 and/or LpHsfA2 could be distorted by overexpression in transient assays. Thus, it remains unclear if the Hsfs involved in the developmental regulation actually hetero-oligomerize in sunflower embryos.
Additional functional analysis of the HSE mutants described here will be performed in transgenic plants and by transient expression. We expect that in the natural context of the Ha hsp17.6 G1 promoter these mutants will help in elucidating other possible interactions among the different types of plant Hsfs.
MATERIALS AND METHODS
Site-Directed Mutagenesis of the Ha hsp17.6 G1 Promoter and Construction of the Chimeric Genes
The WT and Mut0 GUS reporter plasmids were obtained from the previously described binary plasmids, −1,486::GUS and −1,486(m)::GUS, respectively (Carranco et al., 1999). In both cases, the 3.9-kb XhoI-EcoRI DNA fragment was cloned in pBluescript SK+. The resulting WT GUS reporter plasmid was used as the template DNA for site-directed mutagenesis by PCR amplification. We used Pwo DNA polymerase (Roche Molecular Biochemicals, Summerville, NJ), with conditions described by Rojas et al. (1999), downstream primer 5′-ATGGGTCGAATATGTTG-3′ (noncoding strand in Ha hsp17.6 G1 from position +90), and the following mutagenic upstream primer for each mutant gene (denominations following primer sequences correspond to those in Fig. 1): 5′-AAAAAAGCTTATTCTCTATCTGTaCAGATAAG-3′ (Mut1); 5′-AAAAAAGCTTATTCTCTATCTGTCCAtATAAGTAC-3′ (Mut2); 5′-AAAAAAGCTTATTCTCTATCTGTCCAGATAAGTACAtAATATTTC-3′ (Mut3); 5′AAAAAAGCTTATTCTCTATCTGTCCAGATAAGTACAGAATATTTCAtAACACTAC-3′ (Mut5); and 5′AAAAAAGCTTATTCTCTATCTGTCCAGAaAAGTACAGAATATTcTAGAACACTACTACG-3′ (MutP).
In all cases, the coding strand from Ha hsp17.6 G1 is depicted with nucleotide substitutions indicated with lowercase letters. The 222-bp DNA fragments PCR-amplified in each case were purified from 2% (w/v) agarose gels and were digested with HindIII. The resulting 175-bp HindIII fragments substituted, in each case, the original fragment in the WT plasmid. Mutant plasmid denominations correspond to that of the mutagenic primers listed above. For construction of the equivalent lacZ reporter plasmids (see also Fig. 1), DNA from each GUS reporter mutant plasmid was used as template for amplification with the following primers: 5′-TAATAATCCGTcGAcAAAAAAGC-3′ and 5′-GGATATTGtCgACGTAGTAG-3′. These primers (located, respectively, in coding and noncoding strands of Ha hsp17.6 G1 from positions −140 and −54) amplified 88-bp DNA fragments with the WT or mutant HSE sequences from Ha hsp17.6 G1. The SalI sites created by nucleotide substitutions present in these primers allowed for the cloning of the different 64-bp HSE fragments at the XhoI site of vector pZJ (Slater and Craig, 1987). The resulting lacZ chimeric genes were denominated as the equivalent GUS reporter plasmids (Fig. 1). The sequence and orientation of amplified fragments in all the WT and mutant reporter plasmids were verified by sequencing both strands of DNA. All Hsf expression constructs used in this work have been described before (Treuter et al., 1993; Boscheinen et al., 1997; Lyck et al., 1997).
Transient Expression in Sunflower (Helianthus annuus) Embryos, Yeast (Saccharomyces cerevisiae) Strains, and Tobacco (Nicotiana tabacum) Protoplasts
We used the procedures previously described by Prieto-Dapena et al. (1999) and Rojas et al. (1999) for particle bombardment and transient expression in sunflower embryos. Normalization of GUS activity with luciferase and statistical analysis of data was also as reported by Rojas et al. (1999). Transient expression assays in tobacco protoplasts was as described by Treuter et al. (1993) and Döring et al. (2000). The RSY4 yeast strains and the protocols for β-galactosidase activity assays in yeast were described by Boscheinen et al. (1997). Hsf immunoblots were performed as previously described by Lyck et al. (1997) by using the anti-LpHsfA1 and anti-LpHsfA2 immune sera.
DNA-Binding Experiments
Electrophoretical gel retardation assays were performed in 2% (w/v) agarose gels run in the conditions described by Rojas et al. (1999) except that temperature was maintained at 4°C. The yeast total protein extracts used in the binding reactions were prepared as previously described for samples from tomato suspension cultures (Scharf et al., 1990). Binding reactions were assembled in a total volume of 20 μL and proceeded for 20 min at 20°C under the conditions described by Almoguera et al. (1998) except that the binding reaction buffer was buffer B (described by Scharf et al., 1990). The WT HSE probe used in the binding assays contained the Ha hsp17.6 G1 sequences between positions −140 and −54 (Carranco et al., 1997). This probe was prepared by labeling, with γ-ATP32 and T4 DNA kinase (Rojas et al., 1999), the 88-bp DNA fragment used to construct the WT HSE lacZ reporter plasmid described above. The synthetic oHSE and oHSEm oligonucleotide DNA fragments used as binding probes and as specific (oHSE) or unspecific (oHSEm) binding competitors have been previously described (Rojas et al., 1999). Unlabeled competitor fragments were present at 50-fold molar excess in the binding reactions. Binding was started by the addition of the protein extract(s) after premixing the rest of components, and was stopped by loading the samples in agarose gels and performing electrophoresis.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Prof. Lutz Nover for critical reading of the manuscript and helpful discussions. We also thank Pilar Bazaga for excellent technical work.
Footnotes
This work was supported by the Spanish “Ministerio de Ciencia y Tecnología: Plan Nacional de I+D+I” (grant no. BIO99–794 to J.J.). This work was also supported by the Spanish Ministerio de Educación y Cultura (PhD fellowship to A.R.), by the Deutsche Forschungsgemeinschaft (grant no. SFB 474), and by the Fonds der Chemischen Industrie (to K.-D.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010927.
LITERATURE CITED
- Almoguera C, Prieto-Dapena P, Jordano J. Dual regulation of a heat shock promoter during embryogenesis: stage-dependent role of heat shock elements. Plant J. 1998;13:437–446. doi: 10.1046/j.1365-313x.1998.00044.x. [DOI] [PubMed] [Google Scholar]
- Amin J, Ananthan J, Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol. 1988;8:3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti K, Schmidt E, Lyck R, Heerklotz D, Bublak D, Scharf KD. Isolation and characterization of HsfA3, a new heat stress transcription factor of Lycopersicon peruvianum. Plant J. 2000;22:355–365. doi: 10.1046/j.1365-313x.2000.00746.x. [DOI] [PubMed] [Google Scholar]
- Bonner JJ, Ballou C, Fackenthal DL. Interactions between DNA-bound trimers of the yeast heat shock factor. Mol Cell Biol. 1994;14:501–508. doi: 10.1128/mcb.14.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscheinen O, Lyck R, Queitsch C, Treuter E, Zimarino V, Scharf KD. Heat stress transcription factors from tomato can functionally replace Hsf1 in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1997;255:322–331. doi: 10.1007/s004380050503. [DOI] [PubMed] [Google Scholar]
- Carranco R, Almoguera C, Jordano J. A plant small heat shock protein gene expressed during zygotic embryogenesis but noninducible by heat stress. J Biol Chem. 1997;272:27470–27475. doi: 10.1074/jbc.272.43.27470. [DOI] [PubMed] [Google Scholar]
- Carranco R, Almoguera C, Jordano J. An imperfect heat shock element and different upstream sequences are required for the seed-specific expression of a small heat shock protein gene. Plant Physiol. 1999;121:723–730. doi: 10.1104/pp.121.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coca MA, Almoguera C, Thomas TL, Jordano J. Differential regulation of small heat-shock genes in plants: analysis of a water stress-inducible and developmentally activated sunflower promoter. Plant Mol Biol. 1996;31:863–876. doi: 10.1007/BF00019473. [DOI] [PubMed] [Google Scholar]
- Döring P, Treuter E, Kistner C, Lyck R, Chen A, Nover L. The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell. 2000;12:265–278. [PMC free article] [PubMed] [Google Scholar]
- Fernandes M, Xiao H, Lis JT. Binding of heat shock factor to and transcriptional activation of heat shock genes in Drosophila. Nucleic Acids Res. 1995;23:4799–4804. doi: 10.1093/nar/23.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz D, Döring P, Bonzelius F, Winkelhaus S, Nover L. The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol Cell Biol. 2001;21:1759–1768. doi: 10.1128/MCB.21.5.1759-1768.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger PE, Morimoto RI. Selection of new HSF1 and HSF2 DNA-binding sites reveals difference in trimer cooperativity. Mol Cell Biol. 1994;14:7592–7603. doi: 10.1128/mcb.14.11.7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger PE, Sarge KD, Morimoto RI. Mouse heat shock transcription factors 1 and 2 prefer a trimeric binding site but interact differently with the HSP70 heat shock element. Mol Cell Biol. 1993;13:3370–3383. doi: 10.1128/mcb.13.6.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyck R, Harmening U, Höhfeld I, Treuter E, Scharf KD, Nover L. Intracellular distribution and identification of the nuclear localization signals of two plant heat-stress transcription factors. Planta. 1997;202:117–125. doi: 10.1007/s004250050110. [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra S, Ganguli A, Scharf KD. Arabidopsis and the Hsf world: How many heat stress transcription factors do we need? Cell Stress Chaperones. 2001;6:177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Gagliardi D, Vergne P, Czarnecka VE, Gurley WB. The Hsf world: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones. 1996;1:215–223. doi: 10.1379/1466-1268(1996)001<0215:thwcap>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisic O, Xiao H, Lis JT. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5-bp recognition unit. Cell. 1989;59:797–806. doi: 10.1016/0092-8674(89)90603-x. [DOI] [PubMed] [Google Scholar]
- Prändl R, Schöffl F. Heat shock elements are involved in heat shock promoter activation during tobacco seed maturation. Plant Mol Biol. 1996;31:157–162. doi: 10.1007/BF00020615. [DOI] [PubMed] [Google Scholar]
- Prieto-Dapena P, Almoguera C, Rojas A, Jordano J. Seed-specific expression patterns and regulation by ABI3 of an unusual late embryogenesis-abundant gene in sunflower. Plant Mol Biol. 1999;39:615–627. doi: 10.1023/a:1006178220289. [DOI] [PubMed] [Google Scholar]
- Rojas A, Almoguera C, Jordano J. Transcriptional activation of a heat shock gene promoter in sunflower embryos: synergism between ABI3 and heat shock factors. Plant J. 1999;20:601–610. doi: 10.1046/j.1365-313x.1999.00635.x. [DOI] [PubMed] [Google Scholar]
- Santoro N, Johansson N, Thiele DJ. Heat shock element architecture is an important determinant in the temperature and transactivation domain requirements for heat shock transcription factor. Mol Cell Biol. 1998;18:6340–6352. doi: 10.1128/mcb.18.11.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L. The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol. 1998;18:2240–2251. doi: 10.1128/mcb.18.4.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Rose S, Zott W, Schöffl F, Nover L. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast Hsf. EMBO J. 1990;9:4495–4501. doi: 10.1002/j.1460-2075.1990.tb07900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater MR, Craig EA. Transcriptional regulation of an hsp70 heat shock gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1906–1916. doi: 10.1128/mcb.7.5.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuter E, Nover L, Ohme K, Scharf KD. Promoter specificity and deletion analysis of three heat stress transcription factors of tomato. Mol Gen Genet. 1993;240:113–125. doi: 10.1007/BF00276890. [DOI] [PubMed] [Google Scholar]
- Waters ER, Lee GJ, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot. 1996;47:325–338. [Google Scholar]
- Wehmeyer N, Vierling E. The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol. 2000;122:1099–1108. doi: 10.1104/pp.122.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Perisic O, Lis JT. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5-bp unit. Cell. 1991;64:585–593. doi: 10.1016/0092-8674(91)90242-q. [DOI] [PubMed] [Google Scholar]